Wheat ABA Receptor TaPYL5 Constitutes a Signaling Module with Its Downstream Partners TaPP2C53/TaSnRK2.1/TaABI1 to Modulate Plant Drought Response
Abstract
:1. Introduction
2. Results
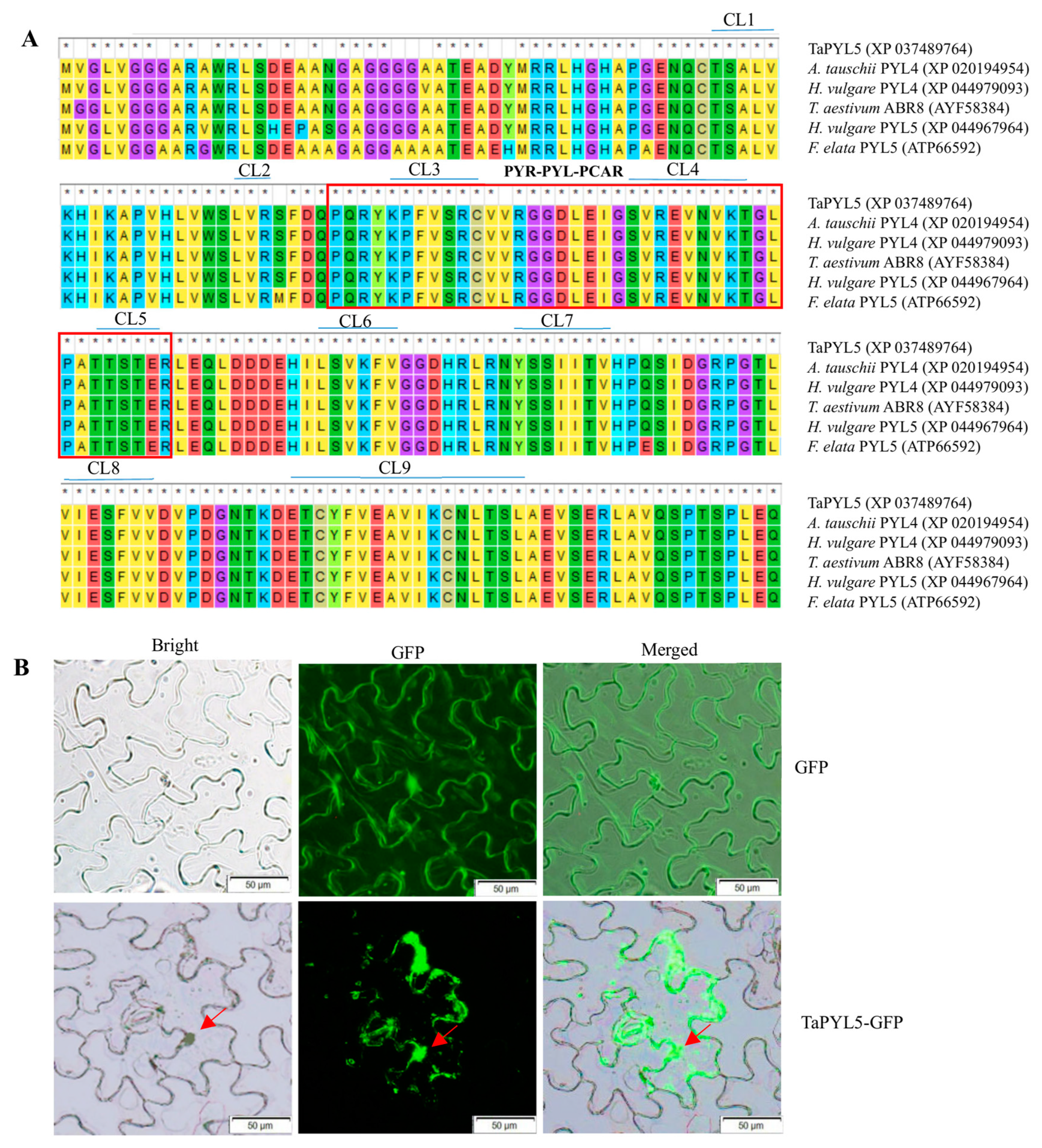
2.1. Molecular Characterization of TaPYL5
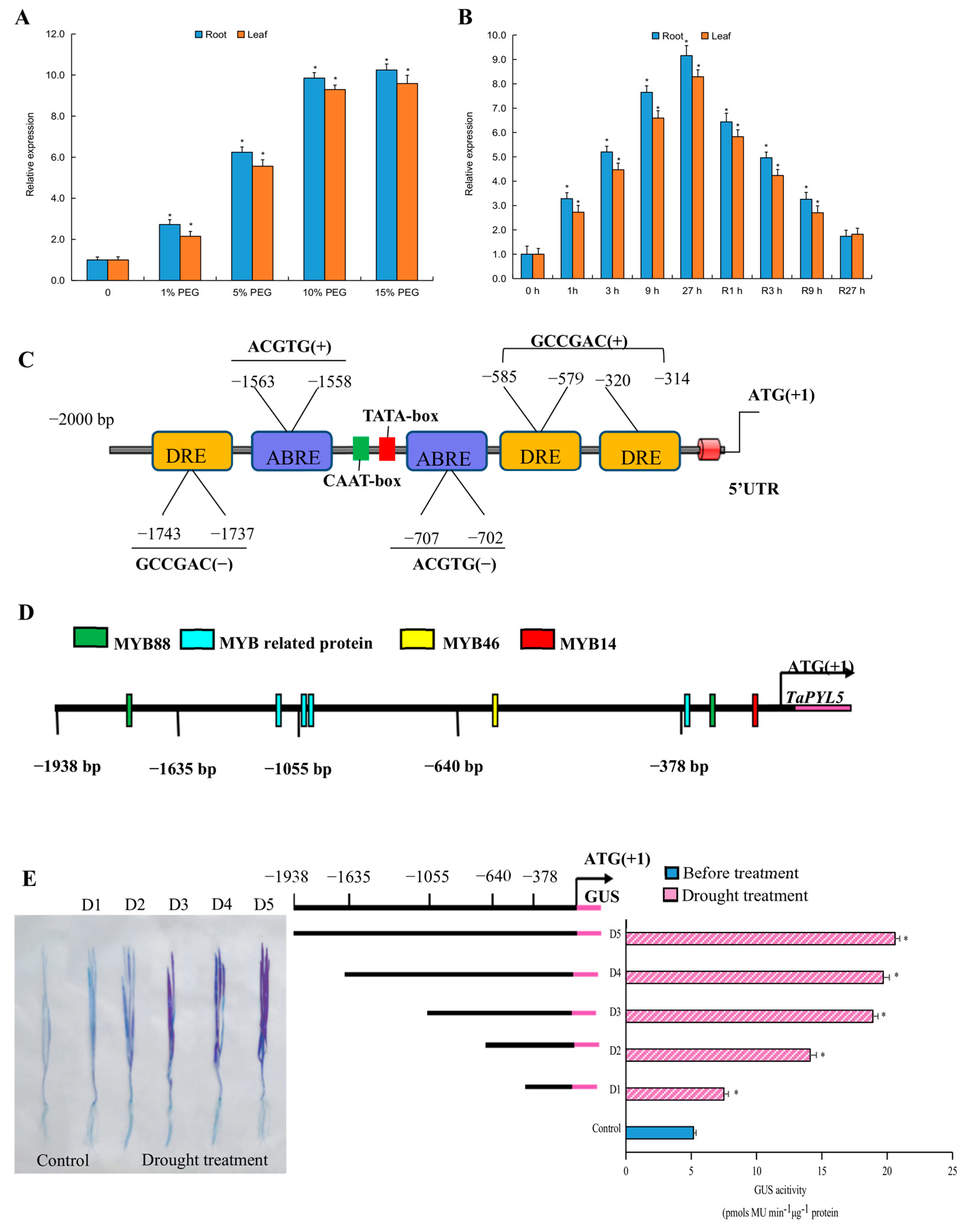
2.2. TaPYL5 Expression Is Sensitive in Response to Drought Stress
2.3. A Suite of Cis-Regulatory Elements in TaPYL5 Promoter Impacts Gene Responses to Drought Stress
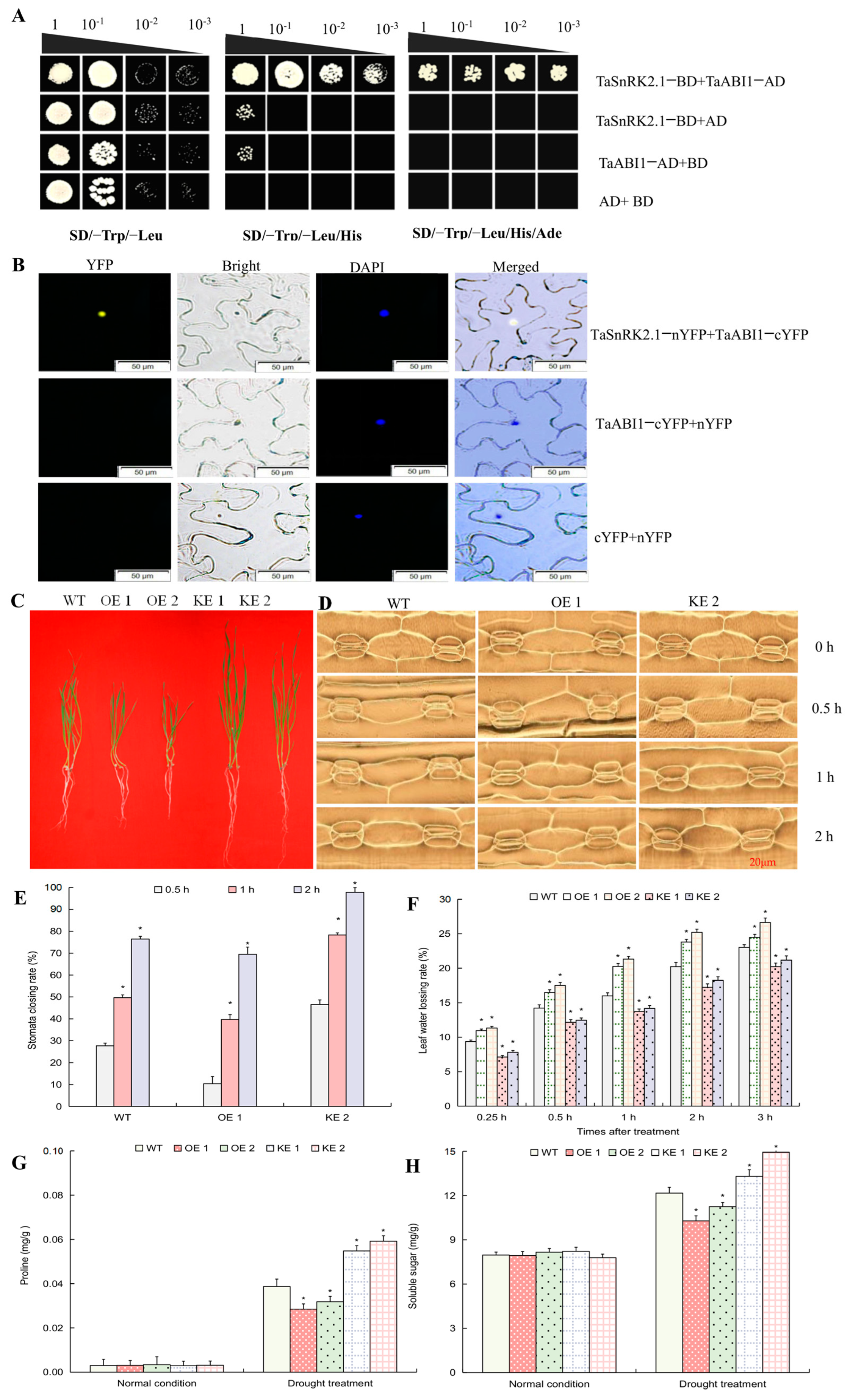
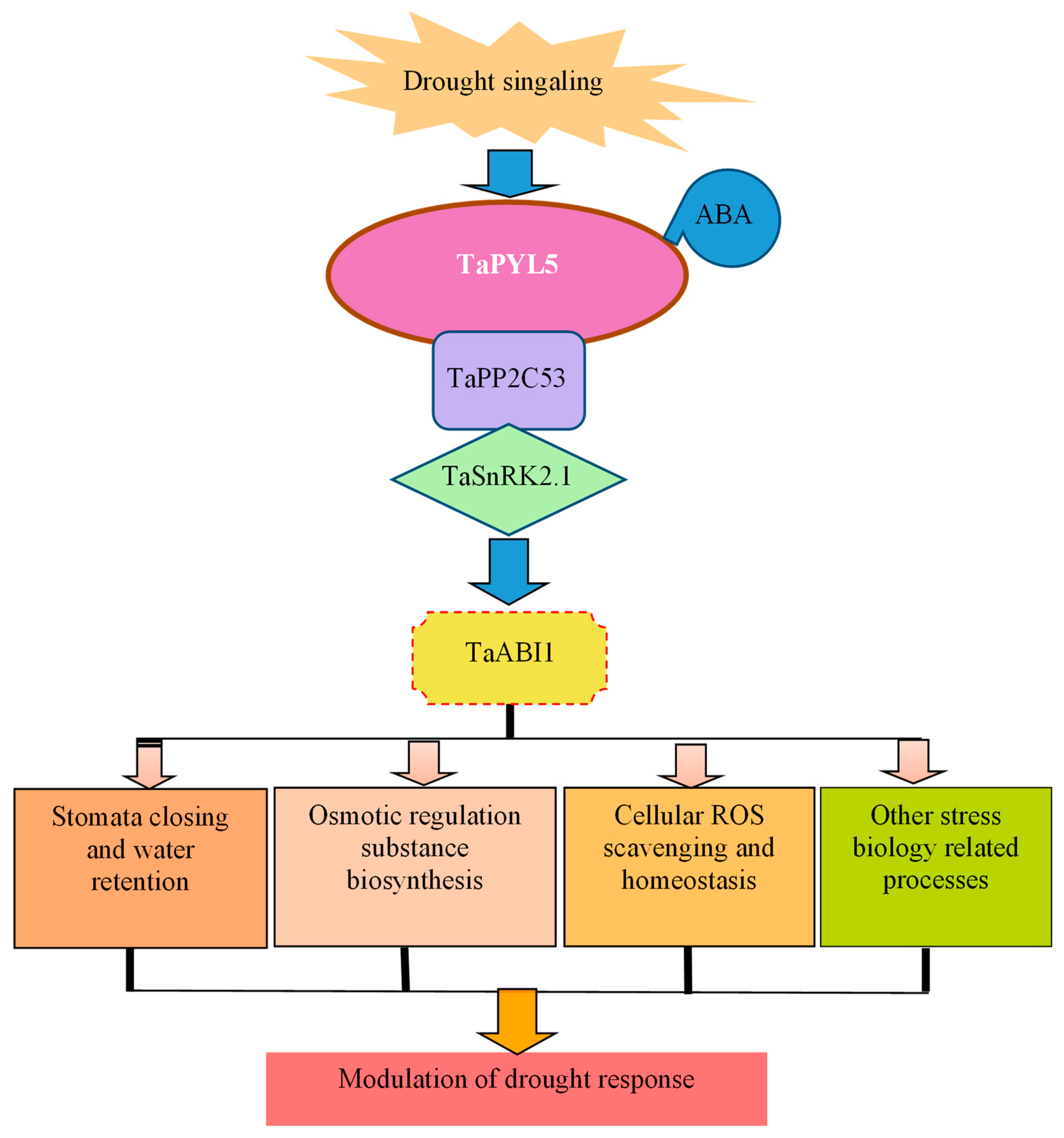
2.4. TaPYL5 Involves the Constitution of an ABA Core Signaling Module with PP2C and SnRK2 Members
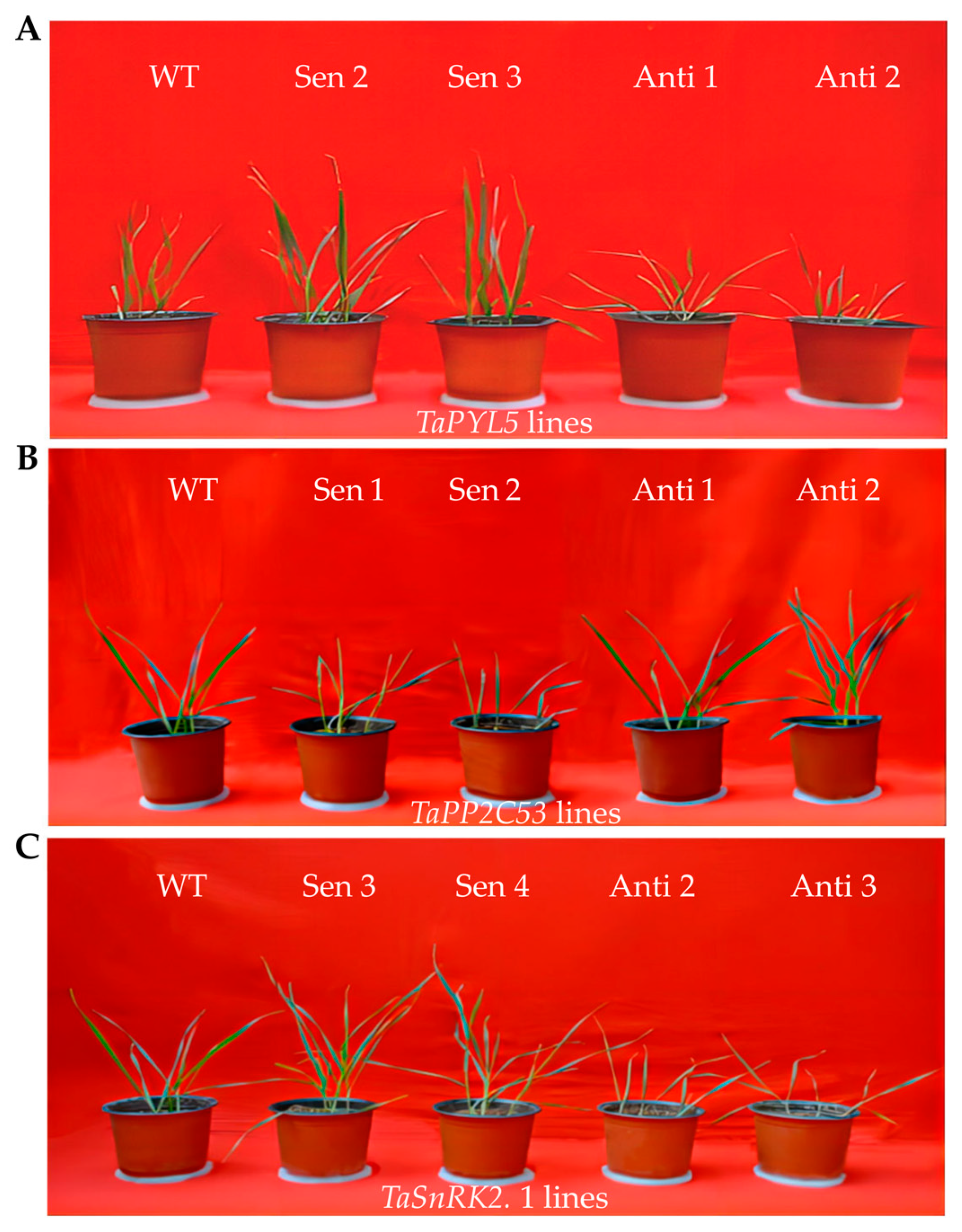
2.5. Transgenic Lines of Genes Encoding TaPYL5 and Its Downstream Partners Modify Plant Drought Response
2.6. TaPYL5-Mediated Drought Response Is Associated with the Role in Modulating Stomata Movement, Osmolyte Accumulation, and ROS Homeostasis
2.7. TaSnRK2.1 Protein Interacts with ABA-Responsive Transcription Factor TaABI1 to Negatively Impact Plant Drought Resistance
2.8. Transcriptome Profile Mediated by TaPYL5 upon Drought Signaling
3. Discussion
3.1. TaPYL5 Response to Drought Signaling Associates with a Set of Cis-Elements Situated in Promoter
3.2. TaPYL5 Is Involved in Establishing an ABA Signaling Module with Distinct PP2C and SnRK2 Members to Contribute to Plant Drought Response
3.3. TaPYL5 and Genes Encoding Its Partners Contribute Largely to Plant Drought Response
3.4. TaABI1 Is a Downstream Component of TaPYL5 Signaling Module and Negatively Regulates Drought Adaptation
3.5. TaPYL5 Modifies Gene Transcription Globally to Impact Plant Drought Response-Associated Physiological Processes
4. Materials and Methods
4.1. Characterization Analysis on TaPYL5
4.2. Analysis of TaPYL5 Protein Localization at the Subcellular Level
4.3. Expression Analysis of TaPYL5 upon Drought and Characterization of Cis-Acting Regulatory Elements in the TaPYL5 Promoter
4.4. Yeast Two-Hybrid and BiFC Assays to Identify Protein-Protein Interactions
4.5. Assessment for Growth Traits of Transgenic Lines
4.6. Determination of Physiological Parameters Associated with Osmotic Stress Responses in Transgenic Lines
