Clinical Validation of a Size-Based Microfluidic Device for Circulating Tumor Cell Isolation and Analysis in Renal Cell Carcinoma
Abstract
1. Introduction
2. Results
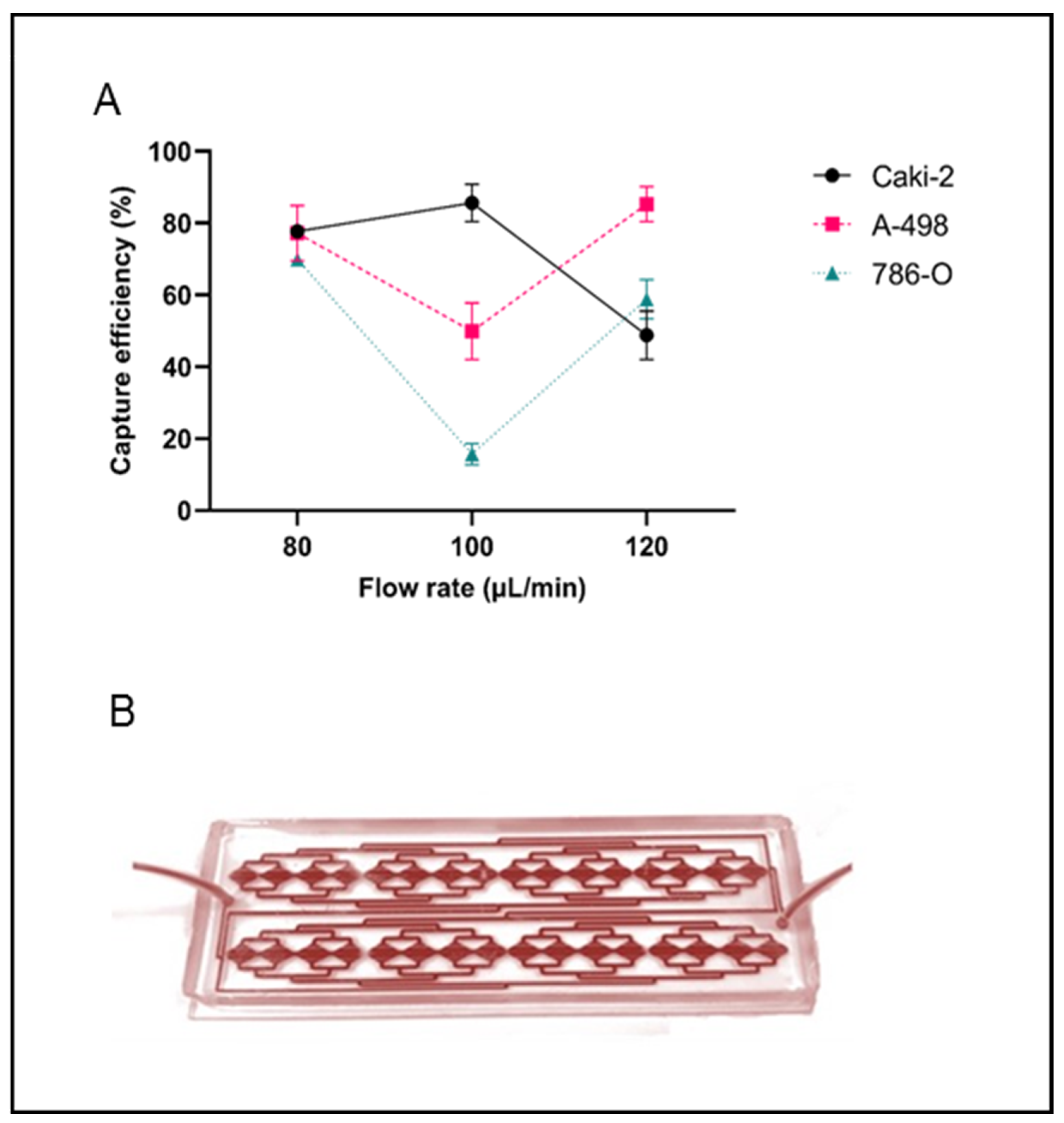
2.1. CTC Isolation Efficiency
2.2. Characteristics of the Study Cohort
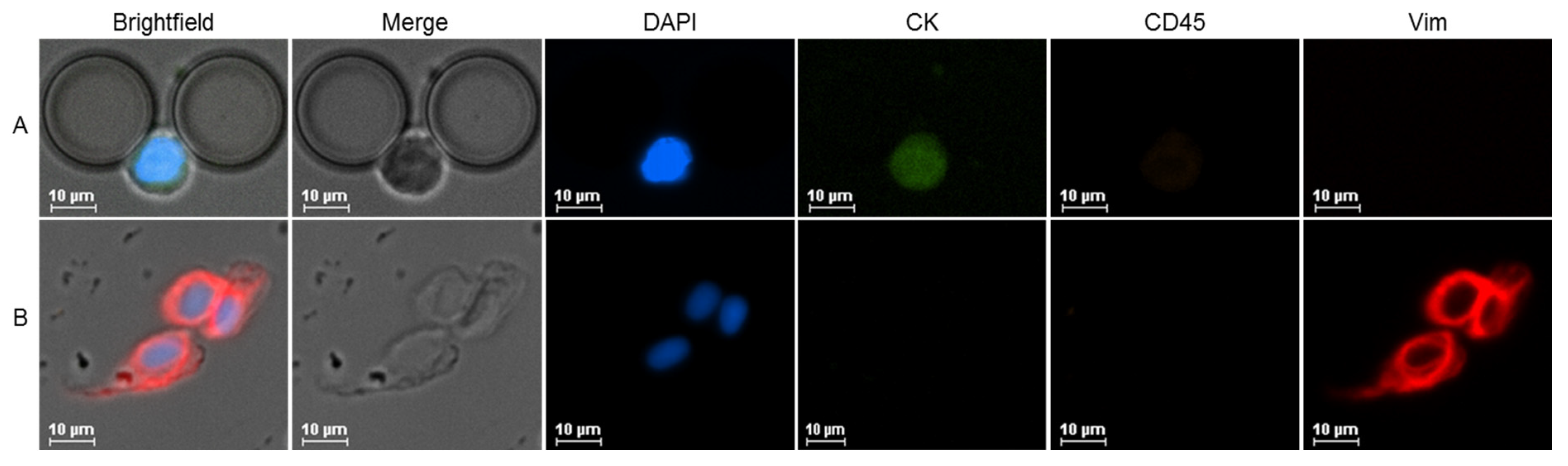
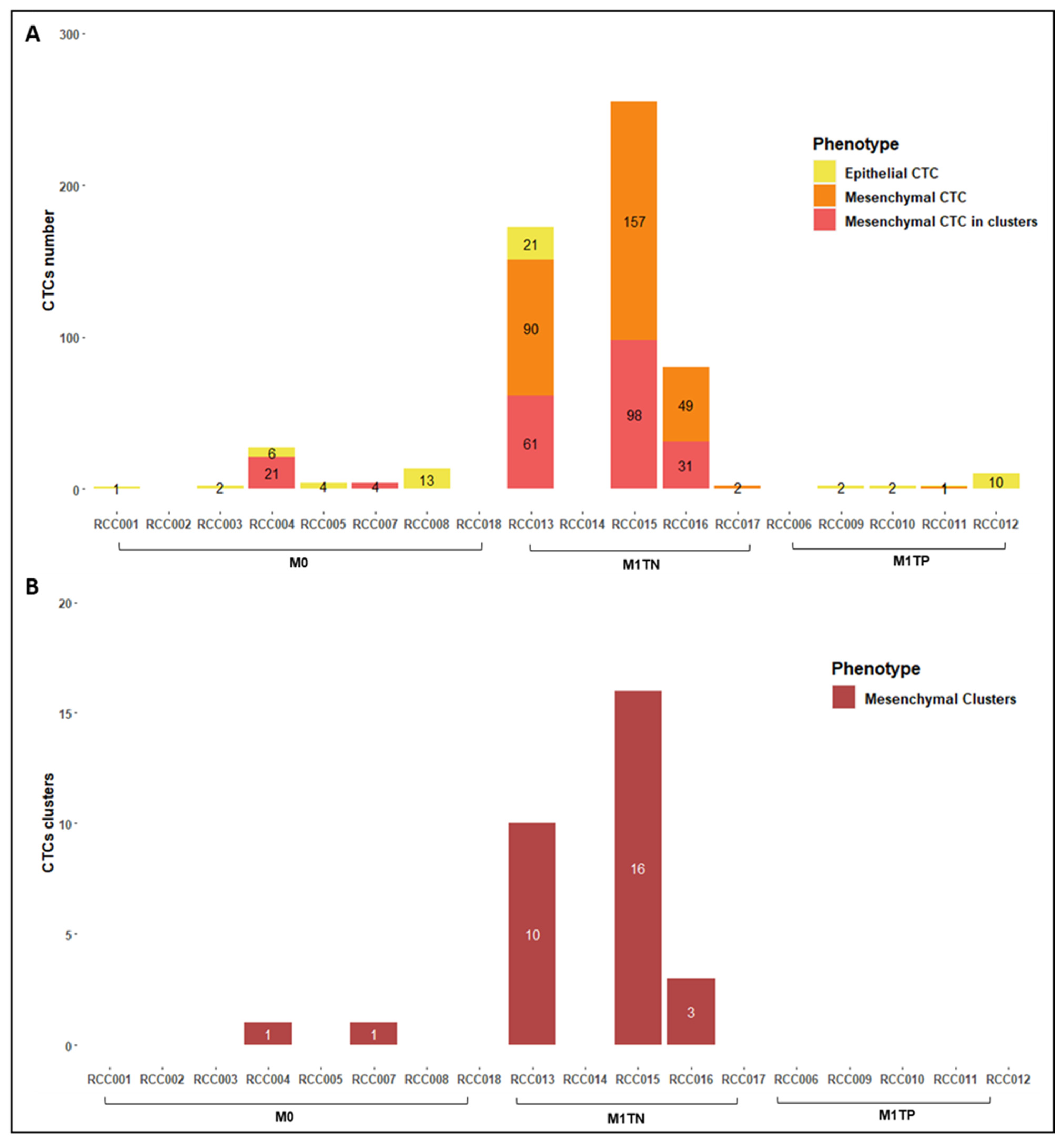
2.3. CTC Counts and Characterization
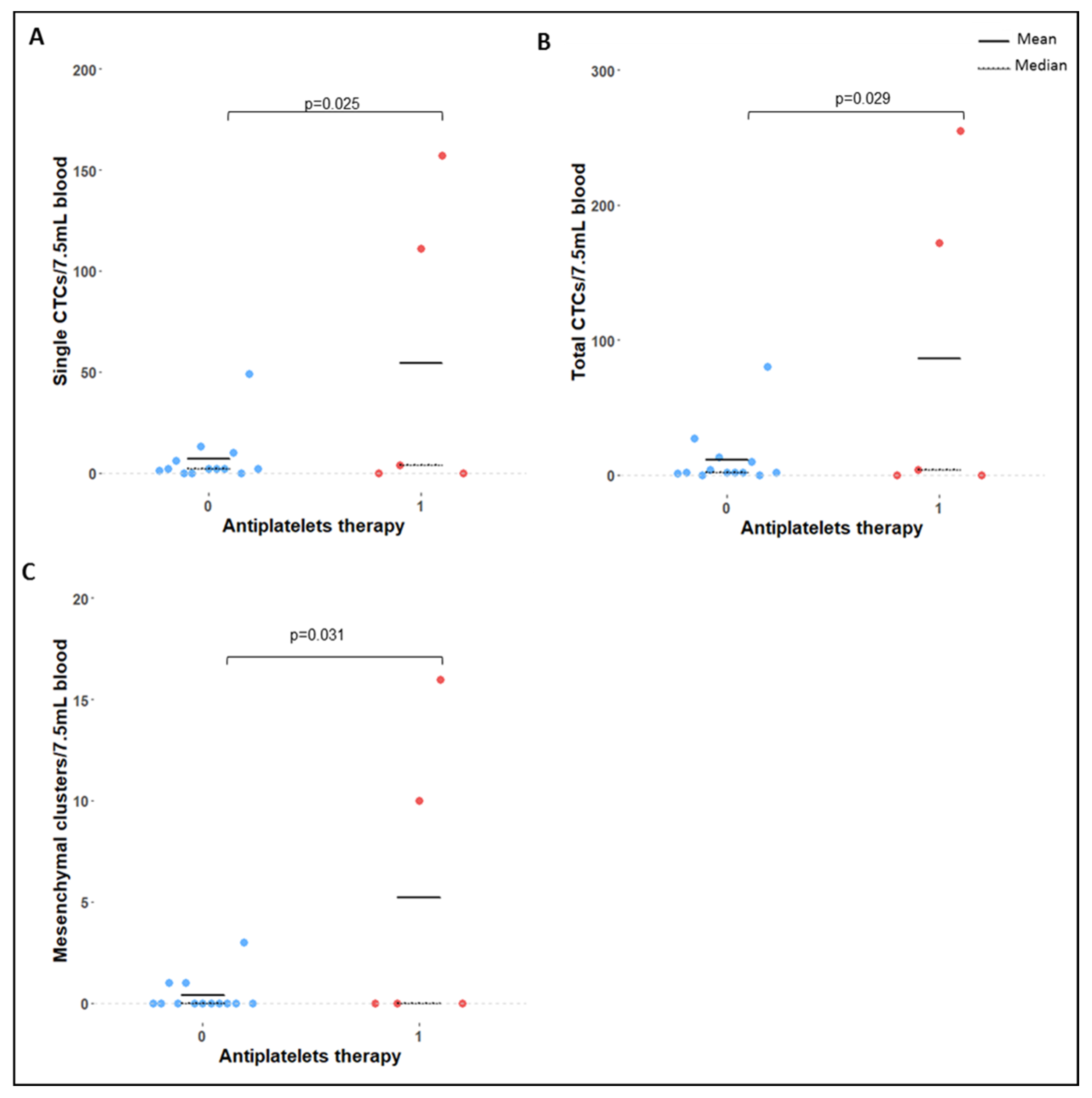
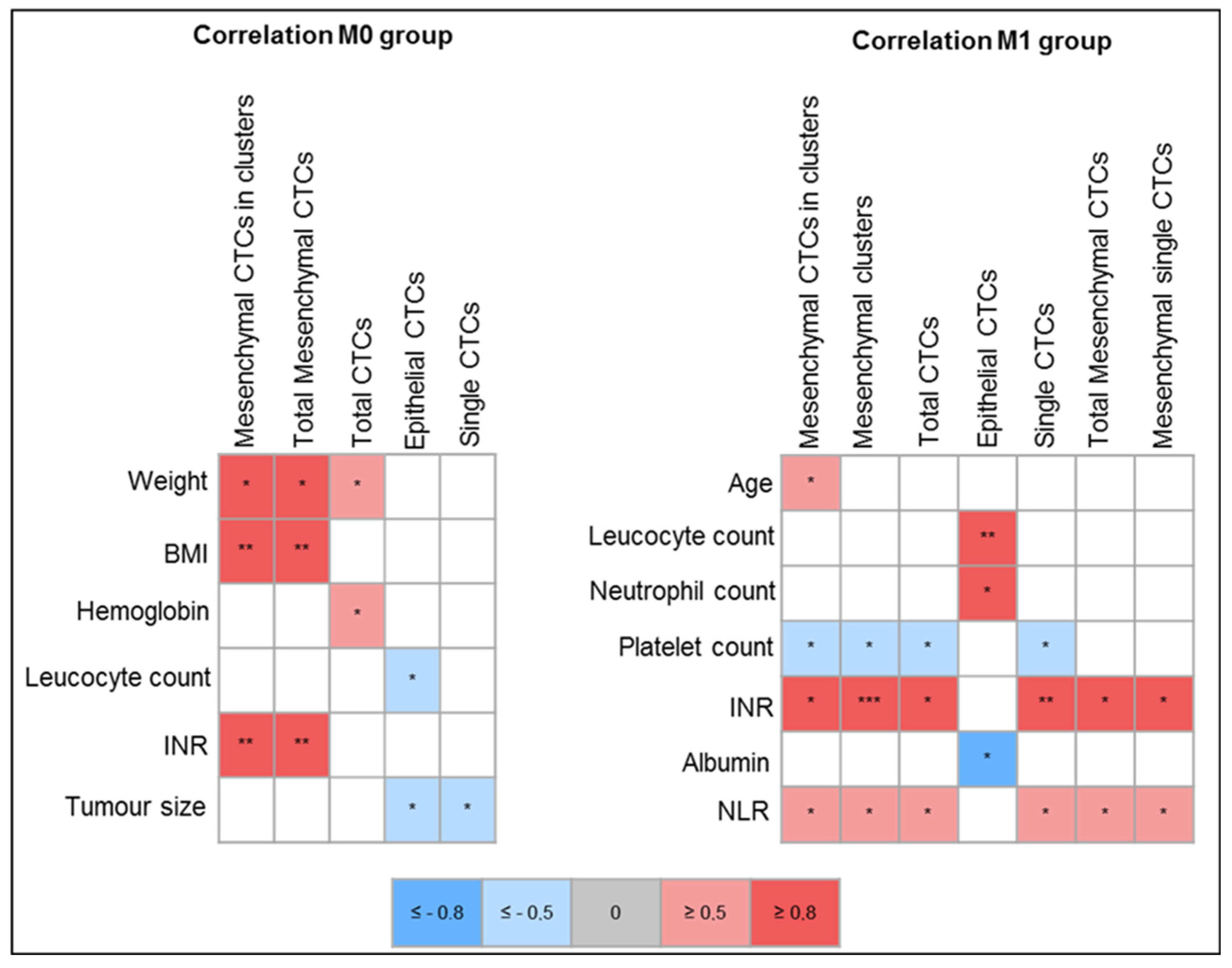
2.4. Correlation of Clinical Variables with CTC Count and Phenotype
2.5. Survival Analysis
3. Discussion
3.1. Detection Rates
3.2. CTC Count
3.3. CTC Clusters
3.4. CTCs and Survival Outcomes
3.5. Correlation of CTC Counts with Clinical Variables
3.6. Final Remarks, Study Limitations and Future Directions
4. Materials and Methods
4.1. Microfluidic Device
4.2. Cell Culture
4.3. Spiking Experiments
4.4. Immunocytochemistry Protocol and Immunofluorescence Imaging
4.5. Patient Recruitment and Sample Collection
4.6. CTC Isolation and Characterization
4.7. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bukavina, L.; Bensalah, K.; Bray, F.; Carlo, M.; Challacombe, B.; Karam, J.A.; Kassouf, W.; Mitchell, T.; Montironi, R.; O’Brien, T.; et al. Epidemiology of Renal Cell Carcinoma: 2022 Update. Eur. Urol. 2022, 82, 529–542. [Google Scholar] [CrossRef]
- World Health Organization: Regional Office for Europe. World Cancer Report: Cancer Research for Cancer Development; IARC: Lyon, France, 2020; ISBN 9789283204473. [Google Scholar]
- Palmela Leitão, T.; Miranda, M.; Polido, J.; Morais, J.; Corredeira, P.; Alves, P.; Oliveira, T.; Pereira e Silva, R.; Fernandes, R.; Ferreira, J.; et al. Circulating Tumor Cell Detection Methods in Renal Cell Carcinoma: A Systematic Review. Crit. Rev. Oncol. Hematol. 2021, 161, 103331. [Google Scholar] [CrossRef]
- Ionescu-Zanetti, C.; Brobey, R.; Rosenblatt, K.; Dehghani, M.; Schwartz, M.; Amato, R. Somatic Mutation Detection from Liquid Biopsies via NGS: Urological Cancers. J. Mol. Diagn. 2015, 17, 815. [Google Scholar]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pantel, K.; Alix-Panabières, C. Real-Time Liquid Biopsy in Cancer Patients: Fact or Fiction? Cancer Res. 2013, 73, 6384–6388. [Google Scholar] [CrossRef][Green Version]
- Domínguez-Vigil, I.G.; Moreno-Martínez, A.K.; Wang, J.Y.; Roehrl, M.H.A.; Barrera-Saldaña, H.A. The Dawn of the Liquid Biopsy in the Fight against Cancer. Oncotarget 2018, 9, 2912–2922. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liberko, M.; Kolostova, K.; Bobek, V. Essentials of Circulating Tumor Cells for Clinical Research and Practice. Crit. Rev. Oncol. Hematol. 2013, 88, 338–356. [Google Scholar] [CrossRef] [PubMed]
- Bluemke, K.; Bilkenroth, U.; Meye, A.; Fuessel, S.; Lautenschlaeger, C.; Goebel, S.; Melchior, A.; Heynemann, H.; Fornara, P.; Taubert, H. Detection of Circulating Tumor Cells in Peripheral Blood of Patients with Renal Cell Carcinoma Correlates with Prognosis. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2190–2194. [Google Scholar] [CrossRef][Green Version]
- Nel, I.; Gauler, T.C.; Bublitz, K.; Lazaridis, L.; Goergens, A.; Giebel, B.; Schuler, M.; Hoffmann, A.-C. Circulating Tumor Cell Composition in Renal Cell Carcinoma. PLoS ONE 2016, 11, e0153018. [Google Scholar] [CrossRef]
- Basso, U.; Facchinetti, A.; Rossi, E.; Maruzzo, M.; Conteduca, V.; Aieta, M.; Massari, F.; Fraccon, A.P.; Mucciarini, C.; Sava, T.; et al. Prognostic Role of Circulating Tumor Cells-CTCs in Metastatic Renal Cell Carcinoma. J. Clin. Oncol. 2017, 35. [Google Scholar] [CrossRef]
