5-HT3 Receptors on Mitochondria Influence Mitochondrial Function
Abstract
1. Introduction
2. Results
2.1. 5-HT3 Receptor Subunit Protein Localization Signal Predictions
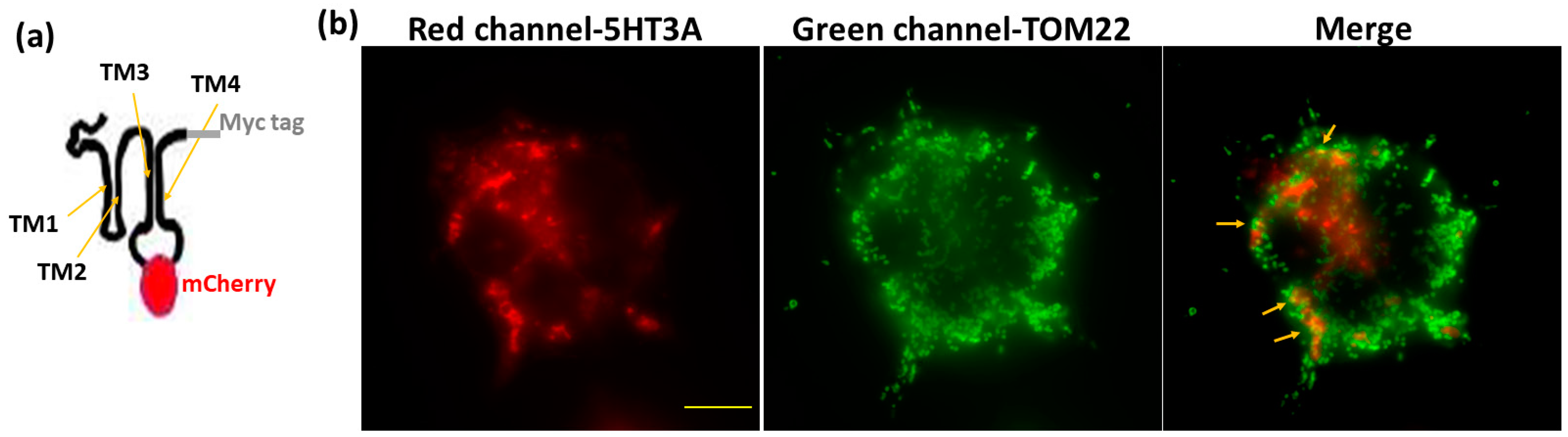
2.2. 5-HT3A and 5HT3E Subunits Localize to Mitochondria
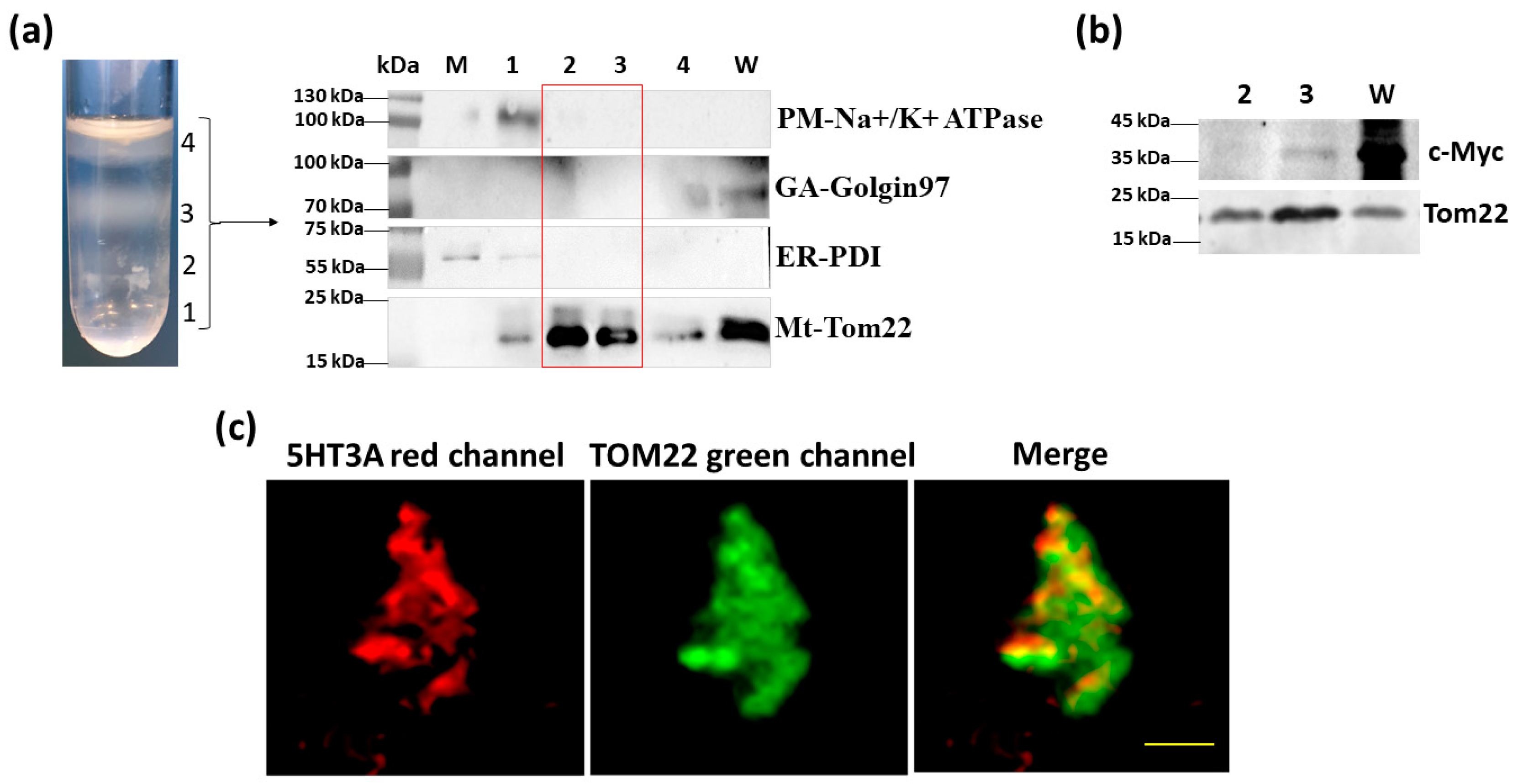
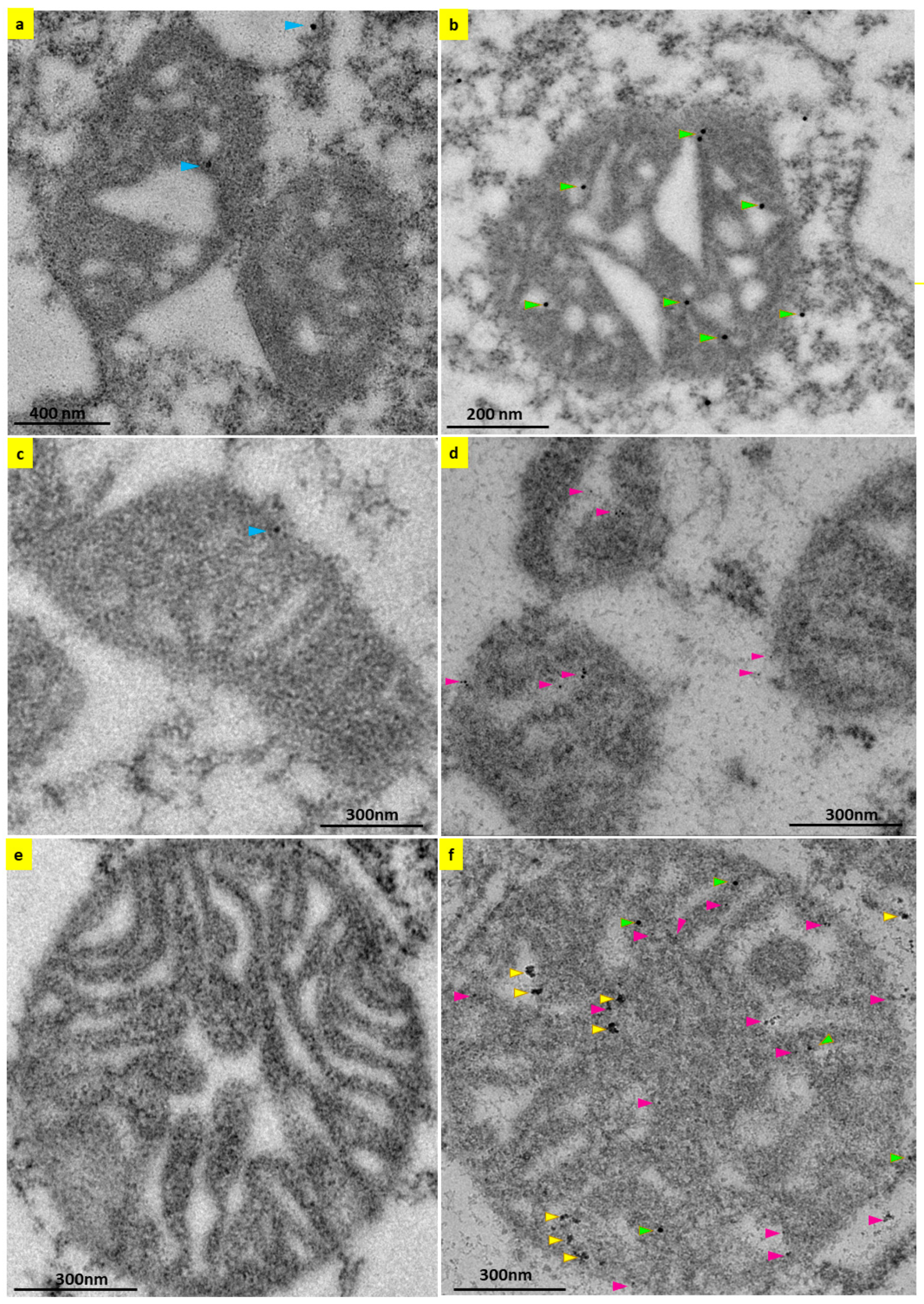
2.3. Localization of 5-HT3 Receptor Subunits to the Inner Mitochondrial Membrane
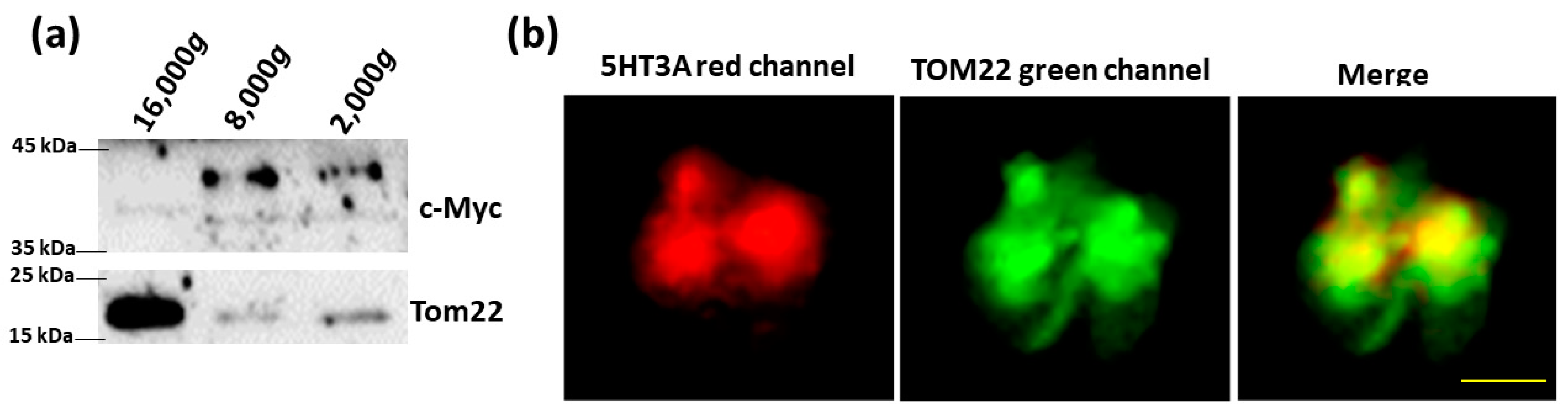
2.4. Cell-Free Mitochondria Contain 5HT3A Subunits
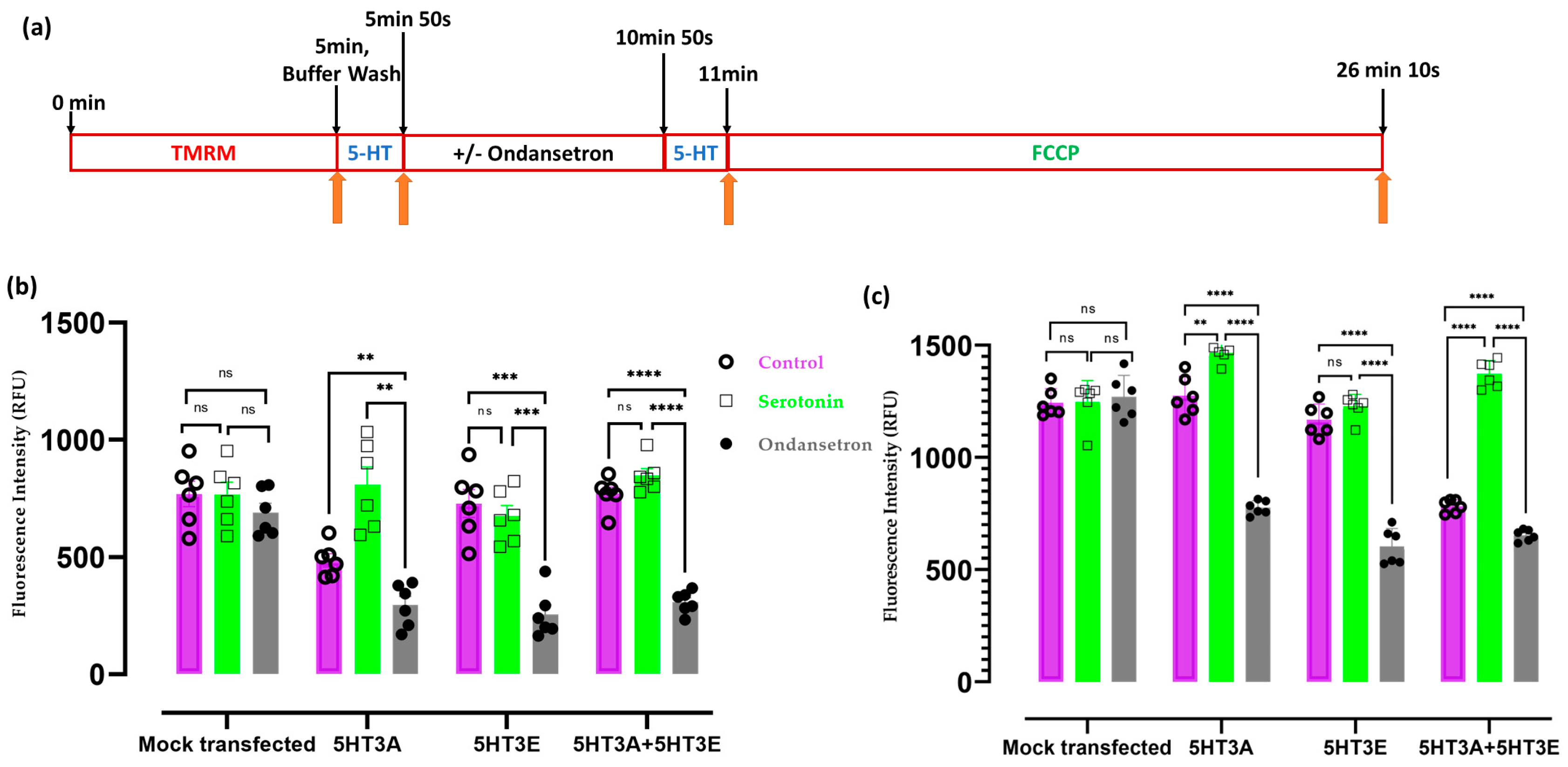
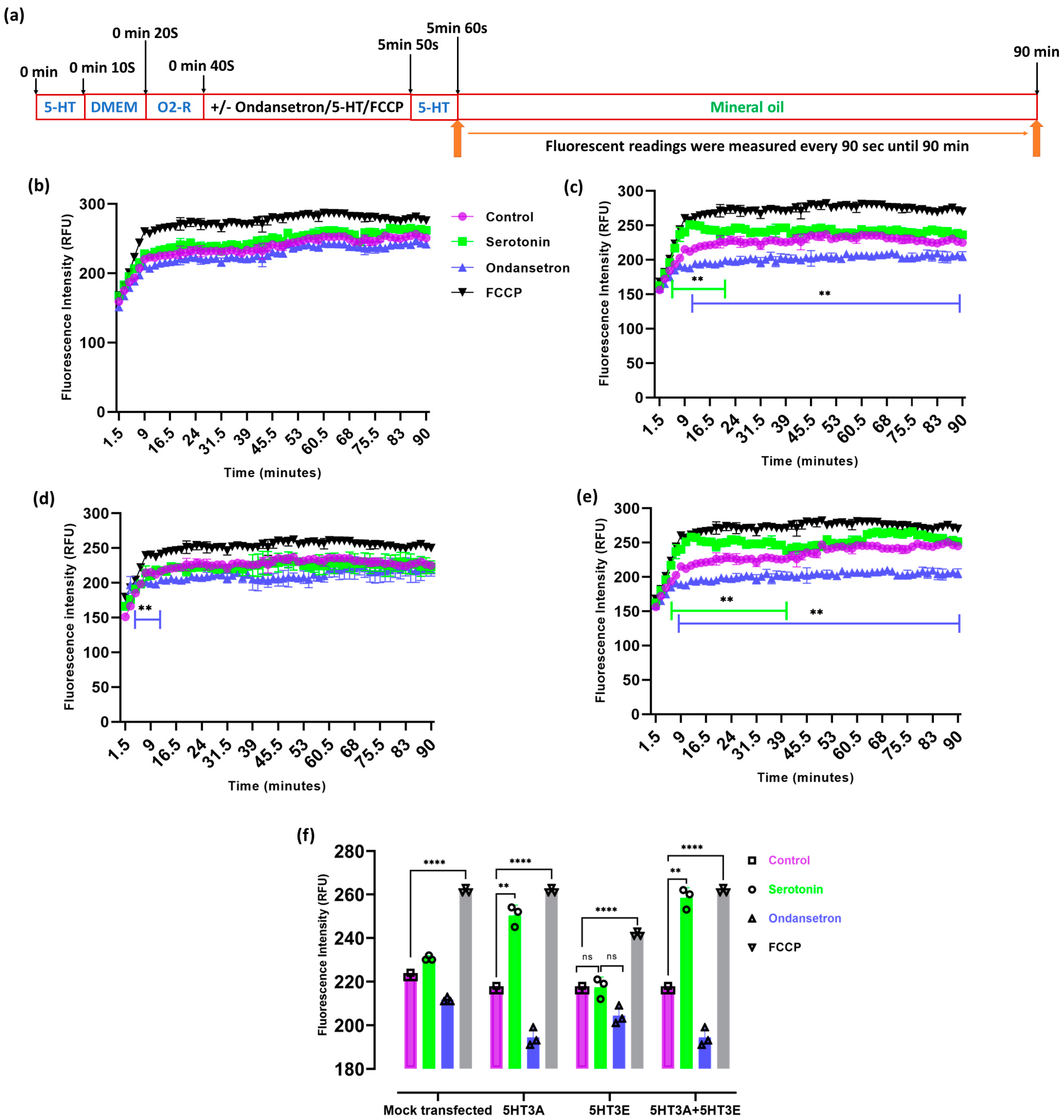
2.5. 5-HT3 Receptors Modulate Mitochondrial Membrane Potential
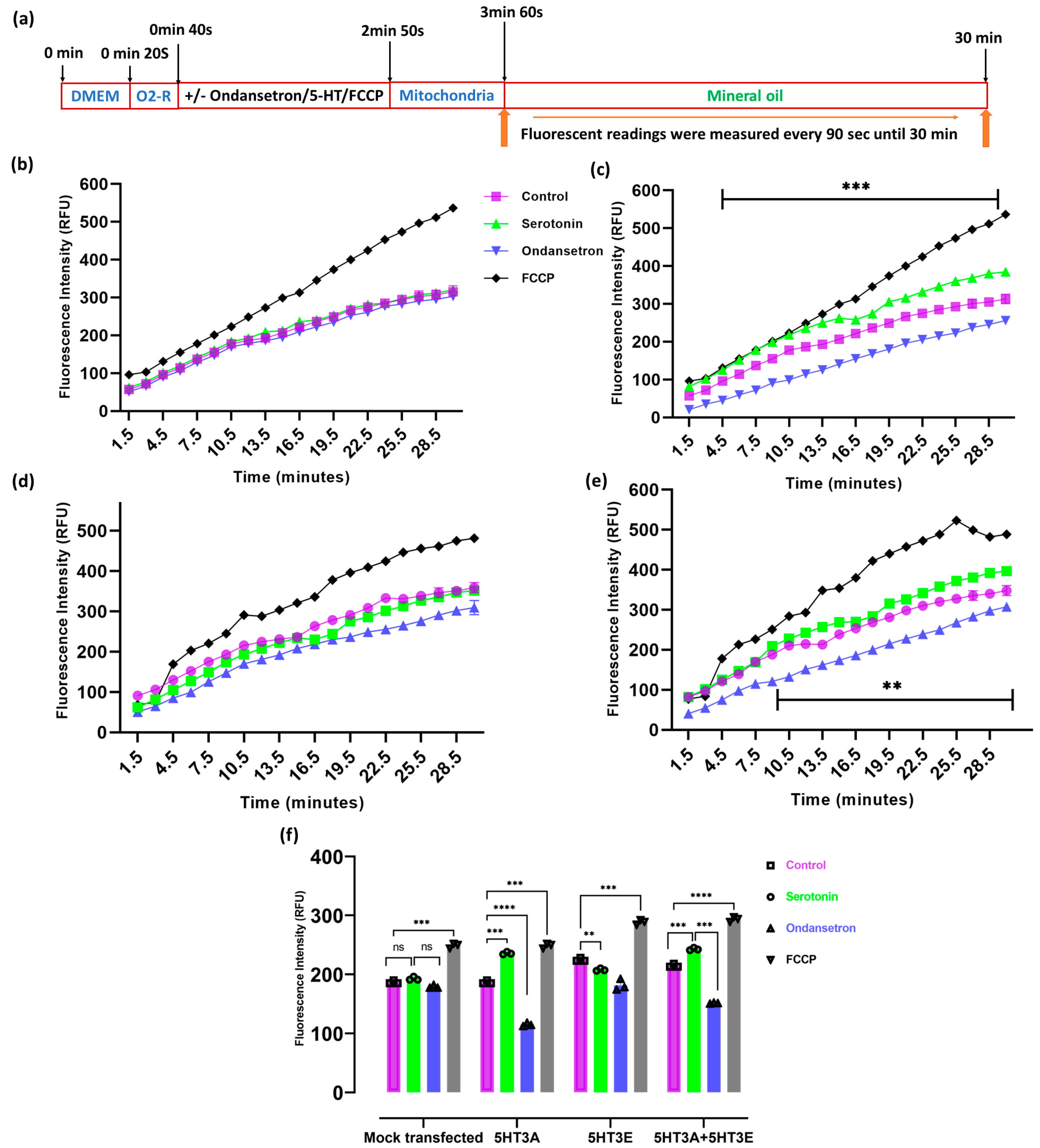
2.6. 5-HT3 Receptor Alters the Oxygen Consumption Rate of Mitochondria
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Media
5.2. In Silico Analyses of Localization Signals Present in 5-HT3 Receptor Subunits
5.3. 5-HT3 Receptor Subunit Constructs
5.4. Cell Culture and Transfections
5.5. Isolation of Mitochondrial Fractions
5.6. Isolation of Cell-Free Mitochondria
5.7. Imaging of the Cells and Purified Mitochondria
5.7.1. Sample Preparation for Fluorescence Imaging
5.7.2. Transmission Electron Microscopy Sample Preparation
5.8. Mitochondrial Membrane Potential Assays
5.9. Oxygen Consumption Rate (OCR) Measurements
5.10. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barnes, N.M.; Hales, T.G.; Lummis, S.C.; Peters, J.A. The 5-HT3 receptor—The relationship between structure and function. Neuropharmacology 2009, 56, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Niesler, B.; Frank, B.; Kapeller, J.; Rappold, G.A. Cloning, physical mapping and expression analysis of the human 5-HT3 serotonin receptor-like genes HTR3C, HTR3D and HTR3E. Gene 2003, 310, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, J.D.; Gill, C.H.; Zebda, N.; Spencer, J.P.; Leyland, R.; Rance, K.H.; Trinh, H.; Balmer, G.; Kelly, F.M.; Yusaf, S.P.; et al. Characterisation of 5-HT3C, 5-HT3D and 5-HT3E receptor subunits: Evolution, distribution and function. J. Neurochem. 2009, 108, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Lummis, S.C.R. 5-HT(3) receptors. J. Biol. Chem. 2012, 287, 40239–40245. [Google Scholar] [CrossRef] [PubMed]
- Miyake, A.; Mochizuki, S.; Takemoto, Y.; Akuzawa, S. Molecular cloning of human 5-hydroxytryptamine3 receptor: Heterogeneity in distribution and function among species. Mol. Pharmacol. 1995, 48, 407–416. [Google Scholar] [PubMed]
- Hassaine, G.; Deluz, C.; Grasso, L.; Wyss, R.; Tol, M.B.; Hovius, R.; Graff, A.; Stahlberg, H.; Tomizaki, T.; Desmyter, A.; et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 2014, 512, 276–281. [Google Scholar] [CrossRef]
- Basak, S.; Gicheru, Y.; Samanta, A.; Molugu, S.K.; Huang, W.; la de Fuente, M.; Hughes, T.; Taylor, D.J.; Nieman, M.T.; Moiseenkova-Bell, V.; et al. Cryo-EM structure of 5-HT(3A) receptor in its resting conformation. Nat. Commun. 2018, 9, 514. [Google Scholar] [CrossRef]
- Polovinkin, L.; Hassaine, G.; Perot, J.; Neumann, E.; Jensen, A.A.; Lefebvre, S.N.; Corringer, P.-J.; Neyton, J.; Chipot, C.; Dehez, F.; et al. Conformational transitions of the serotonin 5-HT(3) receptor. Nature 2018, 563, 275–279. [Google Scholar] [CrossRef]
- Basak, S.; Gicheru, Y.; Kapoor, A.; Mayer, M.L.; Filizola, M.; Chakrapani, S. Molecular mechanism of setron-mediated inhibition of full-length 5-HT(3A) receptor. Nat. Commun. 2019, 10, 3225. [Google Scholar] [CrossRef]
