B-Cell Receptor Signaling and Beyond: The Role of Igα (CD79a)/Igβ (CD79b) in Normal and Malignant B Cells
Abstract
:1. Introduction
2. BCR Signaling in Normal B Cells
3. B-Cell Derived Malignancies and BCR Signaling
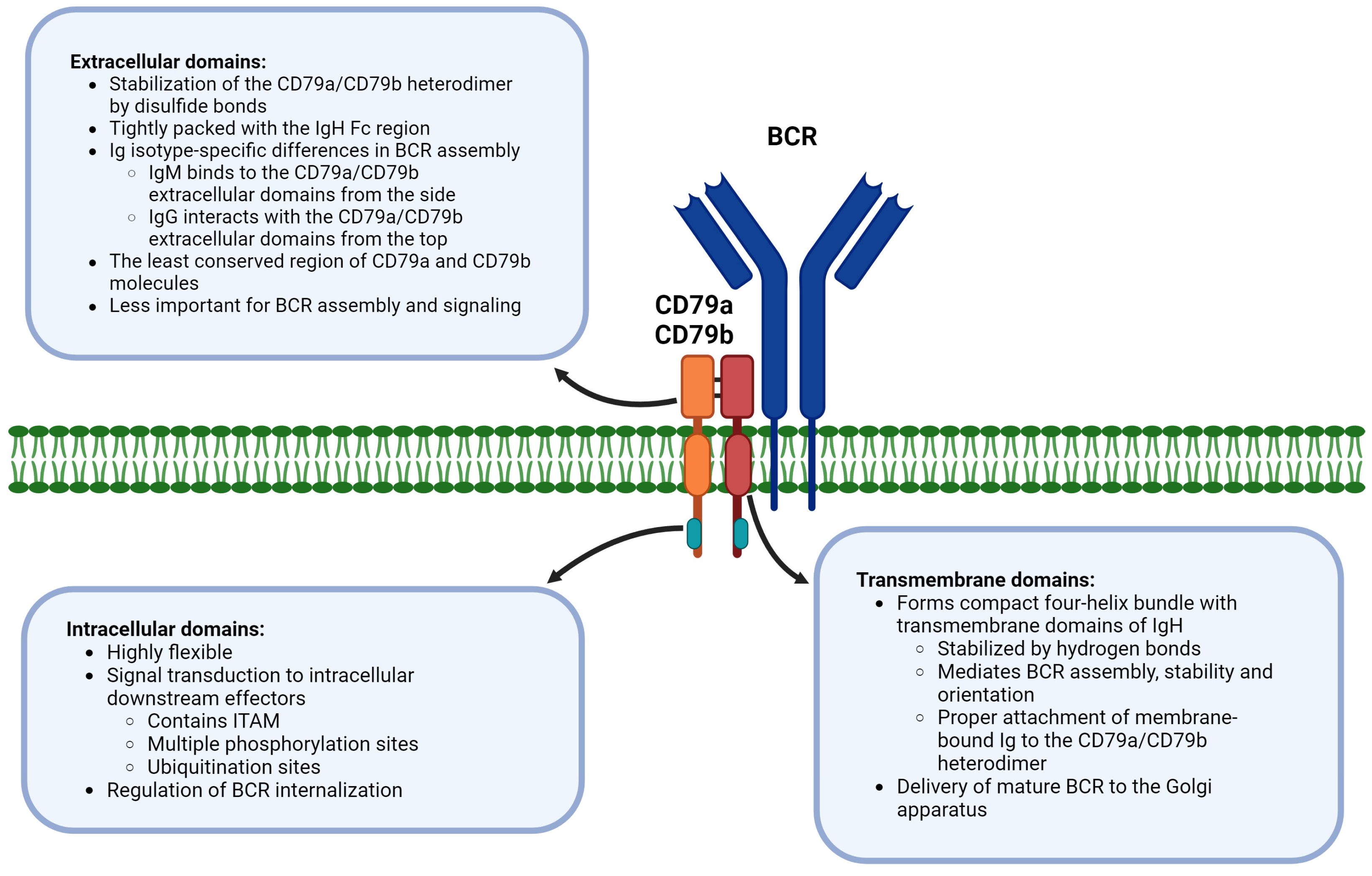
4. Extracellular, Transmembrane, and Intracellular Domains of CD79a/CD79b Are Functionally Distinct in BCR
5. The CD79a/CD79b Heterodimer Is Critically Important for BCR Functionality
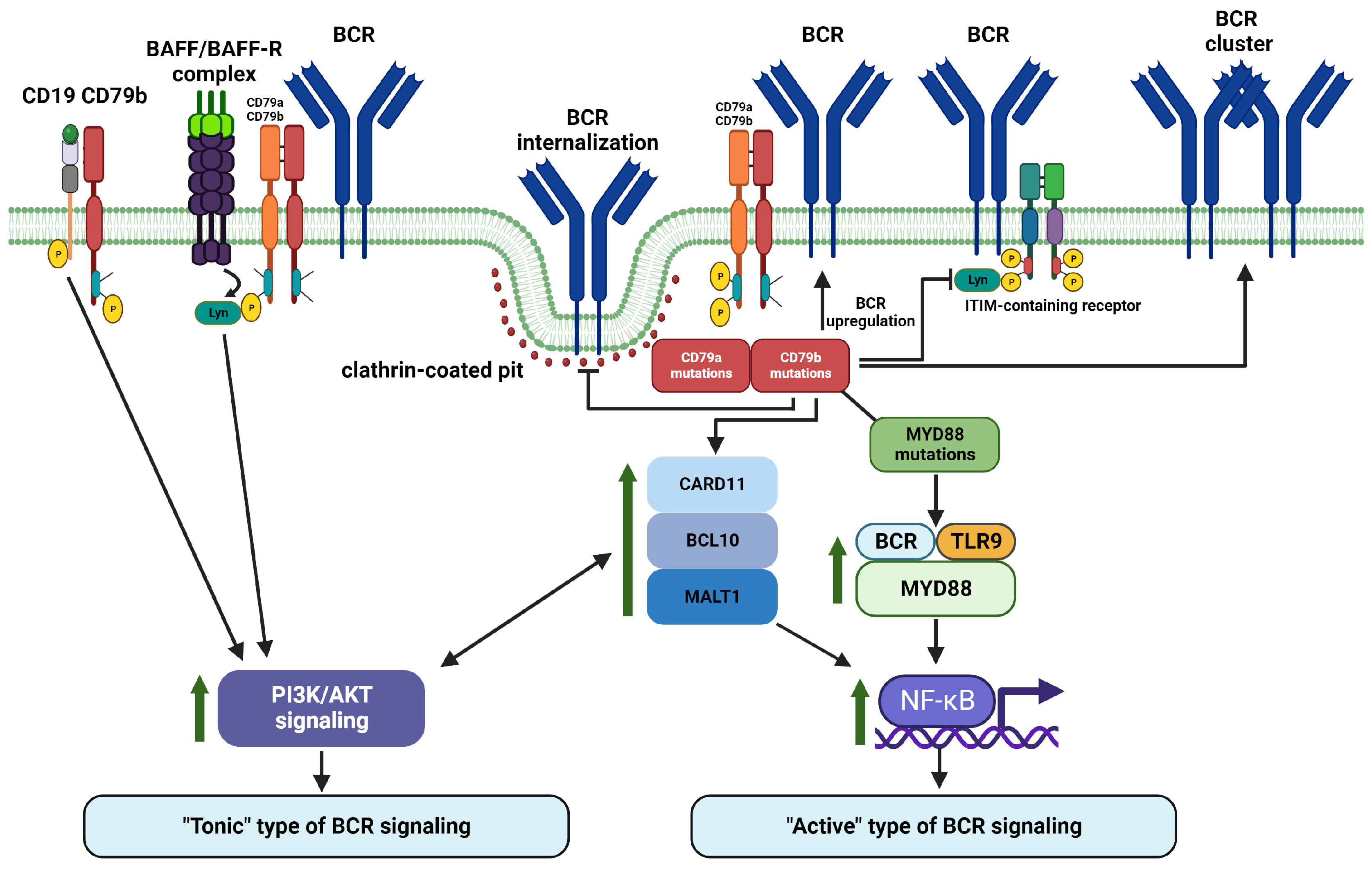
6. BCR Signaling Is Regulated by Phosphorylation, Ubiquitination, and Glycosylation of CD79a and CD79b
7. BCR Signaling Is Regulated at the Level of CD79a/CD79b Heterodimer by Physical Interactions with Regulatory Molecules
8. CD79a and CD79b Are Important Regulators of Proximal and Distal BCR Signaling in Malignant B Cells
9. Open Questions and Future Directions
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tanaka, S.; Baba, Y. B Cell Receptor Signaling. Adv. Exp. Med. Biol. 2020, 1254, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Huse, K.; Bai, B.; Hilden, V.I.; Bollum, L.K.; Våtsveen, T.K.; Munthe, L.A.; Smeland, E.B.; Irish, J.M.; Wälchli, S.; Myklebust, J.H. Mechanism of CD79A and CD79B Support for IgM+ B Cell Fitness through B Cell Receptor Surface Expression. J. Immunol. 2022, 209, 2042–2053. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhou, D.; Wang, L.; Zhu, L.; Ye, X. MYD88(L265P) and CD79B double mutations type (MCD type) of diffuse large B-cell lymphoma: Mechanism, clinical characteristics, and targeted therapy. Ther. Adv. Hematol. 2022, 13, 20406207211072839. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, Y.; Zhang, S.; Haneef, K.; Liu, W. Structural and immunogenomic insights into B-cell receptor activation. J. Genet. Genom. 2020, 47, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Avalos, A.M.; Ploegh, H.L. Early BCR Events and Antigen Capture, Processing, and Loading on MHC Class II on B Cells. Front. Immunol. 2014, 5, 92. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, L.; Sasaki, Y.; Calado, D.P.; Zhang, B.; Paik, J.H.; DePinho, R.A.; Kutok, J.L.; Kearney, J.F.; Otipoby, K.L.; Rajewsky, K. PI3 kinase signals BCR-dependent mature B cell survival. Cell 2009, 139, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Wiestner, A. Targeting B cell receptor signalling in cancer: Preclinical and clinical advances. Nat. Rev. Cancer 2018, 18, 148–167. [Google Scholar] [CrossRef]
- Young, R.M.; Staudt, L.M. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat. Rev. Drug Discov. 2013, 12, 229–243. [Google Scholar] [CrossRef]
- Puri, K.D.; Di Paolo, J.A.; Gold, M.R. B-cell receptor signaling inhibitors for treatment of autoimmune inflammatory diseases and B-cell malignancies. Int. Rev. Immunol. 2013, 32, 397–427. [Google Scholar] [CrossRef]
- Rawlings, D.J.; Metzler, G.; Wray-Dutra, M.; Jackson, S.W. Altered B cell signalling in autoimmunity. Nat. Rev. Immunol. 2017, 17, 421–436. [Google Scholar] [CrossRef]
- Liu, W.; Tolar, P.; Song, W.; Kim, T.J. Editorial: BCR Signaling and B Cell Activation. Front. Immunol. 2020, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Köhrer, S.; Havranek, O.; Seyfried, F.; Hurtz, C.; Coffey, G.P.; Kim, E.; Ten Hacken, E.; Jäger, U.; Vanura, K.; O’Brien, S.; et al. Pre-BCR signaling in precursor B-cell acute lymphoblastic leukemia regulates PI3K/AKT, FOXO1 and MYC, and can be targeted by SYK inhibition. Leukemia 2016, 30, 1246–1254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Srivastava, G.; Lu, L. The pre-B cell receptor and its function during B cell development. Cell Mol. Immunol. 2004, 1, 89–94. [Google Scholar] [PubMed]
- Winkler, T.H.; Mårtensson, I.-L. The Role of the Pre-B Cell Receptor in B Cell Development, Repertoire Selection, and Tolerance. Front. Immunol. 2018, 9, 2423. [Google Scholar] [CrossRef] [PubMed]
- Keren, Z.; Melamed, D. Antigen receptor signaling competence and the determination of B cell fate in B-lymphopoiesis. Histol. Histopathol. 2005, 20, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Neys, S.F.H.; Heukels, P.; van Hulst, J.A.C.; Rip, J.; Wijsenbeek, M.S.; Hendriks, R.W.; Corneth, O.B.J. Aberrant B Cell Receptor Signaling in Naïve B Cells from Patients with Idiopathic Pulmonary Fibrosis. Cells 2021, 10, 1321. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Jing, Y.; Yang, L.; Kang, D.; Jiang, P.; Li, N.; Cheng, J.; Li, J.; Li, X.; Peng, Z.; et al. The regulators of BCR signaling during B cell activation. Blood Sci. 2019, 1, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Berry, C.T.; Liu, X.; Myles, A.; Nandi, S.; Chen, Y.H.; Hershberg, U.; Brodsky, I.E.; Cancro, M.P.; Lengner, C.J.; May, M.J.; et al. BCR-Induced Ca2+ Signals Dynamically Tune Survival, Metabolic Reprogramming, and Proliferation of Naive B Cells. Cell Rep. 2020, 31, 107474. [Google Scholar] [CrossRef]
- McShane, A.N.; Malinova, D. The Ins and Outs of Antigen Uptake in B cells. Front. Immunol. 2022, 13, 892169. [Google Scholar] [CrossRef]
- Rastogi, I.; Jeon, D.; Moseman, J.E.; Muralidhar, A.; Potluri, H.K.; McNeel, D.G. Role of B cells as antigen presenting cells. Front. Immunol. 2022, 13, 954936. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, J.H. How the Signaling Crosstalk of B Cell Receptor (BCR) and Co-Receptors Regulates Antibody Class Switch Recombination: A New Perspective of Checkpoints of BCR Signaling. Front. Immunol. 2021, 12, 663443. [Google Scholar] [CrossRef] [PubMed]
- Vlachiotis, S.; Abolhassani, H. Transcriptional regulation of B cell class-switch recombination: The role in development of noninfectious complications. Expert Rev. Clin. Immunol. 2022, 18, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Tsai, D.-Y.; Hung, K.-H.; Chang, C.-W.; Lin, K.-I. Regulatory mechanisms of B cell responses and the implication in B cell-related diseases. J. Biomed. Sci. 2019, 26, 64. [Google Scholar] [CrossRef]
