Upside-Down Preference in the Forskolin-Induced In Vitro Differentiation of 50B11 Sensory Neurons: A Morphological Investigation by Label-Free Non-Linear Microscopy
Abstract
1. Introduction
2. Results
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. 50B11 Cell Line
5.2. Custom-Build Multi-Photon Microscope
5.3. Mitochondrial Morphology
5.4. Morphological Assessment under Microscopy
5.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodrigues, A.C.; Kang, P.B. Neuropathic and Myopathic Pain. Semin. Pediatr. Neurol. 2016, 23, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Delatycki, M.B.; Corben, L.A. Clinical Features of Friedreich Ataxia. J. Child Neurol. 2012, 27, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Züchner, S. MFN2 Hereditary Motor and Sensory Neuropathy. In GeneReviews®; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J., Gripp, K.W., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, WA, USA, 1993. [Google Scholar]
- Carroll, A.S.; Burns, J.; Nicholson, G.; Kiernan, M.C.; Vucic, S. Inherited Neuropathies. Semin. Neurol. 2019, 39, 620–639. [Google Scholar] [CrossRef]
- Louraki, M.; Karayianni, C.; Kanaka-Gantenbein, C.; Katsalouli, M.; Karavanaki, K. Peripheral Neuropathy in Children with Type 1 Diabetes. Diabetes Metab. 2012, 38, 281–289. [Google Scholar] [CrossRef]
- McMillan, H.J.; Ryan, M.M. Overview of Pediatric Peripheral Neuropathies. In Neuromuscular Disorders of Infancy, Childhood, and Adolescence; Elsevier: Amsterdam, The Netherlands, 2015; pp. 274–288. ISBN 978-0-12-417044-5. [Google Scholar]
- Haberberger, R.V.; Barry, C.; Matusica, D. Immortalized Dorsal Root Ganglion Neuron Cell Lines. Front. Cell Neurosci. 2020, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Mi, R.; Haughey, N.; Oz, M.; Höke, A. Immortalization and Characterization of a Nociceptive Dorsal Root Ganglion Sensory Neuronal Line. J. Peripher. Nerv. Syst. 2007, 12, 121–130. [Google Scholar] [CrossRef]