- Basak, S.; Kumar, A.; Ramsey, S.; Gibbs, E.; Kapoor, A.; Filizola, M.; Chakrapani, S. High-resolution structures of multiple 5-HT(3A)R-setron complexes reveal a novel mechanism of competitive inhibition. eLife 2020, 9, e57870. [Google Scholar] [CrossRef]
- Zarkadas, E.; Zhang, H.; Cai, W.; Effantin, G.; Perot, J.; Neyton, J.; Chipot, C.; Schoehn, G.; Dehez, F.; Nury, H. The Binding of Palonosetron and Other Antiemetic Drugs to the Serotonin 5-HT3 Receptor. Structure 2020, 28, 1131–1140.e4. [Google Scholar] [CrossRef] [PubMed]
- Walstab, J.; Rappold, G.; Niesler, B. 5-HT3 receptors: Role in disease and target of drugs. Pharmacol. Therapeut. 2010, 128, 146–169. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.; Smidt, M.; Van Hooft, J. The serotonin 5-HT3 receptor: A novel neurodevelopmental target. Front. Cell. Neurosci. 2013, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Mawe, G.M.; Hoffman, J.M. Serotonin signalling in the gut—Functions, dysfunctions and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 473. [Google Scholar] [CrossRef]
- Thompson, A.J.; Lummis, S.C.R. 5-HT3 receptors. Curr. Pharm. Des. 2006, 12, 3615–3630. [Google Scholar] [CrossRef]
- Dubin, A.E.; Huvar, R.; D’Andrea, M.R.; Pyati, J.; Zhu, J.Y.; Joy, K.C.; Wilson, S.J.; Galindo, J.E.; Glass, C.A.; Luo, L.; et al. The pharmacological and functional characteristics of the serotonin 5-HT3A receptor are specifically modified by a 5-HT3B receptor subunit. J. Biol. Chem. 1999, 274, 30799–30810. [Google Scholar] [CrossRef]
- Davies, P.A.; Pistis, M.; Hanna, M.C.; Peters, J.A.; Lambert, J.J.; Hales, T.G.; Kirkness, E.F.J.N. The 5-HT 3B subunit is a major determinant of serotonin-receptor function. Nature 1999, 397, 359–363. [Google Scholar] [CrossRef]
- Chetty, N.; Coupar, I.M.; Tan, Y.Y.; Desmond, P.V.; Irving, H.R. Distribution of serotonin receptors and interacting proteins in the human sigmoid colon. Neurogastroenterol. Motil. 2009, 21, 551–558, e14–e15. [Google Scholar] [CrossRef]
- Walstab, J.; Hammer, C.; Lasitschka, F.; Möller, D.; Connolly, C.N.; Rappold, G.; Brüss, M.; Bönisch, H.; Niesler, B. RIC-3 exclusively enhances the surface expression of human homomeric 5-hydroxytryptamine type 3A (5-HT3A) receptors despite direct interactions with 5-HT3A, -C, -D, and -E subunits. J. Biol. Chem. 2010, 285, 26956–26965. [Google Scholar] [CrossRef]
- Yaakob, N.S.; Chinkwo, K.A.; Chetty, N.; Coupar, I.M.; Irving, H.R. Distribution of 5-HT3, 5-HT4, and 5-HT7 Receptors Along the Human Colon. J. Neurogastroenterol. Motil. 2015, 21, 361–369. [Google Scholar] [CrossRef]
- Pick, H.; Preuss, A.K.; Mayer, M.; Wohland, T.; Hovius, R.; Vogel, H. Monitoring expression and clustering of the ionotropic 5HT3 receptor in plasma membranes of live biological cells. Biochemistry 2003, 42, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Ilegems, E.; Pick, H.M.; Deluz, C.; Kellenberger, S.; Vogel, H. Noninvasive imaging of 5-HT3 receptor trafficking in live cells: From biosynthesis to endocytosis. J. Biol. Chem. 2004, 279, 53346–53352. [Google Scholar] [CrossRef] [PubMed]
- Miles, T.F.; Dougherty, D.A.; Lester, H.A. The 5-HT3AB receptor shows an A3B2 stoichiometry at the plasma membrane. Biophys. J. 2013, 105, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Abad, I.P.L.; Fam, R.L.; Nguyen, D.-T.; Nowell, C.J.; Trinh, P.N.; Manallack, D.T.; Freihat, L.A.; Chakrabarti, J.; Jamil, A.; Exintaris, B.; et al. Visualising functional 5-HT3 receptors containing A and C subunits at or near the cell surface. Biomed. Pharmacotherap. 2020, 132, 110860. [Google Scholar] [CrossRef] [PubMed]