- Luo, W.; Mayeux, J.; Gutierrez, T.; Russell, L.; Getahun, A.; Müller, J.; Tedder, T.; Parnes, J.; Rickert, R.; Nitschke, L.; et al. A balance between B cell receptor and inhibitory receptor signaling controls plasma cell differentiation by maintaining optimal Ets1 levels. J. Immunol. 2014, 193, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Kluckova, K.; D’Avola, A.; Riches, J.C. Advances in Understanding of Metabolism of B-Cell Lymphoma: Implications for Therapy. Cancers 2022, 14, 5552. [Google Scholar] [CrossRef] [PubMed]
- Doughty, C.A.; Bleiman, B.F.; Wagner, D.J.; Dufort, F.J.; Mataraza, J.M.; Roberts, M.F.; Chiles, T.C. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: Role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood 2006, 107, 4458–4465. [Google Scholar] [CrossRef]
- Iperi, C.; Bordron, A.; Dueymes, M.; Pers, J.-O.; Jamin, C. Metabolic Program of Regulatory B Lymphocytes and Influence in the Control of Malignant and Autoimmune Situations. Front. Immunol. 2021, 12, 735463. [Google Scholar] [CrossRef]
- Raza, I.G.A.; Clarke, A.J. B Cell Metabolism and Autophagy in Autoimmunity. Front. Immunol. 2021, 12, 681105. [Google Scholar] [CrossRef]
- Watanabe, K.; Tsubata, T. Autophagy connects antigen receptor signaling to costimulatory signaling in B lymphocytes. Autophagy 2009, 5, 108–110. [Google Scholar] [CrossRef]
- Eeva, J.; Pelkonen, J. Mechanisms of B cell receptor induced apoptosis. Apoptosis 2004, 9, 525–531. [Google Scholar] [CrossRef]
- Nemazee, D. Mechanisms of central tolerance for B cells. Nat. Rev. Immunol. 2017, 17, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Yam-Puc, J.C.; Zhang, L.; Maqueda-Alfaro, R.A.; Garcia-Ibanez, L.; Zhang, Y.; Davies, J.; Senis, Y.A.; Snaith, M.; Toellner, K.-M. Enhanced BCR signaling inflicts early plasmablast and germinal center B cell death. iScience 2021, 24, 102038. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Susanibar-Adaniya, S.; Barta, S.K. 2021 Update on Diffuse large B cell lymphoma: A review of current data and potential applications on risk stratification and management. Am. J. Hematol. 2021, 96, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Barta, S.K. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2019, 94, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.; Garg, V.; Mallick, S.; Gogia, A. Current trends in diagnosis and management of follicular lymphoma. Am. J. Blood Res. 2022, 12, 105–124. [Google Scholar] [PubMed]
- Carbone, A.; Roulland, S.; Gloghini, A.; Younes, A.; von Keudell, G.; López-Guillermo, A.; Fitzgibbon, J. Follicular lymphoma. Nat. Rev. Dis. Primers 2019, 5, 83. [Google Scholar] [CrossRef]
- Profitós-Pelejà, N.; Santos, J.C.; Marín-Niebla, A.; Roué, G.; Ribeiro, M.L. Regulation of B-Cell Receptor Signaling and Its Therapeutic Relevance in Aggressive B-Cell Lymphomas. Cancers 2022, 14, 860. [Google Scholar] [CrossRef]
- Niemann, C.U.; Wiestner, A. B-cell receptor signaling as a driver of lymphoma development and evolution. Semin. Cancer Biol. 2013, 23, 410–421. [Google Scholar] [CrossRef]
- Dühren-von Minden, M.; Übelhart, R.; Schneider, D.; Wossning, T.; Bach, M.P.; Buchner, M.; Hofmann, D.; Surova, E.; Follo, M.; Köhler, F.; et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 2012, 489, 309–312. [Google Scholar] [CrossRef]
- Young, R.M.; Phelan, J.D.; Wilson, W.H.; Staudt, L.M. Pathogenic B-cell receptor signaling in lymphoid malignancies: New insights to improve treatment. Immunol. Rev. 2019, 291, 190–213. [Google Scholar] [CrossRef] [PubMed]
- Valla, K.; Flowers, C.R.; Koff, J.L. Targeting the B cell receptor pathway in non-Hodgkin lymphoma. Expert Opin. Investig. Drugs 2018, 27, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, M.; Dreyling, M.; Binder, M.; Trepel, M. The role of B cell antigen receptors in mantle cell lymphoma. J. Hematol. Oncol. 2017, 10, 164. [Google Scholar] [CrossRef] [PubMed]
- Sachen, K.L.; Strohman, M.J.; Singletary, J.; Alizadeh, A.A.; Kattah, N.H.; Lossos, C.; Mellins, E.D.; Levy, S.; Levy, R. Self-antigen recognition by follicular lymphoma B-cell receptors. Blood 2012, 120, 4182–4190. [Google Scholar] [CrossRef] [PubMed]
- Corso, J.; Pan, K.T.; Walter, R.; Doebele, C.; Mohr, S.; Bohnenberger, H.; Ströbel, P.; Lenz, C.; Slabicki, M.; Hüllein, J.; et al. Elucidation of tonic and activated B-cell receptor signaling in Burkitt’s lymphoma provides insights into regulation of cell survival. Proc. Natl. Acad. Sci. USA 2016, 113, 5688–5693. [Google Scholar] [CrossRef] [PubMed]
- Noy, A.; de Vos, S.; Thieblemont, C.; Martin, P.; Flowers, C.R.; Morschhauser, F.; Collins, G.P.; Ma, S.; Coleman, M.; Peles, S.; et al. Targeting Bruton tyrosine kinase with ibrutinib in relapsed/refractory marginal zone lymphoma. Blood 2017, 129, 2224–2232. [Google Scholar] [CrossRef] [PubMed]
- Bogusz, A.M.; Baxter, R.H.; Currie, T.; Sinha, P.; Sohani, A.R.; Kutok, J.L.; Rodig, S.J. Quantitative immunofluorescence reveals the signature of active B-cell receptor signaling in diffuse large B-cell lymphoma. Clin. Cancer Res. 2012, 18, 6122–6135. [Google Scholar] [CrossRef]
- Davis, R.E.; Ngo, V.N.; Lenz, G.; Tolar, P.; Young, R.M.; Romesser, P.B.; Kohlhammer, H.; Lamy, L.; Zhao, H.; Yang, Y.; et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010, 463, 88–92. [Google Scholar] [CrossRef]
- Phelan, J.D.; Young, R.M.; Webster, D.E.; Roulland, S.; Wright, G.W.; Kasbekar, M.; Shaffer, A.L., 3rd; Ceribelli, M.; Wang, J.Q.; Schmitz, R.; et al. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature 2018, 560, 387–391. [Google Scholar] [CrossRef]
- Havranek, O.; Xu, J.; Köhrer, S.; Wang, Z.; Becker, L.; Comer, J.M.; Henderson, J.; Ma, W.; Man Chun Ma, J.; Westin, J.R.; et al. Tonic B-cell receptor signaling in diffuse large B-cell lymphoma. Blood 2017, 130, 995–1006. [Google Scholar] [CrossRef]