- Basso, U.; Facchinetti, A.; Rossi, E.; Maruzzo, M.; Conteduca, V.; Aieta, M.; Massari, F.; Fraccon, A.P.; Mucciarini, C.; Sava, T.; et al. Prognostic Role of Circulating Tumor Cells in Metastatic Renal Cell Carcinoma: A Large, Multicenter, Prospective Trial. Oncologist 2021, 26, 740–750. [Google Scholar] [CrossRef]
- Gilbert, S.M.; Whitson, J.M.; Mansukhani, M.; Buttyan, R.; Benson, M.C.; Olsson, C.A.; Sawczuk, I.S.; McKiernan, J.M. Detection of Carbonic Anhydrase-9 Gene Expression in Peripheral Blood Cells Predicts Risk of Disease Recurrence in Patients with Renal Cortical Tumors. Urology 2006, 67, 942–945. [Google Scholar] [CrossRef]
- Liu, S.; Tian, Z.; Zhang, L.; Hou, S.; Hu, S.; Wu, J.; Jing, Y.; Sun, H.; Yu, F.; Zhao, L.; et al. Combined Cell Surface Carbonic Anhydrase 9 and CD147 Antigens Enable High-Efficiency Capture of Circulating Tumor Cells in Clear Cell Renal Cell Carcinoma Patients. Oncotarget 2016, 7, 59877–59891. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maheswaran, S.; Haber, D.A. Circulating Tumor Cells: A Window into Cancer Biology and Metastasis. Curr. Opin. Genet. Dev. 2010, 20, 96–99. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Andree, K.C.; van Dalum, G.; Terstappen, L.W.M.M. Challenges in Circulating Tumor Cell Detection by the CellSearch System. Mol. Oncol. 2016, 10, 395–407. [Google Scholar] [CrossRef][Green Version]
- Bilkenroth, U.; Taubert, H.; Riemann, D.; Rebmann, U.; Heynemann, H.; Meye, A. Detection and Enrichment of Disseminated Renal Carcinoma Cells from Peripheral Blood by Immunomagnetic Cell Separation. Int. J. Cancer 2001, 92, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Meye, A.; Bilkenroth, U.; Schmidt, U.; Fussel, S.; Robel, K.; Melchior, A.M.; Blumke, K.; Pinkert, D.; Bartel, F.; Linne, C.; et al. Isolation and Enrichment of Urologic Tumor Cells in Blood Samples by a Semi-Automated CD45 Depletion autoMACS Protocol. Int. J. Oncol. 2002, 21, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating Tumor Cell Isolation, Culture, and Downstream Molecular Analysis. Biotechnol. Adv. 2018, 36, 1063–1078. [Google Scholar] [CrossRef]
- Wang, Z.-L.; Zhang, P.; Li, H.-C.; Yang, X.-J.; Zhang, Y.-P.; Li, Z.-L.; Xue, L.; Xue, Y.-Q.; Li, H.-L.; Chen, Q.; et al. Dynamic Changes of Different Phenotypic and Genetic Circulating Tumor Cells as a Biomarker for Evaluating the Prognosis of RCC. Cancer Biol. Ther. 2019, 20, 505–512. [Google Scholar] [CrossRef]
- Bai, M.; Zou, B.; Wang, Z.; Li, P.; Wang, H.; Ou, Y.; Cui, K.; Bian, J.; Li, S.; Xu, X. Comparison of Two Detection Systems for Circulating Tumor Cells among Patients with Renal Cell Carcinoma. Int. Urol. Nephrol. 2018, 50, 1801–1809. [Google Scholar] [CrossRef]
- Mittal, K.; Williams, A.; Rawal, S.; Ao, Z.; Lu, B.; Torres-Munoz, J.; Borden, E.; O’Malley, M.; Wood, L.; Zheng, S.; et al. Detection of Circulating Tumor Cells in Advanced Renal Cell Carcinoma Patients Using Microfilter-Based Capture. BJU Int. 2012, 110, 9. [Google Scholar] [CrossRef]
- Mittal, K.; Williams, A.; Rawal, S.; Venur, V.A.; Lu, B.; Torres-Munoz, J.; Borden, E.C.; Ao, Z.; O’Malley, M.; Wood, L.S.; et al. Circulating Tumor Cell Kinetics in mRCC Patients Treated with Sunitinib. J. Clin. Oncol. 2014, 32, 481. [Google Scholar] [CrossRef]