- Maricq, A.V.; Peterson, A.S.; Brake, A.J.; Myers, R.M.; Julius, D. Primary structure and functional expression of the 5HT3 receptor, a serotonin-gated ion channel. Science 1991, 254, 432–437. [Google Scholar] [CrossRef]
- Boess, F.G.; Beroukhim, R.; Martin, I.L. Ultrastructure of the 5-Hydroxytryptamine3 Receptor. J. Neurochem. 1995, 64, 1401–1405. [Google Scholar] [CrossRef]
- Green, T.; Stauffer, K.A.; Lummis, S.C. Expression of recombinant homo-oligomeric 5-hydroxytryptamine receptors provides new insights into their maturation and structure. J. Biol. Chem. 1995, 270, 6056–6061. [Google Scholar] [CrossRef]
- Boyd, G.W.; Doward, A.I.; Kirkness, E.F.; Millar, N.S.; Connolly, C.N. Cell surface expression of 5-hydroxytryptamine type 3 receptors is controlled by an endoplasmic reticulum retention signal. J. Biol. Chem. 2003, 278, 27681–27687. [Google Scholar] [CrossRef]
- Price, K.L.; Hirayama, Y.; Lummis, S.C. Subtle differences among 5-HT3AC, 5-HT3AD, and 5-HT3AE receptors are revealed by partial agonists. ACS Chem. Neurosci. 2017, 8, 1085–1091. [Google Scholar] [CrossRef]
- Yaakob, N.S.; Nguyen, D.T.; Exintaris, B.; Irving, H.R. The C and E subunits of the serotonin 5-HT(3) receptor subtly modulate electrical properties of the receptor. Biomed. Pharmacotherap. 2018, 97, 1701–1709. [Google Scholar] [CrossRef]
- Niesler, B.; Walstab, J.; Combrink, S.; Möller, D.; Kapeller, J.; Rietdorf, J.; Bönisch, H.; Göthert, M.; Rappold, G.; Brüss, M. Characterization of the novel human serotonin receptor subunits 5-HT3C,5-HT3D, and 5-HT3E. Mol. Pharmacol. 2007, 72, 8. [Google Scholar] [CrossRef] [PubMed]
- Kapeller, J.; Möller, D.; Lasitschka, F.; Autschbach, F.; Hovius, R.; Rappold, G.; Brüss, M.; Gershon, M.D.; Niesler, B. Serotonin receptor diversity in the human colon: Expression of serotonin type 3 receptor subunits 5-HT3C, 5-HT3D, and 5-HT3E. J. Comp. Neurol. 2011, 519, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Vasioukhin, V.; Bauer, C.; Yin, M.; Fuchs, E. Directed actin polymerization is the driving force for epithelial cell–cell adhesion. Cell 2000, 100, 209–219. [Google Scholar] [CrossRef]
- Emerit, M.B.; Doucet, E.; Darmon, M.; Hamon, M. Native and cloned 5-HT3A (S) receptors are anchored to F-actin in clonal cells and neurons. Mol. Cell. Neurosci. 2002, 20, 110–124. [Google Scholar] [CrossRef]
- Boyd, G.W.; Low, P.; Dunlop, J.I.; Robertson, L.A.; Vardy, A.; Lambert, J.J.; Peters, J.A.; Connolly, C.N. Assembly and cell surface expression of homomeric and heteromeric 5-HT3 receptors: The role of oligomerization and chaperone proteins. Mol. Cell. Neurosci. 2002, 21, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, H.; Xu, H.; Guo, D.; Shi, H.; Li, Y.; Zhang, W.; Gu, Y. 5-HTR3 and 5-HTR4 located on the mitochondrial membrane and functionally regulated mitochondrial functions. Sci. Rep. 2016, 6, 37336. [Google Scholar] [CrossRef]
- Gouw, M.; Michael, S.; Sanchez, H.C.S.; Kumar, M.; Zeke, A.; Lang, B.; Bely, B.; Chemes, L.B.; E Davey, N.; Deng, Z.; et al. The eukaryotic linear motif resource—2018 update. Nucleic Acids Res. 2017, 46, D428–D434. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Casadio, R. TPpred2: Improving the prediction of mitochondrial targeting peptide cleavage sites by exploiting sequence motifs. Bioinformatics 2014, 30, 2973–2974. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Tsuji, J.; Fu, S.-C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom. 2015, 14, 1113–1126. [Google Scholar] [CrossRef]