- Myers, D.R.; Zikherman, J.; Roose, J.P. Tonic Signals: Why Do Lymphocytes Bother? Trends Immunol. 2017, 38, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Frustaci, A.M.; Deodato, M.; Zamprogna, G.; Cairoli, R.; Montillo, M.; Tedeschi, A. Next Generation BTK Inhibitors in CLL: Evolving Challenges and New Opportunities. Cancers 2023, 15, 1504. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, L.; Fernández-Miranda, I.; Pérez-Callejo, D.; Quero, C.; Rodríguez, M.; Martín-Acosta, P.; Gómez, S.; González-Rincón, J.; Santos, A.; Tarin, C.; et al. Proposal and validation of a method to classify genetic subtypes of diffuse large B cell lymphoma. Sci. Rep. 2021, 11, 1886. [Google Scholar] [CrossRef] [PubMed]
- Lacy, S.E.; Barrans, S.L.; Beer, P.A.; Painter, D.; Smith, A.G.; Roman, E.; Cooke, S.L.; Ruiz, C.; Glover, P.; Van Hoppe, S.J.L.; et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: A Haematological Malignancy Research Network report. Blood 2020, 135, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.W.; Huang, D.W.; Phelan, J.D.; Coulibaly, Z.A.; Roulland, S.; Young, R.M.; Wang, J.Q.; Schmitz, R.; Morin, R.D.; Tang, J.; et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications. Cancer Cell 2020, 37, 551–568.e14. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef]
- Ma, X.; Zhu, Y.; Dong, D.; Chen, Y.; Wang, S.; Yang, D.; Ma, Z.; Zhang, A.; Zhang, F.; Guo, C.; et al. Cryo-EM structures of two human B cell receptor isotypes. Science 2022, 377, 880–885. [Google Scholar] [CrossRef]
- Su, Q.; Chen, M.; Shi, Y.; Zhang, X.; Huang, G.; Huang, B.; Liu, D.; Liu, Z.; Shi, Y. Cryo-EM structure of the human IgM B cell receptor. Science 2022, 377, 875–880. [Google Scholar] [CrossRef]
- Dong, Y.; Pi, X.; Bartels-Burgahn, F.; Saltukoglu, D.; Liang, Z.; Yang, J.; Alt, F.W.; Reth, M.; Wu, H. Structural principles of B cell antigen receptor assembly. Nature 2022, 612, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Reth, M.; Nitschke, L.; Hikida, M.; Kurosaki, T. Chapter 10—Structure and Signaling Function of the B-Cell Antigen Receptor and Its Coreceptors. In Molecular Biology of B Cells, 2nd ed.; Alt, F.W., Honjo, T., Radbruch, A., Reth, M., Eds.; Academic Press: London, UK, 2015; pp. 151–170. [Google Scholar]
- Tolar, P.; Pierce, S.K. Unveiling the B cell receptor structure. Science 2022, 377, 819–820. [Google Scholar] [CrossRef] [PubMed]
- Tolar, P.; Sohn, H.W.; Pierce, S.K. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat. Immunol. 2005, 6, 1168–1176. [Google Scholar] [CrossRef] [PubMed]
- Friess, M.D.; Pluhackova, K.; Böckmann, R.A. Structural Model of the mIgM B-Cell Receptor Transmembrane Domain From Self-Association Molecular Dynamics Simulations. Front. Immunol. 2018, 9, 2947. [Google Scholar] [CrossRef] [PubMed]
- Lutz, J.; Dittmann, K.; Bösl, M.R.; Winkler, T.H.; Wienands, J.; Engels, N. Reactivation of IgG-switched memory B cells by BCR-intrinsic signal amplification promotes IgG antibody production. Nat. Commun. 2015, 6, 8575. [Google Scholar] [CrossRef] [PubMed]
- James, L.K. B cells defined by immunoglobulin isotypes. Clin. Exp. Immunol. 2022, 210, 230–239. [Google Scholar] [CrossRef]
- Engels, N.; König, L.M.; Schulze, W.; Radtke, D.; Vanshylla, K.; Lutz, J.; Winkler, T.H.; Nitschke, L.; Wienands, J. The immunoglobulin tail tyrosine motif upgrades memory-type BCRs by incorporating a Grb2-Btk signalling module. Nat. Commun. 2014, 5, 5456. [Google Scholar] [CrossRef]
- Vanshylla, K.; Bartsch, C.; Hitzing, C.; Krümpelmann, L.; Wienands, J.; Engels, N. Grb2 and GRAP connect the B cell antigen receptor to Erk MAP kinase activation in human B cells. Sci. Rep. 2018, 8, 4244. [Google Scholar] [CrossRef]
- Geisberger, R.; Crameri, R.; Achatz, G. Models of signal transduction through the B-cell antigen receptor. Immunology 2003, 110, 401–410. [Google Scholar] [CrossRef]
- Maity, P.C.; Datta, M.; Nicolò, A.; Jumaa, H. Isotype Specific Assembly of B Cell Antigen Receptors and Synergism With Chemokine Receptor CXCR4. Front. Immunol. 2018, 9, 2988. [Google Scholar] [CrossRef]
- Lockey, C.; Young, H.; Brown, J.; Dixon, A.M. Characterization of interactions within the Igα/Igβ transmembrane domains of the human B-cell receptor provides insights into receptor assembly. J. Biol. Chem. 2022, 298, 101843. [Google Scholar] [CrossRef] [PubMed]
- Wemlinger, S.M.; Parker Harp, C.R.; Yu, B.; Hardy, I.R.; Seefeldt, M.; Matsuda, J.; Mingueneau, M.; Spilker, K.A.; Cameron, T.O.; Larrick, J.W.; et al. Preclinical Analysis of Candidate Anti-Human CD79 Therapeutic Antibodies Using a Humanized CD79 Mouse Model. J. Immunol. 2022, 208, 1566–1584. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Wucherpfennig, K.; Patel, D.J. A structural platform for B cell receptor signaling. Cell Res. 2022, 33, 95–96. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, W.; Ma, J.; Xu, Z. A Novel and Validated 8-Pyroptosis-Related Genes Based Risk Prediction Model for Diffuse Large B Cell Lymphoma. Biomolecules 2022, 12, 1835. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Kläsener, K.; Iype, J.M.; Becker, M.; Maity, P.C.; Cavallari, M.; Nielsen, P.J.; Yang, J.; Reth, M. Continuous signaling of CD79b and CD19 is required for the fitness of Burkitt lymphoma B cells. EMBO J. 2018, 37, e97980. [Google Scholar] [CrossRef] [PubMed]
- Luger, D.; Yang, Y.A.; Raviv, A.; Weinberg, D.; Banerjee, S.; Lee, M.J.; Trepel, J.; Yang, L.; Wakefield, L.M. Expression of the B-cell receptor component CD79a on immature myeloid cells contributes to their tumor promoting effects. PLoS ONE 2013, 8, e76115. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.R. The immunobiology of ubiquitin-dependent B cell receptor functions. Mol. Immunol. 2018, 101, 146–154. [Google Scholar] [CrossRef]
- Corneth, O.B.J.; Neys, S.F.H.; Hendriks, R.W. Aberrant B Cell Signaling in Autoimmune Diseases. Cells 2022, 11, 3391. [Google Scholar] [CrossRef]
- Cyster, J.G.; Allen, C.D.C. B Cell Responses: Cell Interaction Dynamics and Decisions. Cell 2019, 177, 524–540. [Google Scholar] [CrossRef]