- El-Heliebi, A.; Kroneis, T.; Zohrer, E.; Haybaeck, J.; Fischereder, K.; Kampel-Kettner, K.; Zigeuner, R.; Pock, H.; Riedl, R.; Stauber, R.; et al. Are Morphological Criteria Sufficient for the Identification of Circulating Tumor Cells in Renal Cancer? J. Transl. Med. 2013, 11, 214. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Williams, A.; Rawal, S.; Ao, Z.; Lu, B.; Torres-Munoz, J.; Rini, B.; Pelley, R.; Budd, G.T.; Borden, E.; Zheng, S.; et al. Capture and Molecular Characterization of CTC in Metastatic Breast, Prostate, Colorectal, and Renal Cancer. Cancer Res. 2012, 72, 2372. [Google Scholar] [CrossRef]
- Broncy, L.; Njima, B.B.; Mejean, A.; Beroud, C.; Romdhane, K.B.; Ilie, M.; Hofman, V.; Muret, J.; Hofman, P.; Bouhamed, H.C.; et al. Single-Cell Genetic Analysis Validates Cytopathological Identification of Circulating Cancer Cells in Patients with Clear Cell Renal Cell Carcinoma. Oncotarget 2018, 9, 20058–20074. [Google Scholar] [CrossRef][Green Version]
- Gutschi, T.; Pachernegg, O.; Heidler, S.; Zigeuner, R.; Pummer, K.; Lackner, K. Detection of Circulating Tumor Cells in Patients with Renal Cell Carcinoma Compared with a Control Group. Eur. Urol. Suppl. 2010, 9, 649. [Google Scholar] [CrossRef]
- Kim, T.H.; Kang, Y.-T.; Cho, Y.-H.; Kim, J.H.; Jeong, B.C.; Seo, S.I.; Jeon, S.S.; Lee, H.M. Detection of Circulating Tumour Cells and Their Potential Use as a Biomarker for Advanced Renal Cell Carcinoma. Can. Urol. Assoc. J. 2019, 13, E285–E291. [Google Scholar] [CrossRef]
- Kang, Y.-T.; Kim, Y.J.; Lee, T.H.; Cho, Y.-H.; Chang, H.J.; Lee, H.-M. Cytopathological Study of the Circulating Tumor Cells Filtered from the Cancer Patients’ Blood Using Hydrogel-Based Cell Block Formation. Sci. Rep. 2018, 8, 15218. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Naoe, M.; Kusaka, C.; Ohta, M.; Hasebe, Y.; Unoki, T.; Shimoyama, H.; Nakasato, T.; Oshinomi, K.; Morita, J.; Fuji, K.; et al. Development of a Highly Sensitive Technique for Capturing Renal Cell Cancer Circulating Tumor Cells. Diagnostics 2019, 9, 96. [Google Scholar] [CrossRef][Green Version]
- Whitesides, G.M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Gwak, H.; Kim, J.; Kashefi-Kheyrabadi, L.; Kwak, B.; Hyun, K.-A.; Jung, H.-I. Progress in Circulating Tumor Cell Research Using Microfluidic Devices. Micromachines 2018, 9, 353. [Google Scholar] [CrossRef][Green Version]
- Carneiro, A.; Piairo, P.; Teixeira, A.; Ferreira, D.; Cotton, S.; Rodrigues, C.; Chícharo, A.; Abalde-Cela, S.; Santos, L.L.; Lima, L.; et al. Discriminating Epithelial to Mesenchymal Transition Phenotypes in Circulating Tumor Cells Isolated from Advanced Gastrointestinal Cancer Patients. Cells 2022, 11, 376. [Google Scholar] [CrossRef]
- Ribeiro-Samy, S.; Oliveira, M.I.; Pereira-Veiga, T.; Muinelo-Romay, L.; Carvalho, S.; Gaspar, J.; Freitas, P.P.; López-López, R.; Costa, C.; Diéguez, L. Fast and Efficient Microfluidic Cell Filter for Isolation of Circulating Tumor Cells from Unprocessed Whole Blood of Colorectal Cancer Patients. Sci. Rep. 2019, 9, 8032. [Google Scholar] [CrossRef][Green Version]
- Lopes, C.; Piairo, P.; Chícharo, A.; Abalde-Cela, S.; Pires, L.R.; Corredeira, P.; Alves, P.; Muinelo-Romay, L.; Costa, L.; Diéguez, L. HER2 Expression in Circulating Tumour Cells Isolated from Metastatic Breast Cancer Patients Using a Size-Based Microfluidic Device. Cancers 2021, 13, 4446. [Google Scholar] [CrossRef]
- Amin, M.B.; Edge, S.B.; Greene, F.L.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Shaw Bagnall, J.; Byun, S.; Begum, S.; Miyamoto, D.T.; Hecht, V.C.; Maheswaran, S.; Stott, S.L.; Toner, M.; Hynes, R.O.; Manalis, S.R. Deformability of Tumor Cells versus Blood Cells. Sci. Rep. 2015, 5, 18542. [Google Scholar] [CrossRef][Green Version]
