Alpha-Deoxyguanosine to Reshape the Alpha-Thrombin Binding Aptamer
Abstract
1. Introduction
2. Results and Discussion
2.1. Design of the TBA Analogues
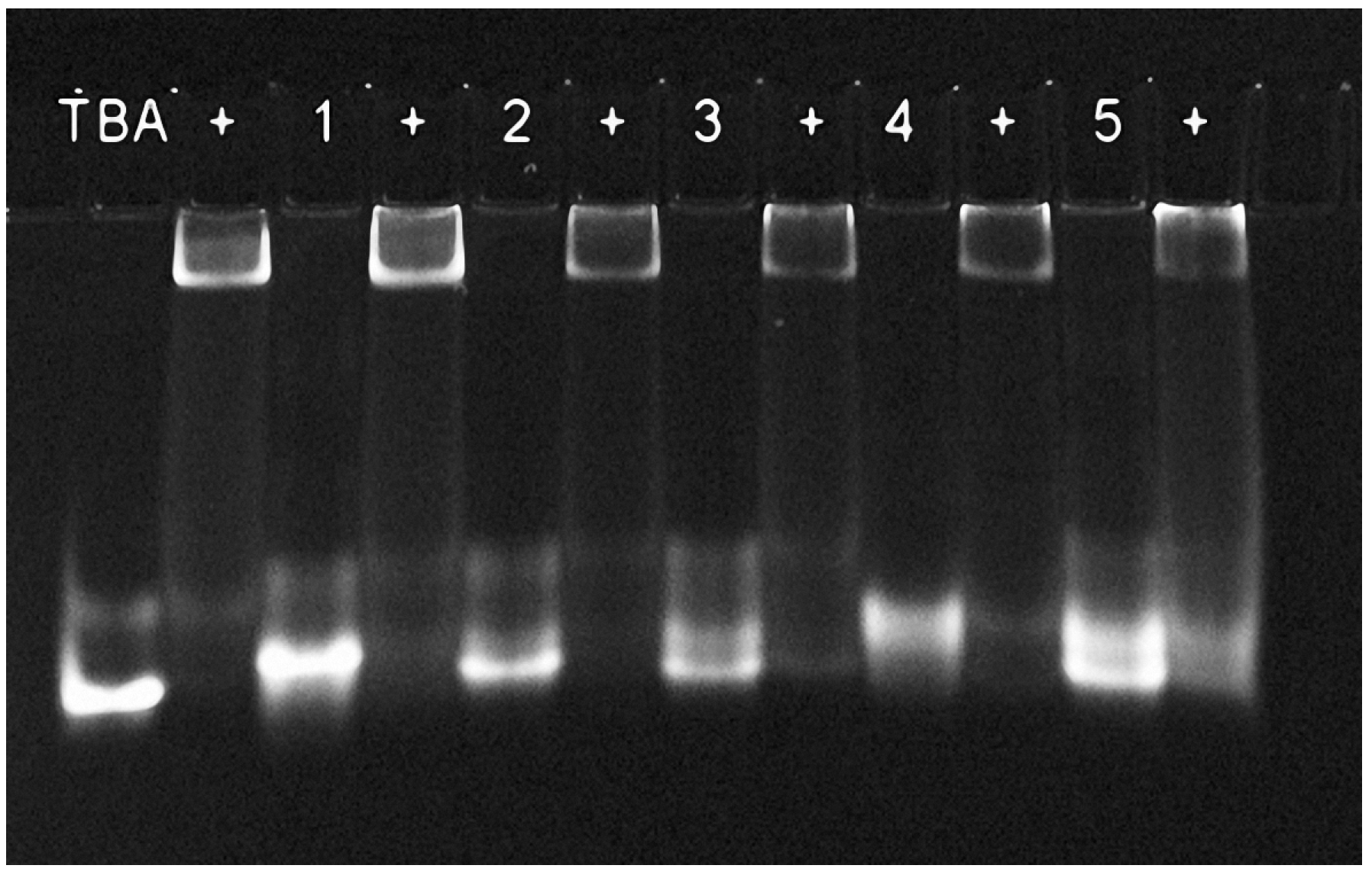
2.2. Binding with Human Alpha-Thrombin
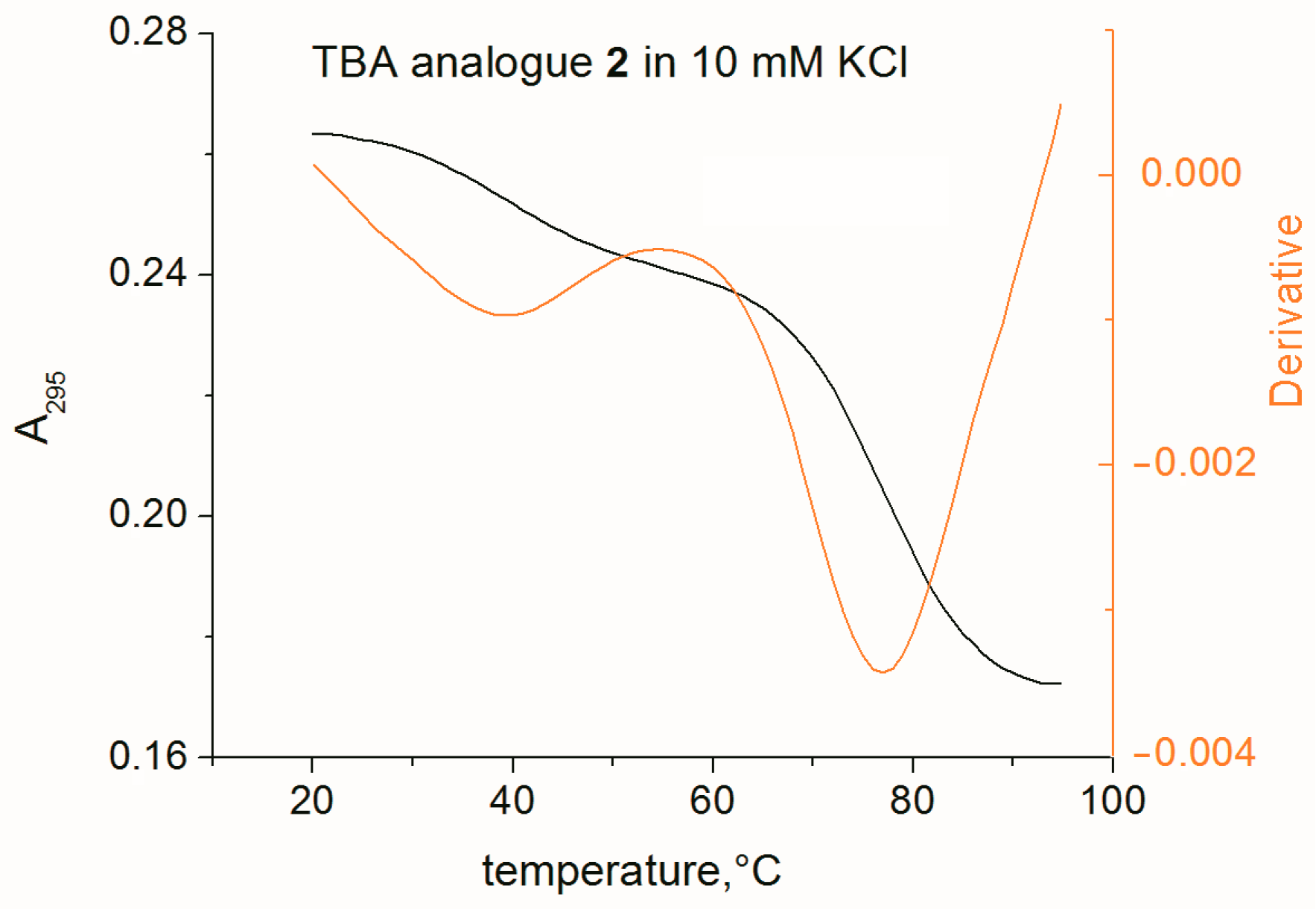
2.3. Thermal Stability and Folding
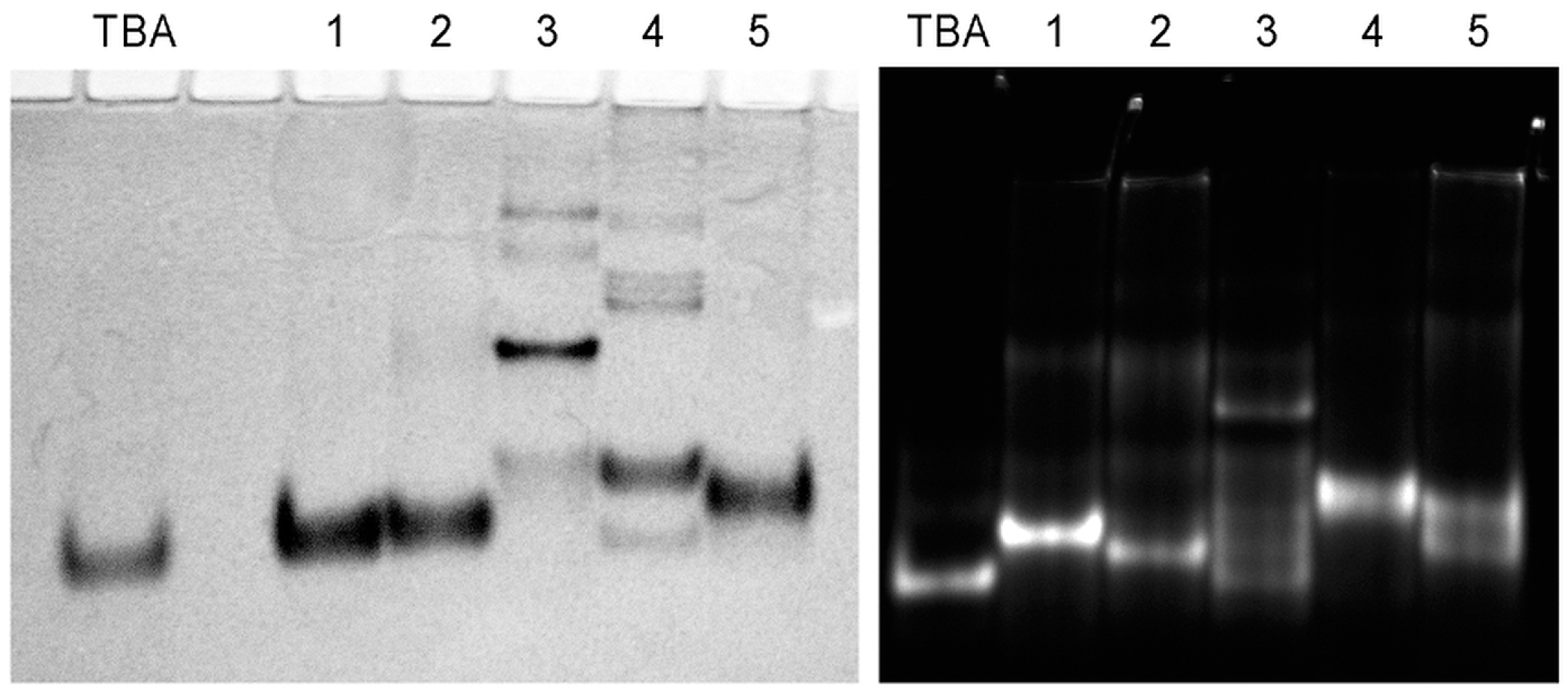
2.4. Structural Details of Modified G-Quadruplexes
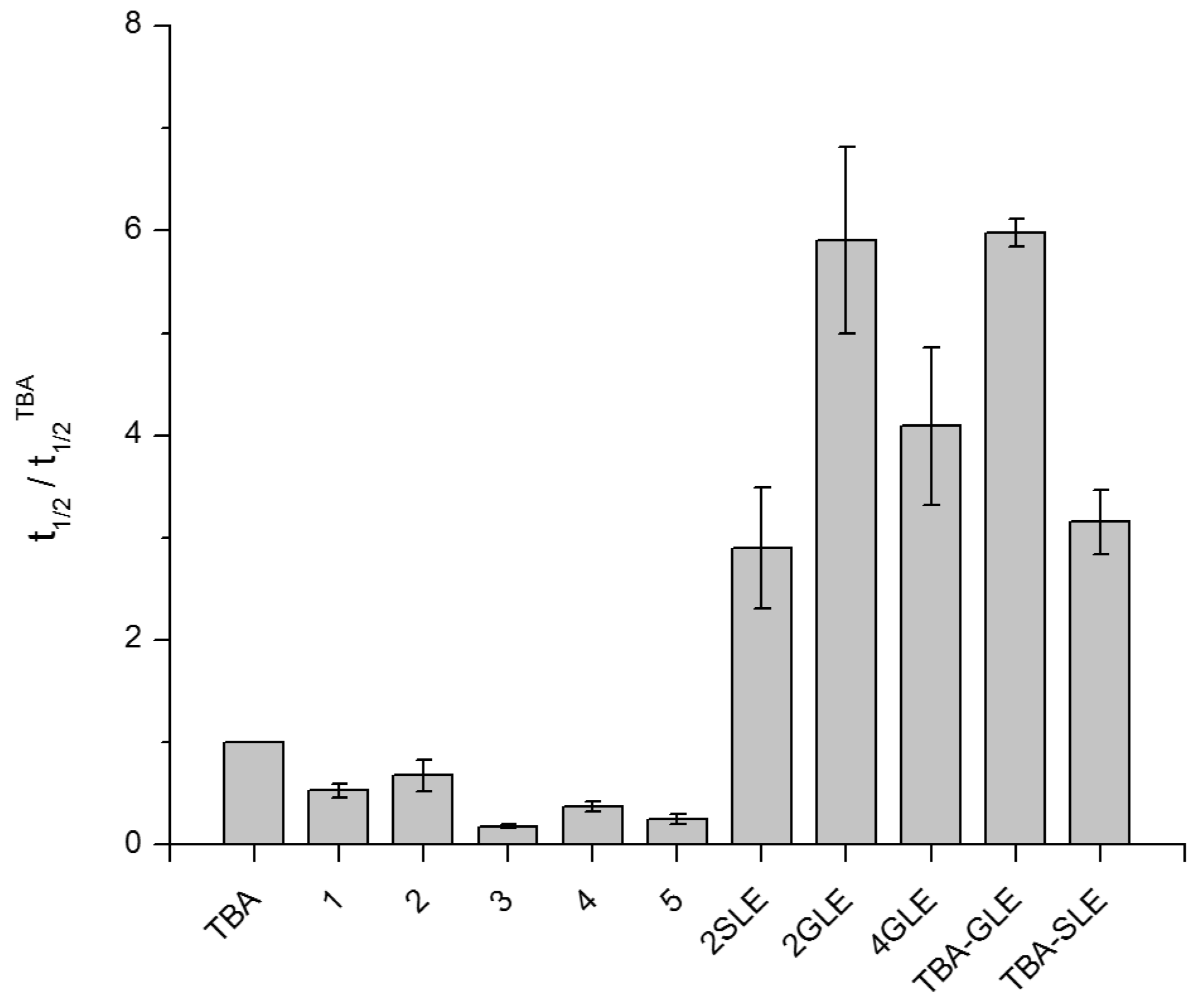
2.5. Anticoagulant Activity of Non-Natural TBA Analogues
2.6. Tuning the Activity of TBA Analogues by Conjugation with Optimized Tripeptides
3. Materials and Methods
3.1. Materials
3.2. Oligonucleotides and Oligonucleotide-Peptide Conjugates
3.3. Ultraviolet Thermal Denaturation
3.4. Circular Dichroism Spectroscopy
3.5. Binding of Aptamers with Thrombin
3.6. Native Gel-Electrophoresis
3.7. Fibrinogen Clotting in the Presence of Aptamers
3.8. Microscale Thermophoresis
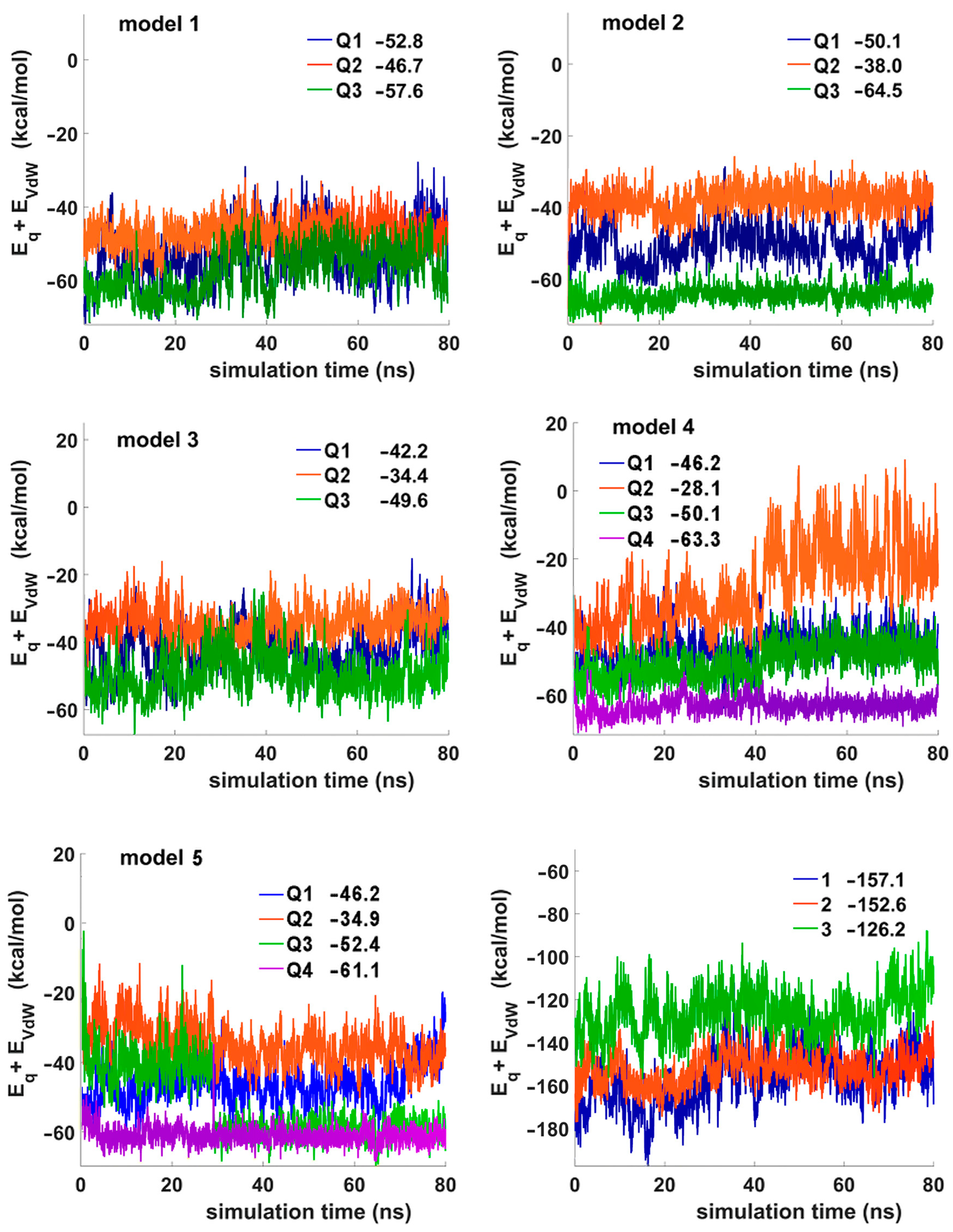
3.9. Molecular Dynamics Simulations of TBA Analogues