- Neupert, W. Protein import into mitochondria. Annu. Rev. Biochem. 1997, 66, 863–917. [Google Scholar] [CrossRef]
- Brüss, M.; Molderings, G.J.; Bönisch, H.; Göthert, M. Pharmacological differences and similarities between the native mouse 5-HT3 receptor in N1E-115 cells and a cloned short splice variant of the mouse 5-HT3 receptor expressed in HEK 293 cells. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1999, 360, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Correia, A.S.; Silva, I.; Oliveira, J.C.; Reguengo, H.; Vale, N. Serotonin Type 3 Receptor Is Potentially Involved in Cellular Stress Induced by Hydrogen Peroxide. Life 2022, 12, 1645. [Google Scholar] [CrossRef] [PubMed]
- Park, S.S.; Jeong, H.; Andreazza, A.C. Circulating cell-free mitochondrial DNA in brain health and disease: A systematic review and meta-analysis. World J. Biol. Psychiatry 2022, 23, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Quirk, P.L.; Rao, S.; Roth, B.L.; Siegel, R.E. Three putative N-glycosylation sites within the murine 5-HT3A receptor sequence affect plasma membrane targeting, ligand binding, and calcium influx in heterologous mammalian cells. J. Neurosci. Res. 2004, 77, 498–506. [Google Scholar] [CrossRef]
- Irving, H.; Turek, I.; Kettle, C.; Yaakob, N. Tapping into 5-HT(3) Receptors to Modify Metabolic and Immune Responses. Int. J. Mol. Sci. 2021, 22, 1910. [Google Scholar] [CrossRef]
- Rapoport, T.A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 2007, 450, 663–669. [Google Scholar] [CrossRef]
- Miquel, M.C.; Emerit, M.B.; Bolaños, F.J.; Schechter, L.E.; Gozlan, H.; Hamon, M. Physicochemical properties of serotonin 5-HT3 binding sites solubilized from membranes of NG 108-15 neuroblastoma-glioma cells. J. Neurochem. 1990, 55, 1526–1536. [Google Scholar] [CrossRef]
- Fletcher, S.; Barnes, N.M. Purification of 5-hydroxytryptamine3 receptors from porcine brain. Br. J. Pharmacol. 1997, 122, 655–662. [Google Scholar] [CrossRef]
- Freeman, S.L.; Glatzle, J.; Robin, C.S.; Valdellon, M.; Sternini, C.; Sharp, J.W.; Raybould, H.E. Ligand-induced 5-HT3 receptor internalization in enteric neurons in rat ileum. Gastroenterology 2006, 131, 97–107. [Google Scholar] [CrossRef]
- Sun, H.; Hu, X.Q.; Emerit, M.B.; Schoenebeck, J.C.; Kimmel, C.E.; Peoples, R.W.; Miko, A.; Zhang, L. Modulation of 5-HT3 receptor desensitization by the light chain of microtubule-associated protein 1B expressed in HEK 293 cells. J. Physiol. 2008, 586, 751–762. [Google Scholar] [CrossRef]
- Rojas, C.; Thomas, A.G.; Alt, J.; Stathis, M.; Zhang, J.; Rubenstein, E.B.; Sebastiani, S.; Cantoreggi, S.; Slusher, B.S. Palonosetron triggers 5-HT(3) receptor internalization and causes prolonged inhibition of receptor function. Eur. J. Pharmacol. 2010, 626, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Stathis, M.; Pietra, C.; Rojas, C.; Slusher, B.S. Inhibition of substance P-mediated responses in NG108-15 cells by netupitant and palonosetron exhibit synergistic effects. Eur. J. Pharmacol. 2012, 689, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Gavel, Y.; von Heijne, G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. Des. Sel. 1990, 4, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef]
- Pirisinu, M.; Blasco, P.; Tian, X.; Sen, Y.; Bode, A.M.; Liu, K.; Dong, Z. Analysis of hydrophobic and hydrophilic moments of short penetrating peptides for enhancing mitochondrial localization: Prediction and validation. FASEB J. 2019, 33, 7970–7984. [Google Scholar] [CrossRef] [PubMed]