- Barriere, G.; Fici, P.; Gallerani, G.; Fabbri, F.; Zoli, W.; Rigaud, M. Circulating Tumor Cells and Epithelial, Mesenchymal and Stemness Markers: Characterization of Cell Subpopulations. Ann. Transl. Med. 2014, 2, 109. [Google Scholar] [CrossRef]
- Small, A.C.; Gong, Y.; Oh, W.K.; Hall, S.J.; van Rijn, C.J.M.; Galsky, M.D. The Emerging Role of Circulating Tumor Cell Detection in Genitourinary Cancer. J. Urol. 2012, 188, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Egami, H.; Kai, M.; Kurusu, Y.; Takano, S.; Ogawa, M. No-Touch Isolation Technique Reduces Intraoperative Shedding of Tumor Cells into the Portal Vein during Resection of Colorectal Cancer. Surgery 1999, 125, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Engilbertsson, H.; Aaltonen, K.E.; Björnsson, S.; Kristmundsson, T.; Patschan, O.; Rydén, L.; Gudjonsson, S. Transurethral Bladder Tumor Resection Can Cause Seeding of Cancer Cells into the Bloodstream. J. Urol. 2015, 193, 53–57. [Google Scholar] [CrossRef]
- Theil, G.; Fischer, K.; Wencker, A.; Mohammed, N.; Luecke, K.; Fornara, P. Verification of a Functionalized Structured Medical Wire for the Isolation of Circulating Tumor Cells (CTC) in Patients with Renal Cell Carcinoma. Eur. Urol. Suppl. 2014, 13, e291. [Google Scholar] [CrossRef]
- Hioki, T.; Sugimura, Y. Detection of Circulating Cancer Cells by Nested Reverse Transcription-Polymerase Chain Reaction of Cytokeratin-19 in Patients with Renal Cell Carcinoma. Hinyokika Kiyo 1999, 45, 577–581. [Google Scholar]
- Shimazui, T.; Yoshikawa, K.; Uemura, H.; Kawamoto, R.; Kawai, K.; Uchida, K.; Hirao, Y.; Saga, S.; Akaza, H. Detection of Cadherin-6 mRNA by Nested RT-PCR as a Potential Marker for Circulating Cancer Cells in Renal Cell Carcinoma. Int. J. Oncol. 2003, 23, 1049–1054. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, R.; Tang, Y.; Chang, J.; Han, R.; Zhang, S.; Jiang, N.; Ma, F. New Applications of the Acridine Orange Fluorescence Staining Method: Screening for Circulating Tumor Cells. Oncol. Lett. 2017, 13, 2221–2229. [Google Scholar] [CrossRef][Green Version]
- Lu, W.; Kang, Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev. Cell 2019, 49, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Mitra, A.; Brownlee, Z.; Xia, X.; Bellister, S.; Overman, M.J.; Kopetz, S.; Ellis, L.M.; Meng, Q.H.; Li, S. Epithelial-Mesenchymal Transitioned Circulating Tumor Cells Capture for Detecting Tumor Progression. Clin. Cancer Res. 2015, 21, 899–906. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nel, I.; Baba, H.A.; Ertle, J.; Weber, F.; Sitek, B.; Eisenacher, M.; Meyer, H.E.; Schlaak, J.F.; Hoffmann, A.-C. Individual Profiling of Circulating Tumor Cell Composition and Therapeutic Outcome in Patients with Hepatocellular Carcinoma. Transl. Oncol. 2013, 6, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Metcalf, R.L.; Carter, L.; Brady, G.; Blackhall, F.H.; Dive, C. Molecular Analysis of Circulating Tumour Cells—Biology and Biomarkers. Nat. Rev. Clin. Oncol. 2014, 11, 129–144. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating Breast Tumor Cells Exhibit Dynamic Changes in Epithelial and Mesenchymal Composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef][Green Version]
- Ao, Z.; Shah, S.H.; Machlin, L.M.; Parajuli, R.; Miller, P.C.; Rawal, S.; Williams, A.J.; Cote, R.J.; Lippman, M.E.; Datar, R.H.; et al. Identification of Cancer-Associated Fibroblasts in Circulating Blood from Patients with Metastatic Breast Cancer Identification of cCAFs from Metastatic Cancer Patients. Cancer Res. 2015, 75, 4681–4687. [Google Scholar] [CrossRef][Green Version]