4.7. Transcriptome Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berdugo, M.; Delgado-Baquerizo, M.; Soliveres, S.; Hernández-Clemente, R.; Zhao, Y.; Gaitán, J.J.; Gross, N.; Saiz, H.; Maire, V.; Lehmann, A.; et al. Global ecosystem thresholds driven by aridity. Science 2020, 367, 787–790. [Google Scholar] [CrossRef] [PubMed]
- Leng, G.; Hall, J. Crop yield sensitivity of global major agricultural countries to droughts and the projected changes in the future. Sci. Total Environ. 2019, 654, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Jogaiah, S.; Govind, S.R.; Tran, L.S. Systems biology-based approaches toward understanding drought tolerance in food crops. Crit. Rev. Biotechnol. 2013, 33, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, G.; Almeida, A.C.; Falvey, M.; Olmedo, G.F.; Taylor, P.; Santibañez, F.; Coops, N.C. Effects of climate change on forest plantation productivity in Chile. Global Change Biol. 2022, 28, 7391–7409. [Google Scholar] [CrossRef]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.; Testerink, C. Roots withstanding their environment: Exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Xiong, H.; Yu, J.; Miao, J.; Li, J.; Zhang, H.; Wang, X.; Liu, P.; Zhao, Y.; Jiang, C.; Yin, Z.; et al. Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol. 2018, 178, 451–467. [Google Scholar] [CrossRef]
- Kang, J.; Voothuluru, P.; Hoyos-Miernyk, E.; Alexander, D.; Oliver, M.J.; Sharp, R.E. Antioxidant metabolism underlies different metabolic strategies for primary root growth maintenance under water stress in cotton and maize. Antioxidants 2022, 11, 820. [Google Scholar] [CrossRef]
- Lynch, J.P. Rightsizing root phenotypes for drought resistance. J. Exp. Bot. 2018, 69, 3279–3292. [Google Scholar] [CrossRef]
- Durand, M.; Porcheron, B.; Hennion, N.; Maurousset, L.; Lemoine, R.; Pourtau, N. Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol. 2016, 170, 1460–1479. [Google Scholar] [CrossRef]