- Del Cadia, M.; De Rienzo, F.; Weston, D.A.; Thompson, A.J.; Menziani, M.C.; Lummis, S.C. Exploring a potential palonosetron allosteric binding site in the 5-HT(3) receptor. Bioorg. Med. Chem. 2013, 21, 7523–7528. [Google Scholar] [CrossRef] [PubMed]
- Lansdell, S.J.; Sathyaprakash, C.; Doward, A.; Millar, N.S. Activation of human 5-hydroxytryptamine type 3 receptors via an allosteric transmembrane site. Mol. Pharmacol. 2015, 87, 87–95. [Google Scholar] [CrossRef]
- Jack, T.; Leuenberger, M.; Ruepp, M.D.; Vernekar, S.K.V.; Thompson, A.J.; Braga-Lagache, S.; Heller, M.; Lochner, M. Mapping the Orthosteric Binding Site of the Human 5-HT(3) Receptor Using Photo-cross-linking Antagonists. ACS Chem. Neurosci. 2019, 10, 438–450. [Google Scholar] [CrossRef]
- Rodriguez Araujo, N.; Fabiani, C.; Mazzarini Dimarco, A.; Bouzat, C.; Corradi, J. Orthosteric and Allosteric Activation of Human 5-HT(3)A Receptors. Biophys. J. 2020, 119, 1670–1682. [Google Scholar] [CrossRef]
- Lochner, M.; Lummis, S.C. Agonists and antagonists bind to an A-A interface in the heteromeric 5-HT3AB receptor. Biophys. J. 2010, 98, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Duffy, N.H.; Lester, H.A.; Dougherty, D.A. Ondansetron and granisetron binding orientation in the 5-HT(3) receptor determined by unnatural amino acid mutagenesis. ACS Chem. Biol. 2012, 7, 1738–1745. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.; Macêdo, D.; Bandeira, I.; Maldonado, I.; Salles, L.; Azevedo, M.F.; Rocha, M.A., Jr.; Fregoneze, J.B.; De Castro-e-Silva, E. Central 5-HT3 receptor stimulation by m-CPBG increases blood glucose in rats. Hormone Metabol. Res. 2002, 34, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Browning, K.N. Glucose increases synaptic transmission from vagal afferent central nerve terminals via modulation of 5-HT3 receptors. Am.J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G1050–G1057. [Google Scholar] [CrossRef]
- Troy, A.E.; Simmonds, S.S.; Stocker, S.D.; Browning, K.N. High fat diet attenuates glucose-dependent facilitation of 5-HT3-mediated responses in rat gastric vagal afferents. J. Physiol. 2016, 594, 99–114. [Google Scholar] [CrossRef]
- Goodarzi, P.; Habibi, M.; Roberts, K.; Sutton, J.; Shili, C.N.; Lin, D.; Pezeshki, A. Dietary Tryptophan Supplementation Alters Fat and Glucose Metabolism in a Low-Birthweight Piglet Model. Nutrients 2021, 13, 2561. [Google Scholar] [CrossRef]
- Babic, T.; Troy, A.E.; Fortna, S.R.; Browning, K.N. Glucose-dependent trafficking of 5-HT3 receptors in rat gastrointestinal vagal afferent neurons. Neurogastroenterol. Motil. 2012, 24, e476–e488. [Google Scholar] [CrossRef]
- Weber, S.; Volynets, V.; Kanuri, G.; Bergheim, I.; Bischoff, S.C. Treatment with the 5-HT3 antagonist tropisetron modulates glucose-induced obesity in mice. Int. J. Obes (2005) 2009, 33, 1339–1347. [Google Scholar] [CrossRef]
- Makhmutova, M.; Weitz, J.; Tamayo, A.; Pereira, E.; Boulina, M.; Almaça, J.; Rodriguez-Diaz, R.; Caicedo, A. Pancreatic β-Cells Communicate With Vagal Sensory Neurons. Gastroenterology 2021, 160, 875–888.e11. [Google Scholar] [CrossRef]
- Utsumi, D.; Matsumoto, K.; Amagase, K.; Horie, S.; Kato, S. 5-HT3 receptors promote colonic inflammation via activation of substance P/neurokinin-1 receptors in dextran sulphate sodium-induced murine colitis. Br. J. Pharmacol. 2016, 173, 1835–1849. [Google Scholar] [CrossRef]