- Duda, D.G.; Duyverman, A.M.M.J.; Kohno, M.; Snuderl, M.; Steller, E.J.A.; Fukumura, D.; Jain, R.K. Malignant Cells Facilitate Lung Metastasis by Bringing Their Own Soil. Proc. Natl. Acad. Sci. USA 2010, 107, 21677–21682. [Google Scholar] [CrossRef][Green Version]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aceto, N.; Toner, M.; Maheswaran, S.; Haber, D.A. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. Trends Cancer Res. 2015, 1, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Bootsma, M.; McKay, R.R.; Emamekhoo, H.; Bade, R.M.; Schehr, J.L.; Mannino, M.C.; Singh, A.; Wolfe, S.K.; Schultz, Z.D.; Sperger, J.; et al. Longitudinal Molecular Profiling of Circulating Tumor Cells in Metastatic Renal Cell Carcinoma. J. Clin. Oncol. 2022, 40, 3633–3641. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, P.; Chong, Y.; Xue, Y.; Yang, X.; Li, H.; Wang, L.; Zhang, Y.; Chen, Q.; Li, Z.; et al. Perioperative Circulating Tumor Cells (CTCs), MCTCs, and CTC-White Blood Cells Detected by a Size-Based Platform Predict Prognosis in Renal Cell Carcinoma. Dis. Mrk. 2021, 2021, 9956142. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Xu, F.; Tian, J.; Gao, K.; Wan, Z.; Wang, Y.; Gao, M.; Wang, Z.; Chong, T. The Prognostic Value of Circulating Tumour Cells (CTCs) and CTC White Blood Cell Clusters in Patients with Renal Cell Carcinoma. BMC Cancer 2021, 21, 826. [Google Scholar] [CrossRef]
- Guan, Y.; Xu, F.; Tian, J.; Wang, Y.; Guo, N.; Wan, Z.; He, M.; Gao, M.; Gao, K.; Chong, T. Prognostic Value of Circulating Tumor Cells and Immune-Inflammatory Cells in Patients with Renal Cell Carcinoma. Urol. Oncol. 2022, 40, 167.e21–167.e32. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Rothé, F.; Chaboteaux, C.; Durbecq, V.; Rouas, G.; Criscitiello, C.; Metallo, J.; Kheddoumi, N.; Singhal, S.K.; Michiels, S.; et al. HER2-Positive Circulating Tumor Cells in Breast Cancer. PLoS ONE 2011, 6, e15624. [Google Scholar] [CrossRef]
- Van der Auwera, I.; Elst, H.J.; Van Laere, S.J.; Maes, H.; Huget, P.; van Dam, P.; Van Marck, E.A.; Vermeulen, P.B.; Dirix, L.Y. The Presence of Circulating Total DNA and Methylated Genes Is Associated with Circulating Tumour Cells in Blood from Breast Cancer Patients. Br. J. Cancer 2009, 100, 1277–1286. [Google Scholar] [CrossRef][Green Version]
- Luo, K.; Wang, X.; Zhang, X.; Liu, Z.; Huang, S.; Li, R. The Value of Circulating Tumor Cells in the Prognosis and Treatment of Pancreatic Cancer. Front. Oncol. 2022, 12, 933645. [Google Scholar] [CrossRef]
- Yu, E.; Allan, A.L.; Sanatani, M.; Lewis, D.; Warner, A.; Dar, A.R.; Yaremko, B.P.; Lowes, L.E.; Palma, D.A.; Raphael, J.; et al. Circulating Tumor Cells Detected in Follow-up Predict Survival Outcomes in Tri-Modality Management of Advanced Non-Metastatic Esophageal Cancer: A Secondary Analysis of the QUINTETT Randomized Trial. BMC Cancer 2022, 22, 746. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.; Wei, J.; Zou, Z.-Y.; Qian, X.-P.; Liu, B.-R. Circulating Tumour Cells Predict Survival in Gastric Cancer Patients: A Meta-Analysis. Contemp. Oncol. 2015, 19, 451–457. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bourcy, M.; Suarez-Carmona, M.; Lambert, J.; Francart, M.-E.; Schroeder, H.; Delierneux, C.; Skrypek, N.; Thompson, E.W.; Jérusalem, G.; Berx, G.; et al. Tissue Factor Induced by Epithelial–Mesenchymal Transition Triggers a Procoagulant State That Drives Metastasis of Circulating Tumor Cells. Cancer Res. 2016, 76, 4270–4282. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mitrugno, A.; Tormoen, G.W.; Kuhn, P.; McCarty, O.J.T. The Prothrombotic Activity of Cancer Cells in the Circulation. Blood Rev. 2016, 30, 11–19. [Google Scholar] [CrossRef][Green Version]