- Lee, D.K.; Chung, P.J.; Jeong, J.S.; Jang, G.; Bang, S.W.; Jung, H.; Kim, Y.S.; Ha, S.H.; Choi, Y.D.; Kim, J.K. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptations and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 2017, 15, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, T.; Miyazawa, Y.; Kobayashi, A.; Takahashi, H. Molecular mechanisms of hydrotropism in seedling roots of Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 2013, 100, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, D. Hydrotropism: How roots search for water. J. Exp. Bot. 2018, 69, 2759–2771. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, U.K.; Islam, M.N.; Siddiqui, M.N.; Cao, X.; Khan, M.A.R. Proline, a multifaceted signalling molecule in plant responses to abiotic stress: Understanding the physiological mechanisms. Plant Biol. 2022, 24, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhu, Y.; Yang, H.; Li, X.; Xu, R.; Zhu, F.; Cheng, Y. CsNIP5;1 acts as a multifunctional regulator to confer water loss tolerance in citrus fruit. Plant Sci. 2022, 316, 111150. [Google Scholar] [CrossRef]
- Xiang, J.; Chen, X.; Hu, W.; Xiang, Y.; Yan, M.; Wang, J. Overexpressing heat-shock protein OsHSP50.2 improves drought tolerance in rice. Plant Cell Rep. 2018, 37, 1585–1595. [Google Scholar] [CrossRef]
- Yengkokpam, P.; Mazumder, P.B. Antioxidant enzymatic activities and profiling of gene expression associated with organophosphate stress tolerance in Solanum melongena L. cv. Longai. 3 Biotech. 2021, 11, 510. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Zinta, G.; Hega, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 2016, 7, 276. [Google Scholar] [CrossRef]
- Avendaño-Vázquez, A.O.; Cordoba, E.; Llamas, E.; San Román, C.; Nisar, N.; De la Torre, S.; Ramos-Vega, M.; Gutiérrez-Nava, M.D.; Cazzonelli, C.I.; Pogson, B.J.; et al. An uncharacterized apocarotenoid-derived signal generated in æ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 2014, 26, 2524–2537. [Google Scholar] [CrossRef]
- Guo, X.; Svane, S.F.; Füchtbauer, W.S.; Andersen, J.R.; Jensen, J.; Thorup-Kristensen, K. Genomic prediction of yield and root development in wheat under changing water availability. Plant Methods 2020, 16, 90. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Song, J.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Abscisic acid and hydrogen peroxide are involved in drought priming-induced drought tolerance in wheat (Triticum aestivum L.). Plant Biol. 2020, 22, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D.; Leyser, O. A plant’s diet, surviving in a variable nutrient environment. Science 2020, 368, eaba0196. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Nie, K.; Zhou, H.; Yan, X.; Zhan, Q.; Zheng, Y.; Song, C.P. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020, 228, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, L.; Li, X.; Kong, X.; Wang, X.; Li, Y.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. AtRAE1 is involved in degradation of ABA receptor RCAR1 and negatively regulates ABA signalling in Arabidopsis. Plant Cell Environ. 2018, 41, 231–244. [Google Scholar] [CrossRef]
- Bishopp, A.; Lynch, J.P. The hidden half of crop yields. Nat. Plants 2015, 1, 15117. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Mao, H.; Jian, C.; Cheng, X.; Chen, B.; Mei, F.; Li, F.; Zhang, Y.; Li, S.; Du, L.; Li, T.; et al. The wheat ABA receptor gene TaPYL1-1B contributes to drought tolerance and grain yield by increasing water-use efficiency. Plant Biotechnol. J. 2022, 20, 846–861. [Google Scholar] [CrossRef]
- Li, L.; Mu, S.; Cheng, Z.; Cheng, Y.; Zhang, Y.; Miao, Y.; Hou, C.; Li, X.; Gao, J. Characterization and expression analysis of the WRKY gene family in moso bamboo. Sci. Rep. 2017, 7, 6675. [Google Scholar] [CrossRef]
- Fan, W.; Zhao, M.; Li, S.; Bai, X.; Li, J.; Meng, H.; Mu, Z. Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 2016, 16, 99. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.; Lim, C.W.; Lee, S.C. Identification and functional expression of the pepper RING type E3 ligase, CaDTR1, involved in drought stress tolerance via ABA-mediated signalling. Sci. Rep. 2016, 6, 30097. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, L.; Wei, N.; Liu, Z.H.; Hu, S.; Li, X.B. Overexpression of cotton PYL genes in Arabidopsis enhances the transgenic plant tolerance to drought stress. Plant Physiol. Biochem. 2017, 115, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Moon, S.J.; Min, M.K.; Choi, E.H.; Kim, J.A.; Koh, E.Y.; Yoon, I.; Byun, M.O.; Yoo, S.D.; Kim, B.G. Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Front. Plant Sci. 2015, 6, 614. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, T.F.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Wei, W.L.; Xu, Z.S. The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants. Int. J. Mol. Sci. 2020, 21, 670. [Google Scholar] [CrossRef]