- Zupin, L.; Barbi, E.; Sagredini, R.; Ottaviani, G.; Crovella, S.; Celsi, F. In Vitro Effects of Photobiomodulation Therapy on 50B11 Sensory Neurons: Evaluation of Cell Metabolism, Oxidative Stress, Mitochondrial Membrane Potential (MMP), and Capsaicin-Induced Calcium Flow. J. Biophotonics 2021, 14, e202000347. [Google Scholar] [CrossRef]
- Gnavi, S.; Fornasari, B.E.; Tonda-Turo, C.; Laurano, R.; Zanetti, M.; Ciardelli, G.; Geuna, S. In Vitro Evaluation of Gelatin and Chitosan Electrospun Fibres as an Artificial Guide in Peripheral Nerve Repair: A Comparative Study. J. Tissue Eng. Regen. Med. 2018, 12, e679–e694. [Google Scholar] [CrossRef]
- Mohiuddin, M.S.; Himeno, T.; Yamada, Y.; Morishita, Y.; Kondo, M.; Tsunekawa, S.; Kato, Y.; Nakamura, J.; Kamiya, H. Glucagon Prevents Cytotoxicity Induced by Methylglyoxal in a Rat Neuronal Cell Line Model. Biomolecules 2021, 11, 287. [Google Scholar] [CrossRef]
- Van Opdenbosch, N.; Van den Broeke, C.; De Regge, N.; Tabarés, E.; Favoreel, H.W. The IE180 Protein of Pseudorabies Virus Suppresses Phosphorylation of Translation Initiation Factor EIF2α. J. Virol. 2012, 86, 7235–7240. [Google Scholar] [CrossRef]
- Chen, W.-S.; Yueh, C.-Y.; Huang, Y.-A.; Hwang, E. An Inverted Method for Culturing Dissociated Mouse Hippocampal Neurons. Neurosci. Res. 2011, 70, 118–123. [Google Scholar] [CrossRef]
- Magidson, V.; Khodjakov, A. Circumventing Photodamage in Live-Cell Microscopy. Methods Cell Biol. 2013, 114, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, W.R.; Williams, R.M.; Webb, W.W. Nonlinear Magic: Multiphoton Microscopy in the Biosciences. Nat. Biotechnol. 2003, 21, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Helmchen, F.; Denk, W. Deep Tissue Two-Photon Microscopy. Nat. Methods 2005, 2, 932–940. [Google Scholar] [CrossRef]