- Dirix, L.Y.; Oeyen, S.; Buys, A.; Liégois, V.; Prové, A.; Van De Mooter, T.; Van Laere, S.; Vermeulen, P.B. Coagulation/fibrinolysis and Circulating Tumor Cells in Patients with Advanced Breast Cancer. Breast Cancer Res. Treat. 2022, 192, 583–591. [Google Scholar] [CrossRef]
- Zhang, J.; Qiao, X.; Shi, H.; Han, X.; Liu, W.; Tian, X.; Zeng, X. Circulating Tumor-Associated Neutrophils (cTAN) Contribute to Circulating Tumor Cell Survival by Suppressing Peripheral Leukocyte Activation. Tumour. Biol. 2016, 37, 5397–5404. [Google Scholar] [CrossRef]
- Fridrichova, I.; Kalinkova, L.; Ciernikova, S. Clinical Relevancy of Circulating Tumor Cells in Breast Cancer: Epithelial or Mesenchymal Characteristics, Single Cells or Clusters? Int. J. Mol. Sci. 2022, 23, 12141. [Google Scholar] [CrossRef]
- Peyton, C.C.; Abel, E.J.; Chipollini, J.; Boulware, D.C.; Azizi, M.; Karam, J.A.; Margulis, V.; Master, V.A.; Matin, S.F.; Raman, J.D.; et al. The Value of Neutrophil to Lymphocyte Ratio in Patients Undergoing Cytoreductive Nephrectomy with Thrombectomy. Eur. Urol Focus 2020, 6, 104–111. [Google Scholar] [CrossRef]
- Ning, D.; Cui, K.; Liu, M.; Ou, Y.; Wang, Z.; Zou, B.; Shen, Y.; Lu, X.; Li, S.; Li, P. Comparison of CellSearch and Circulating Tumor Cells (CTC)-Biopsy Systems in Detecting Peripheral Blood Circulating Tumor Cells in Patients with Gastric Cancer. Med. Sci. Monit. 2021, 27, e926565. [Google Scholar] [CrossRef]
- Obermayr, E.; Braicu, E.I.; Polterauer, S.; Loverix, L.; Concin, N.; Woelber, L.; Mahner, S.; Sehouli, J.; Van Gorp, T.; Vergote, I.; et al. Association of a Combined Cancer Exhaustion Score with Circulating Tumor Cells and Outcome in Ovarian Cancer-A Study of the OVCAD Consortium. Cancers 2021, 13, 5865. [Google Scholar] [CrossRef]
- Beer, T.M.; Lalani, A.S.; Lee, S.; Mori, M.; Eilers, K.M.; Curd, J.G.; Henner, W.D.; Ryan, C.W.; Venner, P.; Ruether, J.D.; et al. C-Reactive Protein as a Prognostic Marker for Men with Androgen-Independent Prostate Cancer: Results from the ASCENT Trial. Cancer 2008, 112, 2377–2383. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Guo, L.; Zhang, W.; Li, Y.; Jiang, W.; Di, X.; Ma, J.; Feng, L.; Zhang, K.; Shou, J. Cooperation between the Inflammation and Coagulation Systems Promotes the Survival of Circulating Tumor Cells in Renal Cell Carcinoma Patients. Front. Oncol. 2019, 9, 504. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhang, H.; Hamad, A.; Huang, H.; Tsung, A. Surgery-Mediated Tumor-Promoting Effects on the Immune Microenvironment. Semin. Cancer Biol. 2022, 86, 408–419. [Google Scholar] [CrossRef]
- Anvari, S.; Osei, E.; Maftoon, N. Interactions of Platelets with Circulating Tumor Cells Contribute to Cancer Metastasis. Sci. Rep. 2021, 11, 15477. [Google Scholar] [CrossRef] [PubMed]
- Egan, K.; Cooke, N.; Kenny, D. Living in Shear: Platelets Protect Cancer Cells from Shear Induced Damage. Clin. Exp. Metastasis 2014, 31, 697–704. [Google Scholar] [CrossRef]