- Ritter, K.E.; Buehler, D.P.; Asher, S.B.; Deal, K.K.; Zhao, S.; Guo, Y.; Southard-Smith, E.M. 5-HT3 Signaling Alters Development of Sacral Neural Crest Derivatives That Innervate the Lower Urinary Tract. Int. J. Mol. Sci. 2021, 22, 6838. [Google Scholar] [CrossRef] [PubMed]
- Giorgi, C.; Marchi, S.; Simoes, I.C.; Ren, Z.; Morciano, G.; Perrone, M.; Patalas-Krawczyk, P.; Borchard, S.; Jędrak, P.; Pierzynowska, K.; et al. Mitochondria and Reactive Oxygen Species in Aging and Age-Related Diseases. Int. Rev. Cell Mol. Biol. 2018, 340, 209–344. [Google Scholar] [CrossRef] [PubMed]
- Andrieux, P.; Chevillard, C.; Cunha-Neto, E.; Nunes, J.P.S. Mitochondria as a Cellular Hub in Infection and Inflammation. Int. J. Mol. Sci. 2021, 22, 11338. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, W.; Tsai, P.J.; Chen, P.H.; Ye, M.; Guo, J.; Su, Z. Mutual interaction between endoplasmic reticulum and mitochondria in nonalcoholic fatty liver disease. Lipids Health Dis. 2020, 19, 72. [Google Scholar] [CrossRef] [PubMed]
- Gheibi, S.; Samsonov, A.P.; Gheibi, S.; Vazquez, A.B.; Kashfi, K. Regulation of carbohydrate metabolism by nitric oxide and hydrogen sulfide: Implications in diabetes. Biochem. Pharmacol. 2020, 176, 113819. [Google Scholar] [CrossRef] [PubMed]
- Curtis, J.J.; Vo, N.T.K.; Seymour, C.B.; Mothersill, C.E. 5-HT(2A) and 5-HT(3) receptors contribute to the exacerbation of targeted and non-targeted effects of ionizing radiation-induced cell death in human colon carcinoma cells. Int. J. Radiat. Biol. 2020, 96, 482–490. [Google Scholar] [CrossRef]
- Watanabe, H.; Saito, R.; Nakano, T.; Takahashi, H.; Takahashi, Y.; Sumiyoshi, K.; Sato, K.; Chen, X.; Okada, N.; Iwasaki, S.; et al. Effect of peripheral 5-HT on glucose and lipid metabolism in wether sheep. PLoS ONE 2014, 9, e88058. [Google Scholar] [CrossRef]
- Krupkova, M.; Sedova, L.; Liska, F.; Krenova, D.; Kren, V.; Seda, O. Differential effects of 5-HT3 receptor antagonist on lipid profile in spontaneously hypertensive rat and chromosome 8 congenic strain. Neuro Endocrinol. Lett. 2012, 33 (Suppl. 2), 43–49. [Google Scholar]
- Al Amir Dache, Z.; Otandault, A.; Tanos, R.; Pastor, B.; Meddeb, R.; Sanchez, C.; Arena, G.; Lasorsa, L.; Bennett, A.; Grange, T.; et al. Blood contains circulating cell-free respiratory competent mitochondria. FASEB J. 2020, 34, 3616–3630. [Google Scholar] [CrossRef]
- Lindqvist, D.; Wolkowitz, O.M.; Picard, M.; Ohlsson, L.; Bersani, F.S.; Fernström, J.; Westrin, Å.; Hough, C.M.; Lin, J.; Reus, V.I.; et al. Circulating cell-free mitochondrial DNA, but not leukocyte mitochondrial DNA copy number, is elevated in major depressive disorder. Neuropsychopharmacology 2018, 43, 1557–1564. [Google Scholar] [CrossRef]
- Park, J.H.; Hayakawa, K. Extracellular Mitochondria Signals in CNS Disorders. Front. Cell Dev. Biol. 2021, 9, 642853. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.H.; Jo, S.; Kim, S.J.; Lee, J.M.; Jeong, J.H.; Kang, J.S.; Cho, N.-J.; Kim, S.S.; Lee, E.Y.; Moon, J.-S. Circulating Cell-Free mtDNA Contributes to AIM2 Inflammasome-Mediated Chronic Inflammation in Patients with Type 2 Diabetes. Cells 2019, 8, 328. [Google Scholar] [CrossRef]
- Nakamura, Y.; Park, J.H.; Hayakawa, K. Therapeutic use of extracellular mitochondria in CNS injury and disease. Exp. Neurol. 2020, 324, 113114. [Google Scholar] [CrossRef] [PubMed]
- Parra, E.; LeGatt, A.; Court, A.; Figueroa, F.E.; Khoury, M. Artificial mitochondria transfer prevents staurosporine-induced apoptosis of human T lymphocytes. Cytotherapy 2019, 21, e8. [Google Scholar] [CrossRef]