- Smith, L.D.; Minton, A.R.; Blunt, M.D.; Karydis, L.I.; Dutton, D.A.; Rogers-Broadway, K.R.; Dobson, R.; Liu, R.; Norster, F.; Hogg, E.; et al. BCR signaling contributes to autophagy regulation in chronic lymphocytic leukemia. Leukemia 2020, 34, 640–644. [Google Scholar] [CrossRef]
- Carter, M.J.; Cox, K.L.; Blakemore, S.J.; Bogdanov, Y.D.; Happo, L.; Scott, C.L.; Strasser, A.; Packham, G.K.; Cragg, M.S. BCR-signaling-induced cell death demonstrates dependency on multiple BH3-only proteins in a murine model of B-cell lymphoma. Cell Death Differ. 2016, 23, 303–312. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.K.; Getahun, A.; Gauld, S.B.; Merrell, K.T.; Tamir, I.; Smith, M.J.; Dal Porto, J.M.; Li, Q.Z.; Cambier, J.C. Monophosphorylation of CD79a and CD79b ITAM motifs initiates a SHIP-1 phosphatase-mediated inhibitory signaling cascade required for B cell anergy. Immunity 2011, 35, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Vuillier, F.; Dumas, G.; Magnac, C.; Prevost, M.C.; Lalanne, A.I.; Oppezzo, P.; Melanitou, E.; Dighiero, G.; Payelle-Brogard, B. Lower levels of surface B-cell-receptor expression in chronic lymphocytic leukemia are associated with glycosylation and folding defects of the mu and CD79a chains. Blood 2005, 105, 2933–2940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Veselits, M.; O’Neill, S.; Hou, P.; Reddi, A.L.; Berlin, I.; Ikeda, M.; Nash, P.D.; Longnecker, R.; Band, H.; et al. Ubiquitinylation of Ig beta dictates the endocytic fate of the B cell antigen receptor. J. Immunol. 2007, 179, 4435–4443. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Iwai, K. Roles of the NF-κB Pathway in B-Lymphocyte Biology. Curr. Top. Microbiol. Immunol. 2016, 393, 177–209. [Google Scholar] [CrossRef] [PubMed]
- Almaden, J.V.; Liu, Y.C.; Yang, E.; Otero, D.C.; Birnbaum, H.; Davis-Turak, J.; Asagiri, M.; David, M.; Goldrath, A.W.; Hoffmann, A. B-cell survival and development controlled by the coordination of NF-κB family members RelB and cRel. Blood 2016, 127, 1276–1286. [Google Scholar] [CrossRef]
- Szydłowski, M.; Jabłońska, E.; Juszczyński, P. FOXO1 transcription factor: A critical effector of the PI3K-AKT axis in B-cell development. Int. Rev. Immunol. 2014, 33, 146–157. [Google Scholar] [CrossRef]
- Chen, J.; Limon, J.J.; Blanc, C.; Peng, S.L.; Fruman, D.A. Foxo1 regulates marginal zone B-cell development. Eur. J. Immunol. 2010, 40, 1890–1896. [Google Scholar] [CrossRef]
- Sevdali, E.; Block, V.; Lataretu, M.; Li, H.; Smulski, C.R.; Briem, J.S.; Heitz, Y.; Fischer, B.; Ramirez, N.J.; Grimbacher, B.; et al. BAFFR activates PI3K/AKT signaling in human naive but not in switched memory B cells through direct interactions with B cell antigen receptors. Cell Rep. 2022, 39, 111019. [Google Scholar] [CrossRef]
- Lang, P.; Stolpa, J.C.; Freiberg, B.A.; Crawford, F.; Kappler, J.; Kupfer, A.; Cambier, J.C. TCR-induced transmembrane signaling by peptide/MHC class II via associated Ig-alpha/beta dimers. Science 2001, 291, 1537–1540. [Google Scholar] [CrossRef]
- Katikaneni, D.S.; Jin, L. B cell MHC class II signaling: A story of life and death. Hum. Immunol. 2019, 80, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Ichinose, S.; Hayashizaki, K.; Tsubata, T. Induction of autophagy by B cell antigen receptor stimulation and its inhibition by costimulation. Biochem. Biophys. Res. Commun. 2008, 374, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Arbogast, F.; Arnold, J.; Hammann, P.; Kuhn, L.; Chicher, J.; Murera, D.; Weishaar, J.; Muller, S.; Fauny, J.D.; Gros, F. ATG5 is required for B cell polarization and presentation of particulate antigens. Autophagy 2019, 15, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Caro-Maldonado, A.; Wang, R.; Nichols, A.G.; Kuraoka, M.; Milasta, S.; Sun, L.D.; Gavin, A.L.; Abel, E.D.; Kelsoe, G.; Green, D.R.; et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J. Immunol. 2014, 192, 3626–3636. [Google Scholar] [CrossRef] [PubMed]
- Jumaa, H.; Caganova, M.; McAllister, E.J.; Hoenig, L.; He, X.; Saltukoglu, D.; Brenker, K.; Köhler, M.; Leben, R.; Hauser, A.E.; et al. Immunoglobulin expression in the endoplasmic reticulum shapes the metabolic fitness of B lymphocytes. Life Sci. Alliance 2020, 3, e202000700. [Google Scholar] [CrossRef] [PubMed]
- Love, P.E.; Hayes, S.M. ITAM-mediated signaling by the T-cell antigen receptor. Cold Spring Harb. Perspect. Biol. 2010, 2, a002485. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, H.W., Jr.; Imboden, J.B.; Torres, R.M. Antigen Receptor Genes, Gene Products, and Coreceptors. In Clinical Immunology: Principles and Practice, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Xu, W.; Berning, P.; Lenz, G. Targeting B-cell receptor and PI3K signaling in diffuse large B-cell lymphoma. Blood 2021, 138, 1110–1119. [Google Scholar] [CrossRef]
- Roche, P.A.; Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015, 15, 203–216. [Google Scholar] [CrossRef]
- Busman-Sahay, K.; Drake, L.; Sitaram, A.; Marks, M.; Drake, J.R. Cis and trans regulatory mechanisms control AP2-mediated B cell receptor endocytosis via select tyrosine-based motifs. PLoS ONE 2013, 8, e54938. [Google Scholar] [CrossRef]
- Crute, B.W.; Sheraden, R.; Ott, V.L.; Harley, I.T.W.; Getahun, A.; Cambier, J.C. Inhibitory Receptor Trap: A Platform for Discovery of Inhibitory Receptors That Utilize Inositol Lipid and Phosphotyrosine Phosphatase Effectors. Front. Immunol. 2020, 11, 592329. [Google Scholar] [CrossRef]
- Franks, S.E.; Cambier, J.C. Putting on the Brakes: Regulatory Kinases and Phosphatases Maintaining B Cell Anergy. Front. Immunol. 2018, 9, 665. [Google Scholar] [CrossRef] [PubMed]