- Placke, T.; Örgel, M.; Schaller, M.; Jung, G.; Rammensee, H.-G.; Kopp, H.-G.; Salih, H.R. Platelet-Derived MHC Class I Confers a Pseudonormal Phenotype to Cancer Cells That Subverts the Antitumor Reactivity of Natural Killer Immune CellsPlatelet MHC Class I Impairs NK Antitumor Reactivity. Cancer Res. 2012, 72, 440–448. [Google Scholar] [CrossRef][Green Version]
- Labelle, M.; Begum, S.; Hynes, R.O. Direct Signaling between Platelets and Cancer Cells Induces an Epithelial-Mesenchymal-like Transition and Promotes Metastasis. Cancer Cell 2011, 20, 576–590. [Google Scholar] [CrossRef][Green Version]
- Thalgott, M.; Rack, B.; Maurer, T.; Souvatzoglou, M.; Eiber, M.; Kreß, V.; Heck, M.M.; Andergassen, U.; Nawroth, R.; Gschwend, J.E.; et al. Detection of Circulating Tumor Cells in Different Stages of Prostate Cancer. J. Cancer Res. Clin. Oncol. 2013, 139, 755–763. [Google Scholar] [CrossRef]
- Goldkorn, A.; Ely, B.; Quinn, D.I.; Tangen, C.M.; Fink, L.M.; Xu, T.; Twardowski, P.; Van Veldhuizen, P.J.; Agarwal, N.; Carducci, M.A.; et al. Circulating Tumor Cell Counts Are Prognostic of Overall Survival in SWOG S0421: A Phase III Trial of Docetaxel with or without Atrasentan for Metastatic Castration-Resistant Prostate Cancer. J. Clin. Oncol. 2014, 32, 1136–1142. [Google Scholar] [CrossRef]
- Chen, B.T.; Loberg, R.D.; Neeley, C.K.; O’Hara, S.M.; Gross, S.; Doyle, G.; Dunn, R.L.; Kalikin, L.M.; Pienta, K.J. Preliminary Study of Immunomagnetic Quantification of Circulating Tumor Cells in Patients with Advanced Disease. Urology 2005, 65, 616–621. [Google Scholar] [CrossRef]
- Fleck, A.; Raines, G.; Hawker, F.; Trotter, J.; Wallace, P.I.; Ledingham, I.M.; Calman, K.C. Increased Vascular Permeability: A Major Cause of Hypoalbuminaemia in Disease and Injury. Lancet 1985, 1, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Seymour, L.; Bogaerts, J.; Perrone, A.; Ford, R.; Schwartz, L.H.; Mandrekar, S.; Lin, N.U.; Litière, S.; Dancey, J.; Chen, A.; et al. iRECIST: Guidelines for Response Criteria for Use in Trials Testing Immunotherapeutics. Lancet Oncol. 2017, 18, e143–e152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, F.; Zhu, J.; Mao, Y.; Li, X.; Hu, B.; Zhang, D. Associations between the Epithelial-Mesenchymal Transition Phenotypes of Circulating Tumor Cells and the Clinicopathological Features of Patients with Colorectal Cancer. Dis. Mrk. 2017, 2017, 9474532. [Google Scholar] [CrossRef] [PubMed][Green Version]


| Clinicopathological Characteristics | Overall | M0 | M1TN | M1TP | p-Value |
|---|---|---|---|---|---|
| Number of patients | 18 | 8 | 5 | 5 | |
| Gender, n (%) | |||||
| Female | 5 (28.0) | 2 (25.0) | 1 (20.0) | 2 (40.0) | 1 |
| Age, years | |||||
| Median (range) | 60 (43–78) | 60 (52–70) | 71 (43–78) | 60 (48–69) | 0.511 |
| Smoking habits, n (%) | 7 (38.9) | 5 (62.5) | 1 (20.0) | 1 (20.0) | 0.252 |
| Obesity, n (%) | |||||
| Overweight/Obesity | 12 (66.7) | 7 (87.5) | 1 (20.0) | 4 (80.0) | 0.05 |
| BMI score (%) | |||||
| Median (range) | 25.5 (18–38.6) | 25.5 (23.6–38.6) | 21.5 (21.0–25.4) | 25.5 (18.0–27.1) | 0.049 |
| Hypertension, n (%) | 12 (66.7) | 6 (75.0) | 3 (60.0) | 3 (60.0) | 1 |
| Diabetes, n (%) | 5 (27.8) | 3 (37.5) | 1 (20.0) | 1 (20.0) | 1 |
| ECOG score, n (%) | 0.384 | ||||
| 0 | 10 (55.6) | 4 (50.0) | 3 (60.0) | 3 (60.0) | |
| 1 | 4 (22.2) | 3 (37.5) | 0 | 1 (20.0) | |
| 2 | 2 (11.1) | 1 (12.5) | 0 | 1 (20.0) | |
| 3 | 2 (11.1) | 0 | 2 (40.0) | 0 | |
| T stage, n (%) | 0.003 | ||||
| T1a | 6 (33.3) | 6 (75.0) | 0 | 0 | |
| T1b | 3 (16.7) | 2 (25.0) | 1 (20.0) | 0 | |
| T2a | 2 (11.1) | 0 | 1 (20.0) | 1 (20.0) | |
| T2b | 2 (11.1) | 0 | 1 (20.0) | 1 (20.0) | |