- Shi, W.; Wang, X.; Liu, H.; Cai, Z.; Lu, C.; Chen, Y. A novel ABA-insensitive mutant in Arabidopsis reveals molecular network of ABA-induced anthocyanin accumulation and abiotic stress tolerance. J. Plant Physiol. 2022, 278, 153810. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Li, T.; Ni, C.; Han, L.; Du, P.; Xiao, K. TaPYL4, an ABA receptor gene of wheat, positively regulates plant drought adaptation through modulating the osmotic stress-associated processes. BMC Plant Biol. 2022, 22, 423. [Google Scholar] [CrossRef]
- Hsu, P.K.; Dubeaux, G.; Takahashi, Y.; Schroeder, J.I. Signaling mechanisms in abscisic acid-mediated stomatal closure. Plant J. 2020, 105, 307–321. [Google Scholar] [CrossRef]
- Wang, Z.; Ren, Z.; Cheng, C.; Wang, T.; Ji, H.; Zhao, Y.; Deng, Z.; Zhi, L.; Lu, J.; Wu, X.; et al. Counteraction of ABA-mediated inhibition of seed germination and seedling establishment by ABA signaling terminator in Arabidopsis. Mol. Plant. 2020, 13, 1284–1297. [Google Scholar] [CrossRef]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef]
- Yadukrishnan, P.; Datta, S. Light and abscisic acid interplay in early seedling development. New Phytol. 2021, 229, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Park, M.Y.; Kim, S.Y. The Arabidopsis J protein AtJ1 is essential for seedling growth, flowering time control and ABA response. Plant Cell Physiol. 2014, 55, 2152–2163. [Google Scholar] [CrossRef]
- Quan, W.; Hu, Y.; Mu, Z.; Shi, H.; Chan, Z. Overexpression of AtPYL5 under the control of guard cell specific promoter improves drought stress tolerance in Arabidopsis. Plant Physiol. Biochem. 2018, 129, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, Z.; Ding, Y.; Liu, L.; Han, X.; Zhan, J.; Wei, X.; Diao, Y.; Qin, W.; Wang, P.; et al. Over-expression of an R2R3 MYB gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 2019, 286, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, M.; Mueller, H.M.; Bauer, H.; Peirats-Llobet, M.; Rodriguez, P.L.; Geilfus, C.M.; Carpentier, S.C.; Al Rasheid, K.A.S.; Kollist, H.; Merilo, E.; et al. The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat. Plants 2019, 5, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, N.; Kim, R.; Han, S.; Song, J.; Lee, G.S.; Lee, S.; Min, M.K.; Kim, B.G. Ectopic expression of OsPYL/RCAR7, an ABA receptor having low signaling activity, improves drought tolerance without growth defects in rice. Int. J. Mol. Sci. 2020, 21, 4163. [Google Scholar] [CrossRef]