- Miliotis, S.; Nicolalde, B.; Ortega, M.; Yepez, J.; Caicedo, A. Forms of extracellular mitochondria and their impact in health. Mitochondrion 2019, 48, 16–30. [Google Scholar] [CrossRef]
- Hummel, E.M.; Piovesan, K.; Berg, F.; Herpertz, S.; Kessler, H.; Kumsta, R.; Moser, D.A. Mitochondrial DNA as a marker for treatment-response in post-traumatic stress disorder. Psychoneuroendocrinology 2023, 148, 105993. [Google Scholar] [CrossRef]
- Gorman, G.S.; McFarland, R.; Stewart, J.; Feeney, C.; Turnbull, D.M. Mitochondrial donation: From test tube to clinic. Lancet 2018, 392, 1191–1192. [Google Scholar] [CrossRef]
- Cree, L.; Loi, P. Mitochondrial replacement: From basic research to assisted reproductive technology portfolio tool-technicalities and possible risks. Mol. Hum. Reprod. 2015, 21, 3–10. [Google Scholar] [CrossRef]
- Santra, S.; Gilkerson, R.W.; Davidson, M.; Schon, E.A. Ketogenic treatment reduces deleted mitochondrial DNAs in cultured human cells. Ann. Neurol. 2004, 56, 662–669. [Google Scholar] [CrossRef]
- Amore, G.; Romagnoli, M.; Carbonelli, M.; Barboni, P.; Carelli, V.; La Morgia, C. Therapeutic Options in Hereditary Optic Neuropathies. Drugs 2021, 81, 57–86. [Google Scholar] [CrossRef]
- Mól, A.R.; Castro, M.S.; Fontes, W. NetWheels: A web application to create high quality peptide helical wheel and net projections. bioRxiv 2018. [Google Scholar] [CrossRef]
- Gieselmann, L.; Kreer, C.; Ercanoglu, M.S.; Lehnen, N.; Zehner, M.; Schommers, P.; Potthoff, J.; Gruell, H.; Klein, F. Effective high-throughput isolation of fully human antibodies targeting infectious pathogens. Nat. Protoc. 2021, 16, 3639–3671. [Google Scholar] [CrossRef] [PubMed]
- Shapovalov, G.; Ritaine, A.; Bidaux, G.; Slomianny, C.; Borowiec, A.S.; Gordienko, D.; Bultynck, G.; Skryma, R.; Prevarskaya, N. Organelle membrane derived patches: Reshaping classical methods for new targets. Sci. Rep. 2017, 7, 14082. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Yamashita, S.; Hirusaki, K.; Katoh, K.; Ohta, Y. Isolation of mitochondria by gentle cell membrane disruption, and their subsequent characterization. Biochem. Biophys. Res. Commun. 2015, 463, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Whelan, D.R.; Bell, T.D.M. Image artifacts in Single Molecule Localization Microscopy: Why optimization of sample preparation protocols matters. Sci. Rep. 2015, 5, 7924. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Aristov, A.; Lelandais, B.; Rensen, E.; Zimmer, C. ZOLA-3D allows flexible 3D localization microscopy over an adjustable axial range. Nat. Commun. 2018, 9, 2409. [Google Scholar] [CrossRef]
- Avogaro, L.; Querido, E.; Dalachi, M.; Jantsch, M.F.; Chartrand, P.; Cusanelli, E. Live-cell imaging reveals the dynamics and function of single-telomere TERRA molecules in cancer cells. RNA Biol. 2018, 15, 787–796. [Google Scholar] [CrossRef]
- Fu, Y.-L.; Zhang, B.; Mu, T.-W. LMAN1 (ERGIC-53) promotes trafficking of neuroreceptors. Biochem. Biophys. Res. Commun. 2019, 511, 356–362. [Google Scholar] [CrossRef]