- Pao, L.I.; Famiglietti, S.J.; Cambier, J.C. Asymmetrical phosphorylation and function of immunoreceptor tyrosine-based activation motif tyrosines in B cell antigen receptor signal transduction. J. Immunol. 1998, 160, 3305–3314. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.R.; Campbell, K.S.; Kazlauskas, A.; Johnson, S.A.; Hertz, M.; Potter, T.A.; Pleiman, C.; Cambier, J.C. The B cell antigen receptor complex: Association of Ig-alpha and Ig-beta with distinct cytoplasmic effectors. Science 1992, 258, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Mkaddem, S.B.; Murua, A.; Flament, H.; Titeca-Beauport, D.; Bounaix, C.; Danelli, L.; Launay, P.; Benhamou, M.; Blank, U.; Daugas, E.; et al. Lyn and Fyn function as molecular switches that control immunoreceptors to direct homeostasis or inflammation. Nat. Commun. 2017, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.J.; Lyandres, J.R.; Panigrahi, A.K.; Prak, E.T.; DeFranco, A.L. Developmental acquisition of the Lyn-CD22-SHP-1 inhibitory pathway promotes B cell tolerance. J. Immunol. 2009, 182, 5382–5392. [Google Scholar] [CrossRef]
- Khan, N.; Rothstein, T.L. The Alternate Pathway for BCR Signaling Induced by IL-4 Requires Lyn Tyrosine Kinase. J. Mol. Biol. 2021, 433, 166667. [Google Scholar] [CrossRef]
- Kohlhas, V.; Hallek, M.; Nguyen, P.H. Constitutive activation of Lyn kinase enhances BCR responsiveness, but not the development of CLL in Eµ-TCL1 mice. Blood Adv. 2020, 4, 6106–6116. [Google Scholar] [CrossRef]
- Alsadeq, A.; Hobeika, E.; Medgyesi, D.; Kläsener, K.; Reth, M. The role of the Syk/Shp-1 kinase-phosphatase equilibrium in B cell development and signaling. J. Immunol. 2014, 193, 268–276. [Google Scholar] [CrossRef]
- Adachi, T.; Wienands, J.; Wakabayashi, C.; Yakura, H.; Reth, M.; Tsubata, T. SHP-1 requires inhibitory co-receptors to down-modulate B cell antigen receptor-mediated phosphorylation of cellular substrates. J. Biol. Chem. 2001, 276, 26648–26655. [Google Scholar] [CrossRef]
- Miyazaki, A.; Yogosawa, S.; Murakami, A.; Kitamura, D. Identification of CMTM7 as a transmembrane linker of BLNK and the B-cell receptor. PLoS ONE 2012, 7, e31829. [Google Scholar] [CrossRef]
- Patterson, H.C.; Kraus, M.; Kim, Y.M.; Ploegh, H.; Rajewsky, K. The B cell receptor promotes B cell activation and proliferation through a non-ITAM tyrosine in the Igalpha cytoplasmic domain. Immunity 2006, 25, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Kabak, S.; Skaggs, B.J.; Gold, M.R.; Affolter, M.; West, K.L.; Foster, M.S.; Siemasko, K.; Chan, A.C.; Aebersold, R.; Clark, M.R. The direct recruitment of BLNK to immunoglobulin alpha couples the B-cell antigen receptor to distal signaling pathways. Mol. Cell Biol. 2002, 22, 2524–2535. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Phelan, J.D.; Wright, G.W.; Häupl, B.; Huang, D.W.; Shaffer, A.L., 3rd; Young, R.M.; Wang, Z.; Zhao, H.; Yu, X.; et al. Regulation of B cell receptor-dependent NF-κB signaling by the tumor suppressor KLHL14. Proc. Natl. Acad. Sci. USA 2020, 117, 6092–6102. [Google Scholar] [CrossRef] [PubMed]
- Veselits, M.; Tanaka, A.; Chen, Y.; Hamel, K.; Mandal, M.; Kandasamy, M.; Manicassamy, B.; O’Neill, S.K.; Wilson, P.; Sciammas, R.; et al. Igβ ubiquitination activates PI3K signals required for endosomal sorting. J. Exp. Med. 2017, 214, 3775–3790. [Google Scholar] [CrossRef] [PubMed]
- Satpathy, S.; Wagner, S.A.; Beli, P.; Gupta, R.; Kristiansen, T.A.; Malinova, D.; Francavilla, C.; Tolar, P.; Bishop, G.A.; Hostager, B.S.; et al. Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol. Syst. Biol. 2015, 11, 810. [Google Scholar] [CrossRef] [PubMed]
- Drake, L.; McGovern-Brindisi, E.M.; Drake, J.R. BCR ubiquitination controls BCR-mediated antigen processing and presentation. Blood 2006, 108, 4086–4093. [Google Scholar] [CrossRef] [PubMed]
- Katkere, B.; Rosa, S.; Drake, J.R. The Syk-binding ubiquitin ligase c-Cbl mediates signaling-dependent B cell receptor ubiquitination and B cell receptor-mediated antigen processing and presentation. J. Biol. Chem. 2012, 287, 16636–16644. [Google Scholar] [CrossRef]
- Fearon, D.T.; Carroll, M.C. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu. Rev. Immunol. 2000, 18, 393–422. [Google Scholar] [CrossRef]
- Whillock, A.L.; Ybarra, T.K.; Bishop, G.A. TNF receptor-associated factor 3 restrains B-cell receptor signaling in normal and malignant B cells. J. Biol. Chem. 2021, 296, 100465. [Google Scholar] [CrossRef]
- Bishop, G.A.; Stunz, L.L.; Hostager, B.S. TRAF3 as a Multifaceted Regulator of B Lymphocyte Survival and Activation. Front. Immunol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.; Matsuzawa, A.; Zhang, W.; Tseng, P.H.; Keats, J.J.; Wang, H.; Vignali, D.A.; Bergsagel, P.L.; Karin, M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol. 2008, 9, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Mosquera Orgueira, A.; Ferreiro Ferro, R.; Díaz Arias, J.; Aliste Santos, C.; Antelo Rodríguez, B.; Bao Pérez, L.; Alonso Vence, N.; Bendaña López, Á.; Abuin Blanco, A.; Melero Valentín, P.; et al. Detection of new drivers of frequent B-cell lymphoid neoplasms using an integrated analysis of whole genomes. PLoS ONE 2021, 16, e0248886. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.H.; Young, R.M.; Schmitz, R.; Yang, Y.; Pittaluga, S.; Wright, G.; Lih, C.J.; Williams, P.M.; Shaffer, A.L.; Gerecitano, J.; et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat. Med. 2015, 21, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Visco, C.; Tanasi, I.; Quaglia, F.M.; Ferrarini, I.; Fraenza, C.; Krampera, M. Oncogenic Mutations of MYD88 and CD79B in Diffuse Large B-Cell Lymphoma and Implications for Clinical Practice. Cancers 2020, 12, 2913. [Google Scholar] [CrossRef] [PubMed]
- Grondona, P.; Bucher, P.; Schulze-Osthoff, K.; Hailfinger, S.; Schmitt, A. NF-κB Activation in Lymphoid Malignancies: Genetics, Signaling, and Targeted Therapy. Biomedicines 2018, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Medeiros, L.J.; Xu-Monette, Z.Y.; Li, J.; Young, K.H. Dysregulation of Cell Survival in Diffuse Large B Cell Lymphoma: Mechanisms and Therapeutic Targets. Front. Oncol. 2019, 9, 107. [Google Scholar] [CrossRef] [PubMed]