- Liang, C.; Liu, Y.; Li, Y.; Meng, Z.; Yan, R.; Zhu, T.; Wang, Y.; Kang, S.; Ali Abid, M.; Malik, W.; et al. Activation of ABA receptors gene GhPYL9-11A is positively correlated with cotton drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 2017, 8, 1453. [Google Scholar] [CrossRef]
- He, Z.; Zhong, J.; Sun, X.; Wang, B.; Terzaghi, W.; Dai, M. The maize ABA receptors ZmPYL8, 9, and 12 facilitate plant drought resistance. Front. Plant Sci. 2018, 9, 422. [Google Scholar] [CrossRef]
- Mega, R.; Abe, F.; Kim, J.S.; Tsuboi, Y.; Tanaka, K.; Kobayashi, H.; Sakata, Y.; Hanada, K.; Tsujimoto, H.; Kikuchi, J.; et al. Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 2019, 5, 153–159. [Google Scholar] [CrossRef]
- Deng, Y.N.; Kashtoh, H.; Wang, Q.; Zhen, G.X.; Li, Q.Y.; Tang, L.H.; Gao, H.L.; Zhang, C.R.; Qin, L.; Su, M.; et al. Structure and activity of SLAC1 channels for stomatal signaling in leaves. Proc. Nat. Acad. Sci. USA 2021, 118, e2015151118. [Google Scholar] [CrossRef]
- Li, Y.; Ding, Y.; Qu, L.; Li, X.; Lai, Q.; Zhao, P.; Gao, Y.; Xiang, C.; Cang, C.; Liu, X.; et al. Structure of the Arabidopsis guard cell anion channel SLAC1 suggests activation mechanism by phosphorylation. Nat. Commun. 2022, 13, 2511. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Chen, H.; Wang, L.; Zhao, Q.; Wang, D.; Zhang, T. Cold acclimation alleviates cold stress-induced PSII inhibition and oxidative damage in tobacco leaves. Plant Sign. Behav. 2022, 17, 2013638. [Google Scholar] [CrossRef] [PubMed]
- Lennicke, C.; Cochemé, H.M. Redox metabolism: ROS as specific molecular regulators of cell signaling and function. Mol. Cell 2021, 81, 3691–3707. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chen, T.; Tian, M.; Rahantaniaina, M.S.; Zhang, L.; Wang, R.; Xuan, W.; Han, Y. Genetic manipulation of reactive oxygen species (ROS) homeostasis utilizing CRISPR/Cas9-based gene editing in rice. Method. Mol. Biol. 2022, 2526, 25–41. [Google Scholar]