- Gao, C.; Leng, Y.; Ma, J.; Rooke, V.; Rodriguez-Gonzalez, S.; Ramakrishnan, C.; Deisseroth, K.; Penzo, M.A. Two genetically, anatomically and functionally distinct cell types segregate across anteroposterior axis of paraventricular thalamus. Nat. Neurosci. 2020, 23, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Gulbranson, D.R.; Crisman, L.; Lee, M.; Ouyang, Y.; Menasche, B.L.; Demmitt, B.A.; Wan, C.; Nomura, T.; Ye, Y.; Yu, H.; et al. AAGAB controls AP2 adaptor assembly in clathrin-mediated endocytosis. Develop. Cell 2019, 50, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Larson, S.M.; Truscott, L.C.; Chiou, T.-T.; Patel, A.; Kao, R.; Tu, A.; Tyagi, T.; Lu, X.; Elashoff, D.; De Oliveira, S.N. Pre-clinical development of gene modification of haematopoietic stem cells with chimeric antigen receptors for cancer immunotherapy. Hum. Vaccines Immunother. 2017, 13, 1094–1104. [Google Scholar] [CrossRef] [PubMed]
- Miyama, A.; Mimura, T.; Noma, H.; Goto, M.; Kamei, Y.; Kondo, A.; Saito, Y.; Okuma, H.; Matsubara, M. Specific IgG for cat allergens in patients with allergic conjunctivitis. Int. Ophthalmol. 2015, 35, 575–586. [Google Scholar] [CrossRef]
- Takematsu, H.; Yamamoto, H.; Naito-Matsui, Y.; Fujinawa, R.; Tanaka, K.; Okuno, Y.; Tanaka, Y.; Kyogashima, M.; Kannagi, R.; Kozutsumi, Y. Quantitative transcriptomic profiling of branching in a glycosphingolipid biosynthetic pathway. J. Biol. Chem. 2011, 286, 27214–27224. [Google Scholar] [CrossRef]

| Software | Subunit | Cleavage Site | Signal Peptide | Prediction Score | Reference |
|---|---|---|---|---|---|
| TPpred2 (3.0) | 5HT3A | - | - | 0.996 | [39] |
| 5HT3B | - | - | 1.000 | ||
| 5HT3C | - | - | 0.954 | ||
| 5HT3D | - | - | 0.973 | ||
| 5HT3E | 43 amino acid | 1–43 amino acid | 0.771 | ||
| MitoFates | 5HT3A | 32 amino acid | 2–6 amino acid | 0.018 | [40] |
| 5HT3B | 49 amino acid | - | 0.000 | ||
| 5HT3C | 25 amino acid | - | 0.006 | ||
| 5HT3D | 39 amino acid | - | 0.000 | ||
| 5HT3E | 71 amino acid | - | 0.023 | ||
| SignalP-5.0 | 5HT3A | 27 amino acid | 1–21 amino acid | 0.916 | [38] |
| 5HT3B | 21 amino acid | 1–21 amino acid | 0.978 | ||
| 5HT3C | 27 amino acid | 1–25 amino acid | 0.993 | ||
| 5HT3D | 24 amino acid | 1–23 amino acid | 0.992 | ||
| 5HT3E | 27 amino acid | 1–25 amino acid | 0.983 | ||
| ELM | 5HT3A | 26 amino acid | 1–24 amino acid | - | [37] |
| 5HT3B | 27 amino acid | 1–26 amino acid | - | ||
| 5HT3C | 32 amino acid | 1–29 amino acid | - | ||
| 5HT3D | 24 amino acid | 1–23 amino acid | - | ||
| 5HT3E | 30 amino acid | 1–28 amino acid | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, S.T.R.B.; Turek, I.; Ratcliffe, J.; Beckham, S.; Cianciarulo, C.; Adil, S.S.B.M.Y.; Kettle, C.; Whelan, D.R.; Irving, H.R. 5-HT3 Receptors on Mitochondria Influence Mitochondrial Function. Int. J. Mol. Sci. 2023, 24, 8301. https://doi.org/10.3390/ijms24098301
Rao STRB, Turek I, Ratcliffe J, Beckham S, Cianciarulo C, Adil SSBMY, Kettle C, Whelan DR, Irving HR. 5-HT3 Receptors on Mitochondria Influence Mitochondrial Function. International Journal of Molecular Sciences. 2023; 24(9):8301. https://doi.org/10.3390/ijms24098301
Chicago/Turabian StyleRao, Santosh T. R. B., Ilona Turek, Julian Ratcliffe, Simone Beckham, Cassandra Cianciarulo, Siti S. B. M. Y. Adil, Christine Kettle, Donna R. Whelan, and Helen R. Irving. 2023. "5-HT3 Receptors on Mitochondria Influence Mitochondrial Function" International Journal of Molecular Sciences 24, no. 9: 8301. https://doi.org/10.3390/ijms24098301
APA StyleRao, S. T. R. B., Turek, I., Ratcliffe, J., Beckham, S., Cianciarulo, C., Adil, S. S. B. M. Y., Kettle, C., Whelan, D. R., & Irving, H. R. (2023). 5-HT3 Receptors on Mitochondria Influence Mitochondrial Function. International Journal of Molecular Sciences, 24(9), 8301. https://doi.org/10.3390/ijms24098301