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalathingal, M.; Rhee, Y.M.J. Molecular mechanism of binding between a therapeutic RNA aptamer and its protein target VEGF: A molecular dynamics study. Comput. Chem. 2023, 44, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Ramasanoff, R.R.; Sokolov, P.A. The binding model of adenosine-specific DNA aptamer: Umbrella sampling study. J. Mol. Graph. 2023, 118, 108338. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Napolitano, E.; Platella, C.; Musumeci, D.; Montesarchio, D. G-quadruplex-based aptamers targeting human thrombin: Discovery, chemical modifications and antithrombotic effects. Pharmacol. Ther. 2021, 217, 107649. [Google Scholar] [CrossRef] [PubMed]
- Troisi, R.; Balasco, N.; Autiero, I.; Vitagliano, L.; Sica, F. Exosite binding in thrombin: A global structural/dynamic overview of complexes with aptamers and other ligands. Int. J. Mol. Sci. 2021, 22, 10803. [Google Scholar] [CrossRef]
- Schultze, P.; Macaya, R.F.; Feigon, J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol. 1994, 235, 1532–1547. [Google Scholar] [CrossRef]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Napolitano, V.; Petraccone, L.; Troisi, R.; Spiridonova, V.; Mattia, C.A.; Sica, F. Duplex/quadruplex oligonucleotides: Role of the duplex domain in the stabilization of a new generation of highly effective anti-thrombin aptamers. Int. J. Biol. Macromol. 2018, 107, 1697–1705. [Google Scholar] [CrossRef]