- Chen, Q.; Bai, L.; Wang, W.; Shi, H.; Ramón Botella, J.; Zhan, Q.; Liu, K.; Yang, H.Q.; Song, C.P. COP1 promotes ABA-induced stomatal closure by modulating the abundance of ABI/HAB and AHG3 phosphatases. New Phytol. 2021, 229, 2035–2049. [Google Scholar] [CrossRef]
- Li, X.; Yu, B.; Wu, Q.; Min, Q.; Zeng, R.; Xie, Z.; Huang, J. OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PLoS Genet. 2021, 17, e1009699. [Google Scholar] [CrossRef]
- Kuromori, T.; Fujita, M.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Inter-tissue and inter-organ signaling in drought stress response and phenotyping of drought tolerance. Plant J. 2022, 109, 342–358. [Google Scholar] [CrossRef]
- Saavedra, X.; Modrego, A.; Rodríguez, D.; González-García, M.P.; Sanz, L.; Nicolás, G.; Lorenzo, O. The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol. 2010, 152, 133–150. [Google Scholar] [CrossRef]
- Li, W.; Li, L.G.; Sun, X.F.; Tang, K.X. An oleosin-fusion protein driven by the CaMV35S promoter is accumulated in Arabidopsis (Brassicaceae) seeds and correctly targeted to oil bodies. Gen. Mol. Res. 2012, 11, 2138–2146. [Google Scholar] [CrossRef]
- Im, J.H.; Yoo, S.D. Transient expression in Arabidopsis leaf mesophyll protoplast system for cell-based functional analysis of MAPK cascades signaling. Methods Mol. Biol. 2014, 1171, 3–12. [Google Scholar]
- Sheerin, D.J. Investigation of light-regulated protein-protein interactions using yeast two-hybrid assays. Methods Mol. Biol. 2019, 2026, 1–19. [Google Scholar] [PubMed]
- Yang, T.; Yao, S.; Hao, L.; Zhao, Y.; Lu, W.; Xiao, K. Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway. Plant Cell Rep. 2016, 35, 2309–2323. [Google Scholar] [CrossRef] [PubMed]
- Cheuk, A.; Ouellet, F.; Houde, M. The barley stripe mosaic virus expression system reveals the wheat C2H2 zinc finger protein TaZFP1B as a key regulator of drought tolerance. BMC Plant Biol. 2020, 20, 44. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Guo, X.; Li, B.; Chen, H.; Qiao, X. Characterization of effects of different tea harvesting seasons on quality components, color and sensory quality of “Yinghong 9” and “Huangyu” large-leaf-variety black tea. Molecule 2022, 27, 8720. [Google Scholar] [CrossRef]