| T3a | 5 (27.8) | 0 | 2 (40.0) | 3 (60.0) | |
| N stage, n (%) | 0.045 | ||||
| N0 | 13 (72.2) | 8 (100.0) | 3 (60.0) | 2 (40.0) | |
| N1 | 5 (27.8) | 0 | 2 (40.0) | 3 (60.0) | |
| Histology, n (%) | 0.515 | ||||
| Clear cell | 8 (72.7) | 4 (80.0) | 1 (50.0) | 3 (75.0) | |
| Chromophobe | 1 (9.1) | 0 | 1 (50.0) | 0 | |
| Papillary | 2 (18.2) | 1 (20.0) | 0 | 1 (25.0) | |
| No biopsy (patient preference or unfit) | 3 | 3 | 3 | 1 | |
| Metastatic site, n (%) | 1 | ||||
| Lung | - | - | 3 | 3 | |
| Bone | - | - | 1 | 1 | |
| Distant lymph nodes | - | - | 1 | 1 | |
| Antiplatelet therapy, n (%) | 5 (27.8) | 3 (37.5) | 2 (40.0) | 0 | 0.416 |
| Systemic therapy, n (%) | |||||
| First line | - | - | 2 | 2 | |
| Second line | - | - | - | 3 | |
| Unfit for treatment | - | - | 3 | - | |
| Treatment, n (%) | |||||
| TKI | 4 (22.2) | - | 1 (20.0) | 3 (75.0) | |
| ICI | 3 (16.7) | - | 1 (20.0) | 2 (40.0) | |
| Radical nephrectomy | 8 (72.7) | 5 (62.5) | 0 | - | |
| Partial nephrectomy | 1 (9.1) | 1 (20.0) | 0 | - | |
| Surveillance | 2 (11.1) | 2 (25.0) | 0 | 0 | |
| Unfit for treatment | 3 (16.7) | 0 | 3 (60.0) | 0 |
| M0 | M1TN | M1TP | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Median | Average | Range | Median | Average | Range | Median | Average | Range | |
| Single CTCs | 1.5 | 3.3 | 0–13 | 49 | 63.8 | 0–157 | 2 | 3.2 | 0–10 |
| Epithelial | 3 | 4.2 | 0–13 | 0 | 5.3 | 0–21 | 2 | 3 | 0–10 |
| Mesenchymal | 0 | 0.0 | — | 49 | 59.6 | 0–157 | 0 | 0.2 | 0–1 |
| CTC clusters | 0 | 0.25 | 0–1 | 3 | 5.8 | 0–16 | 0 | 0.0 | — |
| CTCs in clusters (Mesenchymal) | 0 | 3.1 | 0–21 | 31 | 38.0 | 0–98 | 0 | 0.0 | — |
| Total CTCs * | 3 | 6.4 | 0–27 | 80 | 101.8 | 0–255 | 2 | 3.2 | 0–10 |
| Epithelial | 3 | 4.2 | 0–13 | 0 | 5.3 | 0–21 | 2 | 3 | 0–10 |
| Mesenchymal | 0 | 3.1 | 0–21 | 80 | 97.6 | 0–255 | 0 | 0.2 | 0–1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leitão, T.P.; Corredeira, P.; Kucharczak, S.; Rodrigues, M.; Piairo, P.; Rodrigues, C.; Alves, P.; Cavaco, A.M.; Miranda, M.; Antunes, M.; et al. Clinical Validation of a Size-Based Microfluidic Device for Circulating Tumor Cell Isolation and Analysis in Renal Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 8404. https://doi.org/10.3390/ijms24098404
Leitão TP, Corredeira P, Kucharczak S, Rodrigues M, Piairo P, Rodrigues C, Alves P, Cavaco AM, Miranda M, Antunes M, et al. Clinical Validation of a Size-Based Microfluidic Device for Circulating Tumor Cell Isolation and Analysis in Renal Cell Carcinoma. International Journal of Molecular Sciences. 2023; 24(9):8404. https://doi.org/10.3390/ijms24098404
Chicago/Turabian StyleLeitão, Tito Palmela, Patrícia Corredeira, Sandra Kucharczak, Margarida Rodrigues, Paulina Piairo, Carolina Rodrigues, Patrícia Alves, Ana Martins Cavaco, Miguel Miranda, Marília Antunes, and et al. 2023. "Clinical Validation of a Size-Based Microfluidic Device for Circulating Tumor Cell Isolation and Analysis in Renal Cell Carcinoma" International Journal of Molecular Sciences 24, no. 9: 8404. https://doi.org/10.3390/ijms24098404
APA StyleLeitão, T. P., Corredeira, P., Kucharczak, S., Rodrigues, M., Piairo, P., Rodrigues, C., Alves, P., Cavaco, A. M., Miranda, M., Antunes, M., Ferreira, J., Palma Reis, J., Lopes, T., Diéguez, L., & Costa, L. (2023). Clinical Validation of a Size-Based Microfluidic Device for Circulating Tumor Cell Isolation and Analysis in Renal Cell Carcinoma. International Journal of Molecular Sciences, 24(9), 8404. https://doi.org/10.3390/ijms24098404