- Padmanabhan, K.; Tulinsky, A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D 1996, 52, 272–282. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef]
- Benigno, D.; Virgilio, A.; Bello, I.; La Manna, S.; Vellecco, V.; Bucci, M.; Marasco, D.; Panza, E.; Esposito, V.; Galeone, A. Properties and Potential Antiproliferative Activity of Thrombin-Binding Aptamer (TBA) Derivatives with One or Two Additional G-Tetrads. Int. J. Mol. Sci. 2022, 23, 14921. [Google Scholar] [CrossRef]
- Randazzo, A.; Galeone, A.; Esposito, V.; Varra, M.; Mayol, L. Interaction of distamycin A and netropsin with quadruplex and duplex structures: A comparative 1H-NMR study. Nucleosides Nucleotides Nucleic Acids 2002, 21, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Svetlova, J.; Sardushkin, M.; Kolganova, N.; Timofeev, E. Recognition Interface of the Thrombin Binding Aptamer Requires Antiparallel Topology of the Quadruplex Core. Biomolecules 2021, 11, 1332. [Google Scholar] [CrossRef]
- Latha, Y.S.; Yathindra, N. Stereochemical studies on nucleic acid analogues. I. Conformations of alpha-nucleosides and alpha-nucleotides: Interconversion of sugar puckers via O4′-exo. Biopolymers 1992, 32, 249–269. [Google Scholar] [CrossRef] [PubMed]
- Filitcheva, J.; Edwards, P.J.; Norris, G.E.; Filichev, V.V. α-2′-Deoxyguanosine can switch DNA G-quadruplex topologies from antiparallel to parallel. Org. Biomol. Chem. 2019, 17, 4031–4042. [Google Scholar] [CrossRef] [PubMed]
- Aramini, J.M.; Cleaver, S.H.; Pon, R.T.; Cunningham, R.P.; Germann, M.W. Solution structure of a DNA duplex containing an α-anomeric adenosine: Insights into substrate recognition by endonuclease IV. J. Mol. Biol. 2004, 338, 77–91. [Google Scholar] [CrossRef]
- Bielecki, L.; Skalski, B.; Zagorowska, I.; Verrall, R.E.; Adamiak, R.W. Fluorescent α-anomeric 1, N (6) etheno-deoxyadenosine in DNA duplexes. The α-εdA/dG pair. Nucleosides Nucleotides Nucleic Acids 2000, 19, 1735–1750. [Google Scholar] [CrossRef]
- Haase, L.; Karg, B.; Weisz, K. Manipulating DNA G-Quadruplex Structures by Using Guanosine Analogues. ChemBioChem 2019, 20, 985–993. [Google Scholar] [CrossRef]
- Mohr, S.; Jana, J.; Vianney, Y.M.; Weisz, K. Expanding the Topological Landscape by a G-Column Flip of a Parallel G-Quadruplex. Chem. Eur. J. 2021, 27, 10437–10447. [Google Scholar] [CrossRef]