- Kumar, R.; Mamrutha, H.M.; Kaur, A.; Venkatesh, K.; Sharma, D.; Singh, G.P. Optimization of Agrobacterium-mediated transformation in spring bread wheat using mature and immature embryos. Mol. Biol. Rep. 2019, 46, 1845–1853. [Google Scholar] [CrossRef]
- Shen, Q.; Liu, Z.; Song, F.; Xie, Q.; Hanley-Bowdoin, L.; Zhou, X. Tomato SlSnRK1 protein interacts with and phosphorylates âC1, a pathogenesis protein encoded by a geminivirus â-satellite. Plant Physiol. 2011, 157, 1394–1406. [Google Scholar] [CrossRef]
- Sun, J.; Qi, L.; Li, Y.; Zhai, Q.; Li, C. PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 2013, 25, 2102–2114. [Google Scholar] [CrossRef]
- Imadi, S.R.; Kazi, A.G.; Ahanger, M.A.; Gucel, S.; Ahmad, P. Plant transcriptomics and responses to environmental stress: An overview. J. Genet. 2015, 94, 525–537. [Google Scholar] [CrossRef]
- Liu, L.; Huang, L.; Lin, X.; Sun, C. Hydrogen peroxide alleviates salinity-induced damage through enhancing proline accumulation in wheat seedlings. Plant Cell Rep. 2020, 39, 567–575. [Google Scholar] [CrossRef]
- Huang, X.S.; Liu, J.H.; Chen, X.J. Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol. 2010, 10, 230. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO: TermFinder-open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhao, Y.; Hou, X.; Ni, C.; Han, L.; Du, P.; Xiao, K. Wheat ABA Receptor TaPYL5 Constitutes a Signaling Module with Its Downstream Partners TaPP2C53/TaSnRK2.1/TaABI1 to Modulate Plant Drought Response. Int. J. Mol. Sci. 2023, 24, 7969. https://doi.org/10.3390/ijms24097969
Zhang Y, Zhao Y, Hou X, Ni C, Han L, Du P, Xiao K. Wheat ABA Receptor TaPYL5 Constitutes a Signaling Module with Its Downstream Partners TaPP2C53/TaSnRK2.1/TaABI1 to Modulate Plant Drought Response. International Journal of Molecular Sciences. 2023; 24(9):7969. https://doi.org/10.3390/ijms24097969
Chicago/Turabian StyleZhang, Yanyang, Yingjia Zhao, Xiaoyang Hou, Chenyang Ni, Le Han, Pingping Du, and Kai Xiao. 2023. "Wheat ABA Receptor TaPYL5 Constitutes a Signaling Module with Its Downstream Partners TaPP2C53/TaSnRK2.1/TaABI1 to Modulate Plant Drought Response" International Journal of Molecular Sciences 24, no. 9: 7969. https://doi.org/10.3390/ijms24097969