- Nagel, D.; Bognar, M.; Eitelhuber, A.C.; Kutzner, K.; Vincendeau, M.; Krappmann, D. Combinatorial BTK and MALT1 inhibition augments killing of CD79 mutant diffuse large B cell lymphoma. Oncotarget 2015, 6, 42232–42242. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Klein, U. NF-κB Mutations in Germinal Center B-Cell Lymphomas: Relation to NF-κB Function in Normal B Cells. Biomedicines 2022, 10, 2450. [Google Scholar] [CrossRef]
- Ducharme, O.; Beylot-Barry, M.; Pham-Ledard, A.; Bohers, E.; Viailly, P.J.; Bandres, T.; Faur, N.; Frison, E.; Vergier, B.; Jardin, F.; et al. Mutations of the B-Cell Receptor Pathway Confer Chemoresistance in Primary Cutaneous Diffuse Large B-Cell Lymphoma Leg Type. J. Investig. Dermatol. 2019, 139, 2334–2342.e8. [Google Scholar] [CrossRef]
- Myklebust, J.H.; Brody, J.; Kohrt, H.E.; Kolstad, A.; Czerwinski, D.K.; Wälchli, S.; Green, M.R.; Trøen, G.; Liestøl, K.; Beiske, K.; et al. Distinct patterns of B-cell receptor signaling in non-Hodgkin lymphomas identified by single-cell profiling. Blood 2017, 129, 759–770. [Google Scholar] [CrossRef]
- Thurner, L.; Hartmann, S.; Neumann, F.; Hoth, M.; Stilgenbauer, S.; Küppers, R.; Preuss, K.D.; Bewarder, M. Role of Specific B-Cell Receptor Antigens in Lymphomagenesis. Front. Oncol. 2020, 10, 604685. [Google Scholar] [CrossRef] [PubMed]
- Andrades, A.; Álvarez-Pérez, J.C.; Patiño-Mercau, J.R.; Cuadros, M.; Baliñas-Gavira, C.; Medina, P.P. Recurrent splice site mutations affect key diffuse large B-cell lymphoma genes. Blood 2022, 139, 2406–2410. [Google Scholar] [CrossRef] [PubMed]
- Young, R.M.; Wu, T.; Schmitz, R.; Dawood, M.; Xiao, W.; Phelan, J.D.; Xu, W.; Menard, L.; Meffre, E.; Chan, W.C.; et al. Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc. Natl. Acad. Sci. USA 2015, 112, 13447–13454. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Stojanov, P.; Lawrence, M.S.; Auclair, D.; Chapuy, B.; Sougnez, C.; Cruz-Gordillo, P.; Knoechel, B.; Asmann, Y.W.; Slager, S.L.; et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 3879–3884. [Google Scholar] [CrossRef] [PubMed]
- Payelle-Brogard, B.; Magnac, C.; Mauro, F.R.; Mandelli, F.; Dighiero, G. Analysis of the B-cell receptor B29 (CD79b) gene in familial chronic lymphocytic leukemia. Blood 1999, 94, 3516–3522. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Yamaguchi, M.; Kobayashi, K.; Miyazaki, K.; Tawara, I.; Imai, H.; Ono, R.; Nosaka, T.; Tanaka, K.; Katayama, N. MYD88, CD79B, and CARD11 gene mutations in CD5-positive diffuse large B-cell lymphoma. Cancer 2017, 123, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Bohers, E.; Mareschal, S.; Bouzelfen, A.; Marchand, V.; Ruminy, P.; Maingonnat, C.; Ménard, A.L.; Etancelin, P.; Bertrand, P.; Dubois, S.; et al. Targetable activating mutations are very frequent in GCB and ABC diffuse large B-cell lymphoma. Genes Chromosomes Cancer 2014, 53, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Poulain, S.; Roumier, C.; Galiègue-Zouitina, S.; Daudignon, A.; Herbaux, C.; Aiijou, R.; Lainelle, A.; Broucqsault, N.; Bertrand, E.; Manier, S.; et al. Genome wide SNP array identified multiple mechanisms of genetic changes in Waldenstrom macroglobulinemia. Am. J. Hematol. 2013, 88, 948–954. [Google Scholar] [CrossRef]
- Okosun, J.; Bödör, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014, 46, 176–181. [Google Scholar] [CrossRef]
- Krysiak, K.; Gomez, F.; White, B.S.; Matlock, M.; Miller, C.A.; Trani, L.; Fronick, C.C.; Fulton, R.S.; Kreisel, F.; Cashen, A.F.; et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood 2017, 129, 473–483. [Google Scholar] [CrossRef]
- Trøen, G.; Warsame, A.; Delabie, J. CD79B and MYD88 Mutations in Splenic Marginal Zone Lymphoma. ISRN Oncol. 2013, 2013, 252318. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.A.; Talley, J.A.; Do, H.N.; Kagan, H.L.; Kunkel, L.; Berenson, J.; Cooper, M.D.; Saxon, A.; Wall, R. Aberrations of the B-Cell Receptor B29 (CD79b) Gene in Chronic Lymphocytic Leukemia. Blood 1997, 90, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Cetin, G.O.; Baris, I.C.; Caner, V.; Sarikepe, B.; Sen Turk, N.; Tepeli, E.; Hacioglu, S.; Sari, I.; Bagci, G.; Keskin, A. Mutational status of EZH2 and CD79B hot spots in mature B-cell non-Hodgkin’s lymphomas: Novel CD79B variations have been revealed. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 830–836. [Google Scholar] [PubMed]
- Bruno, A.; Boisselier, B.; Labreche, K.; Marie, Y.; Polivka, M.; Jouvet, A.; Adam, C.; Figarella-Branger, D.; Miquel, C.; Eimer, S.; et al. Mutational analysis of primary central nervous system lymphoma. Oncotarget 2014, 5, 5065–5075. [Google Scholar] [CrossRef]
- Kim, Y.; Ju, H.; Kim, D.H.; Yoo, H.Y.; Kim, S.J.; Kim, W.S.; Ko, Y.H. CD79B and MYD88 mutations in diffuse large B-cell lymphoma. Hum. Pathol. 2014, 45, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Saieg, M.A.; Geddie, W.R.; Boerner, S.L.; Bailey, D.; Crump, M.; da Cunha Santos, G. EZH2 and CD79B mutational status over time in B-cell non-Hodgkin lymphomas detected by high-throughput sequencing using minimal samples. Cancer Cytopathol. 2013, 121, 377–386. [Google Scholar] [CrossRef]
- Hunter, Z.R.; Xu, L.; Yang, G.; Zhou, Y.; Liu, X.; Cao, Y.; Manning, R.J.; Tripsas, C.; Patterson, C.J.; Sheehy, P.; et al. The genomic landscape of Waldenström macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood 2014, 123, 1637–1646. [Google Scholar] [CrossRef]
- Alfarano, A.; Indraccolo, S.; Circosta, P.; Minuzzo, S.; Vallario, A.; Zamarchi, R.; Fregonese, A.; Calderazzo, F.; Faldella, A.; Aragno, M.; et al. An alternatively spliced form of CD79b gene may account for altered B-cell receptor expression in B-chronic lymphocytic leukemia. Blood 1999, 93, 2327–2335. [Google Scholar] [CrossRef]
- Jiménez, C.; Alonso-Álvarez, S.; Alcoceba, M.; Ordóñez, G.R.; García-Álvarez, M.; Prieto-Conde, M.I.; Chillón, M.C.; Balanzategui, A.; Corral, R.; Marín, L.A.; et al. From Waldenström’s macroglobulinemia to aggressive diffuse large B-cell lymphoma: A whole-exome analysis of abnormalities leading to transformation. Blood Cancer J. 2017, 7, e591. [Google Scholar] [CrossRef]