- Hirano, M.; Ando, R.; Shimozono, S.; Sugiyama, M.; Takeda, N.; Kurokawa, H.; Deguchi, R.; Endo, K.; Haga, K.; Takai-Todaka, R.; et al. A Highly Photostable and Bright Green Fluorescent Protein. Nat. Biotechnol. 2022, 40, 1132–1142. [Google Scholar] [CrossRef]
- Psilodimitrakopoulos, S.; Petegnief, V.; Soria, G.; Amat-Roldan, I.; Artigas, D.; Planas, A.M.; Loza-Alvarez, P. Estimation of the Effective Orientation of the SHG Source in Primary Cortical Neurons. Opt. Express 2009, 17, 14418. [Google Scholar] [CrossRef] [PubMed]
- Psilodimitrakopoulos, S.; Petegnief, V.; de Vera, N.; Hernandez, O.; Artigas, D.; Planas, A.M.; Loza-Alvarez, P. Quantitative Imaging of Microtubule Alteration as an Early Marker of Axonal Degeneration after Ischemia in Neurons. Biophys. J. 2013, 104, 968–975. [Google Scholar] [CrossRef]
- Van Steenbergen, V.; Boesmans, W.; Li, Z.; de Coene, Y.; Vints, K.; Baatsen, P.; Dewachter, I.; Ameloot, M.; Clays, K.; Vanden Berghe, P. Molecular Understanding of Label-Free Second Harmonic Imaging of Microtubules. Nat. Commun. 2019, 10, 3530. [Google Scholar] [CrossRef]
- Probes for Tubulin and Other Cytoskeletal Proteins—Section 11.2—IT. Available online: https://www.thermofisher.com/uk/en/home/references/molecular-probes-the-handbook/probes-for-cytoskeletal-proteins/probes-for-tubulin-and-other-cytoskeletal-proteins.html (accessed on 6 July 2022).
- Mitchison, T.; Kirschner, M. Dynamic Instability of Microtubule Growth. Nature 1984, 312, 237–242. [Google Scholar] [CrossRef]
- Pallen, S.; Shetty, Y.; Das, S.; Vaz, J.M.; Mazumder, N. Advances in Nonlinear Optical Microscopy Techniques for in Vivo and in Vitro Neuroimaging. Biophys. Rev. 2021, 13, 1199–1217. [Google Scholar] [CrossRef]
- Allen, C.H.; Ahmed, D.; Raiche-Tanner, O.; Chauhan, V.; Mostaço-Guidolin, L.; Cassol, E.; Murugkar, S. Label-Free Two-Photon Imaging of Mitochondrial Activity in Murine Macrophages Stimulated with Bacterial and Viral Ligands. Sci. Rep. 2021, 11, 14081. [Google Scholar] [CrossRef]