- Dominick, P.K.; Jarstfer, M.B. A conformationally constrained nucleotide analogue controls the folding topology of a DNA G-quadruplex. J. Am. Chem. Soc. 2004, 126, 5050–5051. [Google Scholar] [CrossRef]
- Varizhuk, I.V.; Tsvetkov, V.B.; Toropygin, I.Y.; Stomakhin, A.A.; Kolganova, N.A.; Surzhikov, S.A.; Timofeev, E.N. The Regioselective Conjugation of the 15-nt Thrombin Aptamer with an Optimized Tripeptide Sequence Greatly Increases the Anticoagulant Activity of the Aptamer. Pharmaceutics 2023, 15, 604. [Google Scholar] [CrossRef]
- Tsvetkov, V.B. Modeling of possible quadruplexes and i-motifs formed during DNA contacts: Strategy, classification, most probable shapes, origami based on quadruplexes. bioRxiv 2022. bioRxiv:10.10.511558. [Google Scholar]
- Tsvetkov, V.B.; Varizhuk, I.V.; Kurochkin, N.N.; Surzhikov, S.A.; Smirnov, I.P.; Stomakhin, A.A.; Kolganova, N.A.; Timofeev, E.N. Anticoagulant Oligonucleotide-Peptide Conjugates: Identification of Thrombin Aptamer Conjugates with Improved Characteristics. Int. J. Mol. Sci. 2022, 23, 3820. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G.; et al. AMBER 2020; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Izadi, S.; Onufriev, A.V. Accuracy limit of rigid 3-point water models. J. Chem. Phys. 2016, 145, 074501. [Google Scholar] [CrossRef] [PubMed]
- Zgarbová, M.; Luque, F.J.; Sponer, J.; Cheatham, T.E., III; Otyepka, M.; Jurecka, P. Toward Improved Description of DNA Backbone: Revisiting Epsilon and Zeta Torsion Force Field Parameters. J. Chem. Theory Comput. 2013, 9, 2339–2354. [Google Scholar] [CrossRef]
- Zgarbová, M.; Sponer, J.; Otyepka, M.; Cheatham, T.E., III; Galindo-Murillo, R.; Jurecka, P. Refinement of the Sugar-Phosphate Backbone Torsion Beta for AMBER Force Fields Improves the Description of Z- and B-DNA. J. Chem. Theory Comput. 2015, 11, 5723–5736. [Google Scholar] [CrossRef]


| ID | Sequence 1 | Tm, °C 2 | |||
|---|---|---|---|---|---|
| 100 mM K+ | 10 mM K+ | 100 mM Na+ | 10 mM Na+ | ||
| 1 | 5′-ggGTTGGGTGTggGTTGGG | n.d. 3 | n.d. | 74.5 | 56.4 |
| 2 | 5′-gGGTTGGGTGTgGGTTGGG | >90 4 | 76.9 | 61.3 | 45.1 |
| 3 | 5′-GGGTTGGgTGTGGGTTGGg | 59.1 | 41.8 | 44.2 | 26.5 |
| 4 | 5′-ggGGTTGGGGTGTggGGTTGGGG | n.d. | n.d. | n.d. | 87.8 |