- Wang, J.Q.; Jeelall, Y.S.; Humburg, P.; Batchelor, E.L.; Kaya, S.M.; Yoo, H.M.; Goodnow, C.C.; Horikawa, K. Synergistic cooperation and crosstalk between MYD88(L265P) and mutations that dysregulate CD79B and surface IgM. J. Exp. Med. 2017, 214, 2759–2776. [Google Scholar] [CrossRef]
- Gordon, M.S.; Kato, R.M.; Lansigan, F.; Thompson, A.A.; Wall, R.; Rawlings, D.J. Aberrant B cell receptor signaling from B29 (Igbeta, CD79b) gene mutations of chronic lymphocytic leukemia B cells. Proc. Natl. Acad. Sci. USA 2000, 97, 5504–5509. [Google Scholar] [CrossRef] [PubMed]
- Faumont, N.; Taoui, O.; Collares, D.; Jais, J.P.; Leroy, K.; Prévaud, L.; Jardin, F.; Molina, T.J.; Copie-Bergman, C.; Petit, B.; et al. c-Rel Is the Pivotal NF-κB Subunit in Germinal Center Diffuse Large B-Cell Lymphoma: A LYSA Study. Front. Oncol. 2021, 11, 638897. [Google Scholar] [CrossRef] [PubMed]
- Compagno, M.; Lim, W.K.; Grunn, A.; Nandula, S.V.; Brahmachary, M.; Shen, Q.; Bertoni, F.; Ponzoni, M.; Scandurra, M.; Califano, A.; et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009, 459, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.E.; Brown, K.D.; Siebenlist, U.; Staudt, L.M. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 2001, 194, 1861–1874. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.Y.; Bauman, B.M.; Arjunaraja, S.; Dorjbal, B.; Milner, J.D.; Snow, A.L.; Turvey, S.E. The CBM-opathies-A Rapidly Expanding Spectrum of Human Inborn Errors of Immunity Caused by Mutations in the CARD11-BCL10-MALT1 Complex. Front. Immunol. 2018, 9, 2078. [Google Scholar] [CrossRef] [PubMed]
- Turvey, S.E.; Durandy, A.; Fischer, A.; Fung, S.Y.; Geha, R.S.; Gewies, A.; Giese, T.; Greil, J.; Keller, B.; McKinnon, M.L.; et al. The CARD11-BCL10-MALT1 (CBM) signalosome complex: Stepping into the limelight of human primary immunodeficiency. J. Allergy Clin. Immunol. 2014, 134, 276–284. [Google Scholar] [CrossRef]
- Kloo, B.; Nagel, D.; Pfeifer, M.; Grau, M.; Düwel, M.; Vincendeau, M.; Dörken, B.; Lenz, P.; Lenz, G.; Krappmann, D. Critical role of PI3K signaling for NF-kappaB-dependent survival in a subset of activated B-cell-like diffuse large B-cell lymphoma cells. Proc. Natl. Acad. Sci. USA 2011, 108, 272–277. [Google Scholar] [CrossRef]
- Kim, A.; Seong, K.M.; Kang, H.J.; Park, S.; Lee, S.S. Inhibition of Lyn is a promising treatment for mantle cell lymphoma with bortezomib resistance. Oncotarget 2015, 6, 38225–38238. [Google Scholar] [CrossRef]
- Ezell, S.A.; Wang, S.; Bihani, T.; Lai, Z.; Grosskurth, S.E.; Tepsuporn, S.; Davies, B.R.; Huszar, D.; Byth, K.F. Differential regulation of mTOR signaling determines sensitivity to AKT inhibition in diffuse large B cell lymphoma. Oncotarget 2016, 7, 9163–9174. [Google Scholar] [CrossRef]
- Majchrzak, A.; Witkowska, M.; Smolewski, P. Inhibition of the PI3K/Akt/mTOR signaling pathway in diffuse large B-cell lymphoma: Current knowledge and clinical significance. Molecules 2014, 19, 14304–14315. [Google Scholar] [CrossRef]
- Ma, M.C.J.; Tadros, S.; Bouska, A.; Heavican, T.; Yang, H.; Deng, Q.; Moore, D.; Akhter, A.; Hartert, K.; Jain, N.; et al. Subtype-specific and co-occurring genetic alterations in B-cell non-Hodgkin lymphoma. Haematologica 2022, 107, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.P.; Shen, R.; Shi, Z.Y.; Cheng, S.; Wang, L.; Liu, Y.; Zhang, L.; Huang, R.; Ma, X.; Wu, X.; et al. The Prognostic Significance of CD79B Mutation in Diffuse Large B-Cell Lymphoma: A Meta-analysis and Systematic Literature Review. Clin. Lymphoma Myeloma Leuk. 2022, 22, e1051–e1058.e1. [Google Scholar] [CrossRef] [PubMed]
- de Groen, R.A.L.; Schrader, A.M.R.; Kersten, M.J.; Pals, S.T.; Vermaat, J.S.P. MYD88 in the driver’s seat of B-cell lymphomagenesis: From molecular mechanisms to clinical implications. Haematologica 2019, 104, 2337–2348. [Google Scholar] [CrossRef] [PubMed]
- Sewastianik, T.; Guerrera, M.L.; Adler, K.; Dennis, P.S.; Wright, K.; Shanmugam, V.; Huang, Y.; Tanton, H.; Jiang, M.; Kofides, A.; et al. Human MYD88L265P is insufficient by itself to drive neoplastic transformation in mature mouse B cells. Blood Adv. 2019, 3, 3360–3374. [Google Scholar] [CrossRef] [PubMed]
- Alcoceba, M.; García-Álvarez, M.; Medina, A.; Maldonado, R.; González-Calle, V.; Chillón, M.C.; Sarasquete, M.E.; González, M.; García-Sanz, R.; Jiménez, C. MYD88 Mutations: Transforming the Landscape of IgM Monoclonal Gammopathies. Int. J. Mol. Sci. 2022, 23, 5570. [Google Scholar] [CrossRef] [PubMed]
- Motshwene, P.G.; Moncrieffe, M.C.; Grossmann, J.G.; Kao, C.; Ayaluru, M.; Sandercock, A.M.; Robinson, C.V.; Latz, E.; Gay, N.J. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 2009, 284, 25404–25411. [Google Scholar] [CrossRef] [PubMed]
- Balka, K.R.; De Nardo, D. Understanding early TLR signaling through the Myddosome. J. Leukoc. Biol. 2019, 105, 339–351. [Google Scholar] [CrossRef]
- Cao, F.; Deliz-Aguirre, R.; Gerpott, F.H.; Ziska, E.; Taylor, M.J. Myddosome clustering in IL-1 receptor signaling regulates the formation of an NF-κB activating signalosome. EMBO Rep. 2023, 24, e57233. [Google Scholar] [CrossRef]
- De Nardo, D.; Balka, K.R.; Cardona Gloria, Y.; Rao, V.R.; Latz, E.; Masters, S.L. Interleukin-1 receptor-associated kinase 4 (IRAK4) plays a dual role in myddosome formation and Toll-like receptor signaling. J. Biol. Chem. 2018, 293, 15195–15207. [Google Scholar] [CrossRef]
- Weber, T.; Schmitz, R. Molecular Subgroups of Diffuse Large B Cell Lymphoma: Biology and Implications for Clinical Practice. Curr. Oncol. Rep. 2022, 24, 13–21. [Google Scholar] [CrossRef]