- Bower, A.J.; Renteria, C.; Li, J.; Marjanovic, M.; Barkalifa, R.; Boppart, S.A. High-Speed Label-Free Two-Photon Fluorescence Microscopy of Metabolic Transients during Neuronal Activity. Appl. Phys. Lett. 2021, 118, 081104. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Heikal, A.A. Two-Photon Autofluorescence Dynamics Imaging Reveals Sensitivity of Intracellular NADH Concentration and Conformation to Cell Physiology at the Single-Cell Level. J. Photochem. Photobiol. B Biol. 2009, 95, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.M.; Macko, P.; Palosaari, T.; Whelan, M.P. Autofluorescence Microscopy: A Non-Destructive Tool to Monitor Mitochondrial Toxicity. Toxicol. Lett. 2011, 206, 281–288. [Google Scholar] [CrossRef]
- Chazotte, B. Labeling Mitochondria with MitoTracker Dyes. Cold Spring Harb. Protoc. 2011, 2011, pdb.prot5648. [Google Scholar] [CrossRef] [PubMed]
- Hawrot, E. Cultured Sympathetic Neurons in the Study of Nerve Growth Factor Action. In Cellular and Molecular Biology of Neuronal Development; Black, I.B., Ed.; Springer US: Boston, MA, USA, 1984; pp. 143–163. ISBN 978-1-4612-9686-7. [Google Scholar]
- Letourneau, P.C. Cell-Substratum Adhesion of Neurite Growth Cones, and Its Role in Neurite Elongation. Exp. Cell Res. 1979, 124, 127–138. [Google Scholar] [CrossRef]
- Dent, E.W.; Baas, P.W. Microtubules in Neurons as Information Carriers. J. Neurochem. 2014, 129, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Lukina, M.M.; Shirmanova, M.V.; Sergeeva, T.F.; Zagaynova, E.V. Metabolic Imaging in the Study of Oncological Processes (Review). Sovrem. Tehnol. Med. 2016, 8, 113–126. [Google Scholar] [CrossRef]
- Cardanho-Ramos, C.; Faria-Pereira, A.; Morais, V.A. Orchestrating Mitochondria in Neurons: Cytoskeleton as the Conductor. Cytoskeleton 2020, 77, 65–75. [Google Scholar] [CrossRef]
- Kerstein, P.C.; Nichol IV, R.H.; Gomez, T.M. Mechanochemical Regulation of Growth Cone Motility. Front. Cell Neurosci. 2015, 9, 244. [Google Scholar] [CrossRef]