| 5 | 5′-GGGGTTGGggTGTGGGGTTGGgg | >90 | 79.6 | 65.9 | 43.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolganova, N.A.; Tsvetkov, V.B.; Stomakhin, A.A.; Surzhikov, S.A.; Timofeev, E.N.; Varizhuk, I.V. Alpha-Deoxyguanosine to Reshape the Alpha-Thrombin Binding Aptamer. Int. J. Mol. Sci. 2023, 24, 8406. https://doi.org/10.3390/ijms24098406
Kolganova NA, Tsvetkov VB, Stomakhin AA, Surzhikov SA, Timofeev EN, Varizhuk IV. Alpha-Deoxyguanosine to Reshape the Alpha-Thrombin Binding Aptamer. International Journal of Molecular Sciences. 2023; 24(9):8406. https://doi.org/10.3390/ijms24098406
Chicago/Turabian StyleKolganova, Natalia A., Vladimir B. Tsvetkov, Andrey A. Stomakhin, Sergei A. Surzhikov, Edward N. Timofeev, and Irina V. Varizhuk. 2023. "Alpha-Deoxyguanosine to Reshape the Alpha-Thrombin Binding Aptamer" International Journal of Molecular Sciences 24, no. 9: 8406. https://doi.org/10.3390/ijms24098406
APA StyleKolganova, N. A., Tsvetkov, V. B., Stomakhin, A. A., Surzhikov, S. A., Timofeev, E. N., & Varizhuk, I. V. (2023). Alpha-Deoxyguanosine to Reshape the Alpha-Thrombin Binding Aptamer. International Journal of Molecular Sciences, 24(9), 8406. https://doi.org/10.3390/ijms24098406