- Holt, B.D.; Arnold, A.M.; Sydlik, S.A. The Blanket Effect: How Turning the World Upside Down Reveals the Nature of Graphene Oxide Cytocompatibility. Adv. Healthcare Mater. 2021, 10, 2001761. [Google Scholar] [CrossRef] [PubMed]
- Justus, C.R.; Leffler, N.; Ruiz-Echevarria, M.; Yang, L.V. In Vitro Cell Migration and Invasion Assays. JoVE 2014, 51046. [Google Scholar] [CrossRef]
- Shah, M.; George, R.L.; Evancho-Chapman, M.M.; Zhang, G. Current Challenges in Dedifferentiated Fat Cells Research. Organogenesis 2016, 12, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Husson, T.R.; Issa, N.P. Functional Imaging with Mitochondrial Flavoprotein Autofluorescence: Theory, Practice, and Applications. In In Vivo Optical Imaging of Brain Function; Frostig, R.D., Ed.; Frontiers in Neuroscience; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2009; ISBN 978-1-4200-7684-4. [Google Scholar]
- Psilodimitrakopoulos, S.; Mouchliadis, L.; Paradisanos, I.; Lemonis, A.; Kioseoglou, G.; Stratakis, E. Ultrahigh-Resolution Nonlinear Optical Imaging of the Armchair Orientation in 2D Transition Metal Dichalcogenides. Light Sci. Appl. 2018, 7, 18005. [Google Scholar] [CrossRef] [PubMed]
- Meijering, E.; Jacob, M.; Sarria, J.-C.F.; Steiner, P.; Hirling, H.; Unser, M. Design and Validation of a Tool for Neurite Tracing and Analysis in Fluorescence Microscopy Images. Cytometry 2004, 58A, 167–176. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Alon, N.; Duadi, H.; Cohen, O.; Samet, T.; Zilony, N.; Schori, H.; Shefi, O.; Zalevsky, Z. Promotion of Neural Sprouting Using Low-Level Green Light-Emitting Diode Phototherapy. J. Biomed. Opt. 2015, 20, 020502. [Google Scholar] [CrossRef]
- R Core Team. R Core Team R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2023. [Google Scholar]
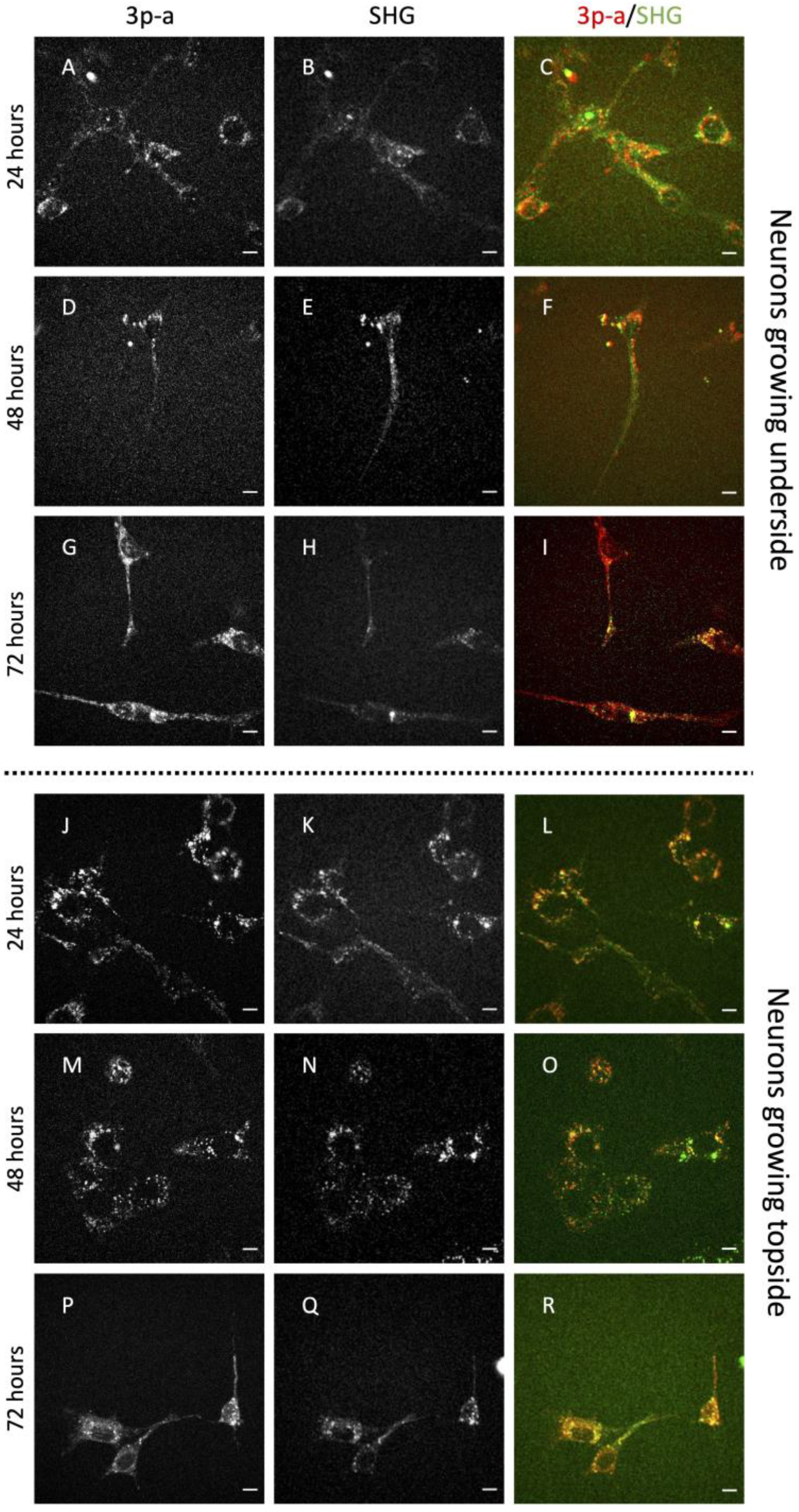
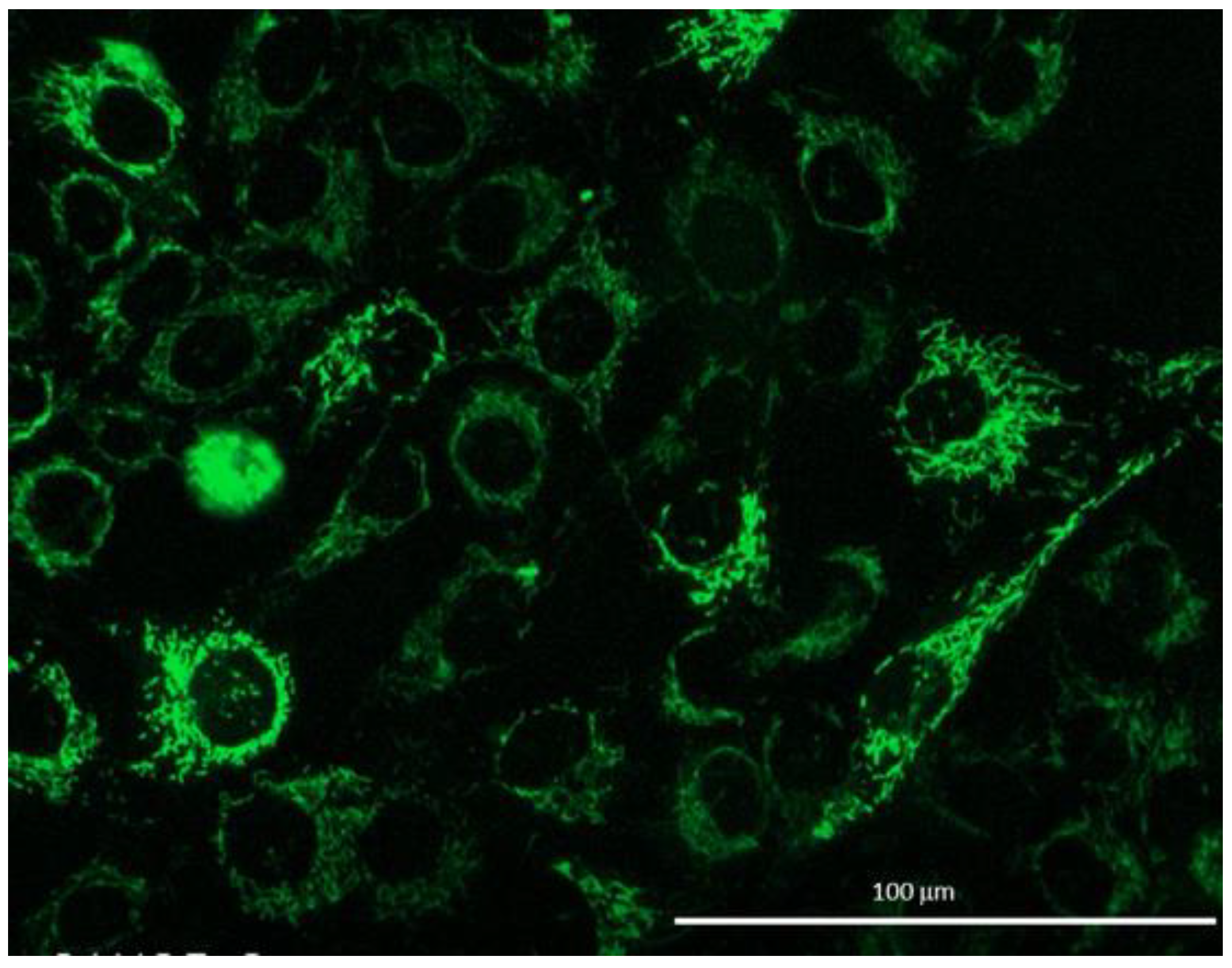
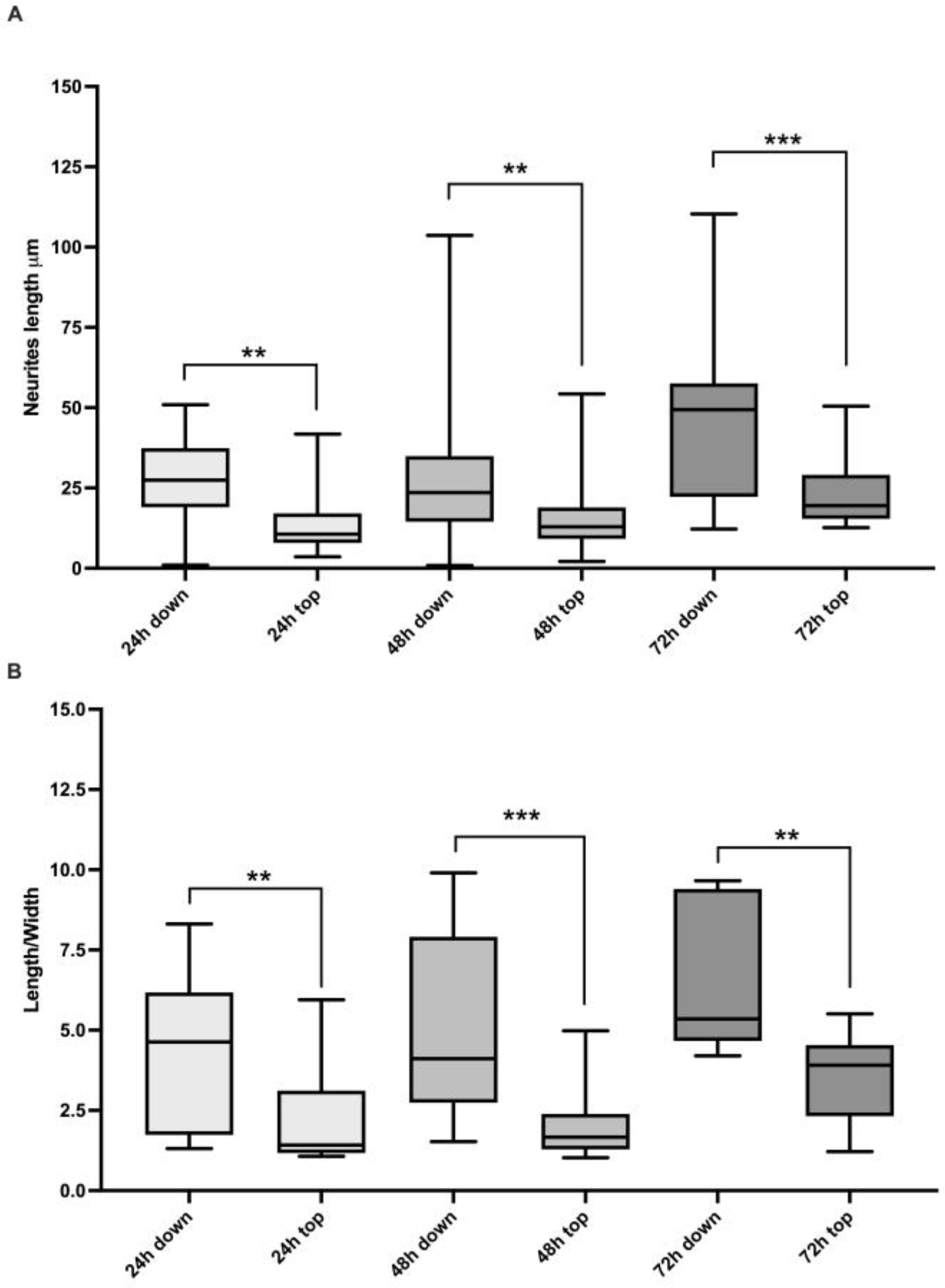
| Condition | n. Analyzed Cells | % of Elongated Cells |
|---|---|---|
| 24 h down | 21 | 52% |
| 24 h top | 19 | 21% |
| 48 h down | 10 | 50% |
| 48 h top | 33 | 6% |
| 72 h down | 7 | 100% |
| 72 h top | 12 | 50% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zupin, L.; Psilodimitrakopoulos, S.; Celsi, F.; Papadimitriou, L.; Ranella, A.; Crovella, S.; Ricci, G.; Stratakis, E.; Pascolo, L. Upside-Down Preference in the Forskolin-Induced In Vitro Differentiation of 50B11 Sensory Neurons: A Morphological Investigation by Label-Free Non-Linear Microscopy. Int. J. Mol. Sci. 2023, 24, 8354. https://doi.org/10.3390/ijms24098354
Zupin L, Psilodimitrakopoulos S, Celsi F, Papadimitriou L, Ranella A, Crovella S, Ricci G, Stratakis E, Pascolo L. Upside-Down Preference in the Forskolin-Induced In Vitro Differentiation of 50B11 Sensory Neurons: A Morphological Investigation by Label-Free Non-Linear Microscopy. International Journal of Molecular Sciences. 2023; 24(9):8354. https://doi.org/10.3390/ijms24098354
Chicago/Turabian StyleZupin, Luisa, Sotiris Psilodimitrakopoulos, Fulvio Celsi, Lina Papadimitriou, Anthi Ranella, Sergio Crovella, Giuseppe Ricci, Emmanuel Stratakis, and Lorella Pascolo. 2023. "Upside-Down Preference in the Forskolin-Induced In Vitro Differentiation of 50B11 Sensory Neurons: A Morphological Investigation by Label-Free Non-Linear Microscopy" International Journal of Molecular Sciences 24, no. 9: 8354. https://doi.org/10.3390/ijms24098354
APA StyleZupin, L., Psilodimitrakopoulos, S., Celsi, F., Papadimitriou, L., Ranella, A., Crovella, S., Ricci, G., Stratakis, E., & Pascolo, L. (2023). Upside-Down Preference in the Forskolin-Induced In Vitro Differentiation of 50B11 Sensory Neurons: A Morphological Investigation by Label-Free Non-Linear Microscopy. International Journal of Molecular Sciences, 24(9), 8354. https://doi.org/10.3390/ijms24098354