- Roschewski, M.; Phelan, J.D.; Wilson, W.H. Molecular Classification and Treatment of Diffuse Large B-Cell Lymphoma and Primary Mediastinal B-Cell Lymphoma. Cancer J. 2020, 26, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Fornecker, L.M.; Muller, L.; Bertrand, F.; Paul, N.; Pichot, A.; Herbrecht, R.; Chenard, M.P.; Mauvieux, L.; Vallat, L.; Bahram, S.; et al. Multi-omics dataset to decipher the complexity of drug resistance in diffuse large B-cell lymphoma. Sci. Rep. 2019, 9, 895. [Google Scholar] [CrossRef] [PubMed]
- George, B.; Chowdhury, S.M.; Hart, A.; Sircar, A.; Singh, S.K.; Nath, U.K.; Mamgain, M.; Singhal, N.K.; Sehgal, L.; Jain, N. Ibrutinib Resistance Mechanisms and Treatment Strategies for B-Cell lymphomas. Cancers 2020, 12, 1328. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, W.S.; Ryu, K.; Kim, S.J.; Park, C. CD79B limits response of diffuse large B cell lymphoma to ibrutinib. Leuk. Lymphoma 2016, 57, 1413–1422. [Google Scholar] [CrossRef] [PubMed]
- Rip, J.; de Bruijn, M.J.W.; Neys, S.F.H.; Singh, S.P.; Willar, J.; van Hulst, J.A.C.; Hendriks, R.W.; Corneth, O.B.J. Bruton’s tyrosine kinase inhibition induces rewiring of proximal and distal B-cell receptor signaling in mice. Eur. J. Immunol. 2021, 51, 2251–2265. [Google Scholar] [CrossRef]
- Domka, K.; Goral, A.; Firczuk, M. cROSsing the Line: Between Beneficial and Harmful Effects of Reactive Oxygen Species in B-Cell Malignancies. Front. Immunol. 2020, 11, 1538. [Google Scholar] [CrossRef]
- Wang, S.S.; Davis, S.; Cerhan, J.R.; Hartge, P.; Severson, R.K.; Cozen, W.; Lan, Q.; Welch, R.; Chanock, S.J.; Rothman, N. Polymorphisms in oxidative stress genes and risk for non-Hodgkin lymphoma. Carcinogenesis 2006, 27, 1828–1834. [Google Scholar] [CrossRef]
- Müller-Winkler, J.; Mitter, R.; Rappe, J.C.F.; Vanes, L.; Schweighoffer, E.; Mohammadi, H.; Wack, A.; Tybulewicz, V.L.J. Critical requirement for BCR, BAFF, and BAFFR in memory B cell survival. J. Exp. Med. 2021, 218, e20191393. [Google Scholar] [CrossRef]


| Frequency of Cases with Any CD79A ITAM Mutation | CD79A Mutation Subtypes | Frequency of Cases with CD79A Mutation Subtypes * | Signaling Consequences of CD79A Mutation Subtypes |
|---|---|---|---|
| 6% in DLBCL [124] 5% in unclassified DLBCL [125] 2.5% in CD5+ DLBCL [138] 0.7% in DLBCL [58] 6.5% in ABC DLBCL [139] 2.9% in ABC DLBCL [48] 0% in ABC DLBCL [125] 0% in GCB DLBCL [139] 6.3% in PCLBCL-LT [131] 5.5% in WM [140] 3% in FL [141] 3% in FL [124] 0% in FL [142] 0.8% in CLL [124] 0% in BL [124] 0% in SMZL [143] 0% in MZL [143] | CD79A ITAM Y188 point mutations | Not detected in multiple studies of DLBCL, FL, and other lymphoid malignancies | Inhibition of tonic BCR signaling, based on a cell line study [50] |
| CD79A ITAM Y199 point mutations | 1.8% in WM [140] | Not reported | |
| Complete ITAM deletions | 1.5% in ABC DLBCL [48] | Increased surface BCR expression [125] Chronic active BCR signal enhancement [139] | |
| Other CD79A mutations affecting ITAM (e.g., deletions, truncations, frameshift, or splice site mutations) | 6% in PCLBCL-LT (ITAM deletions affecting Y188) [131] 6.5% in ABC DLBCL [139] 1.3% in DLBCLs (deletions affecting Y188) [125] 2.9% in ABC DLBCL(splice site mutations) [48] 1.8% in WM (deletion) [140] | Increased surface BCR expression [125] Chronic active BCR signaling enhancement [139] |
| Frequency of Cases with Any CD79B ITAM Mutation | CD79B Mutation Subtypes | Frequency of Cases with CD79B Mutation Subtypes * | Signaling Consequences of CD79B Mutation Subtypes |
|---|---|---|---|
| 65% in CLL [137,144] 56.3% in PCLBCL-LT [131] 33% in MZL [145] 0% in MZL [143] 30% in PCNSL [146] 31% in DLBCL [145] 38% in CD5+ DLBCLs [138] 26.8% in ABC DLBCL [57] 21.1% in ABC DLBCL [48] 10.8% in ABC DLBCL [147] 9% in ABC DLBCL [139] 23% in DLBCL [125] 14.5% in DLBCL [136] 14.3% in DLBCL [54] 14% in DLBCL [58] 12.9% in DLBCL [124] 8.5% in DLBCL [147] 8% in DLBCL [148] 5.6% in DLBCL [139] 5.1% in GCB DLBCL [147] 3.1% in GCB DLBCL [48] 1.9% in GCB DLBCL [57] 1.6% in GCB DLBCL [139] 10% in SMZL [143] 9% in WM [140] 7% in WM [149] 9% in FL [141] 5% in FL [124] 4.9% in FL [142] 0% in FL [148] 0% in FL [145] 0% in CLL [124] 0% in CLL [150] 0% in BL [48,124] 0%in MCL [148] 0% in MCL [145] 0% in marginal zone/MALT lymphoma [148] 0% gastric MALT lymphoma [48] 0% in SLL [148] 0% in CLL/SLL [145] 0% in NMZL [145] | CD79B ITAM Y196 point mutations | 75% in transformed WM [151] 53.1% in PCLBCL-LT [131] 30% in PCNSL [146] 35% in CD5+ DLBCL [138] 18% in ABC DLBCL [48] 1.6% in GCB DLBCL [48] 3.7% in DLBCL [139] 10% in SMZL [143] 9% in WM [140] 8% in DLBCL [148] 3% in FL [141] 7.5% in DLBCL [147] | Surface BCR expression upregulation via inhibition of BCR internalization [48,152] Reduced BCR signaling negative regulation via decreased Lyn binding [48] Enhancement of BCR clustering [3,50] Active BCR signaling enhancement [48] NF-κB activation enhancement [48] |
| CD79B ITAM Y207 point mutations | 4% in CLL [144,153] | Surface BCR expression upregulation [152] Impairment of antigen induced BCR signaling [144,153] | |
| Other CD79B mutations affecting ITAM (e.g., deletions, truncations, frameshift, splice site/intron retention mutations) | 0.7% in ABC DLBCLs (Y196 deletions) [48] 0.6% in DLBCL (deletions affecting Y207) [139] 0.5% in DLBCL (E197stop) [147] 17% in B-CLL (deletions) [144] 1.8% in DLBCL (splice site mutations and intron retention) [134] | Similar consequences as Y196 mutations | |
| CD79B ITAM mutations not affecting ITAM tyrosines | 2.5% in CD5+ DLBCL (deletion in ITAM region before the first tyrosine) [138] 1.6% in GCB DLBCLs (L199Q) [48] 6% in FL (point mutations in ITAM) [141] 3.1% in PCLBCL-LT (E197D point mutation) [131] 31% in DLBCL [145] 1.2% in DLBCLs [139] 12.7% in DLBCL (point mutations and one frameshift) [136] 25% in CLL [137] | Not reported | |
| Non-ITAM mutations | Over 30% in CLL [137] 17% in CLL [144] 1.8% in DLBCL [136] | Decreased BCR expression and signaling in CLL [153] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tkachenko, A.; Kupcova, K.; Havranek, O. B-Cell Receptor Signaling and Beyond: The Role of Igα (CD79a)/Igβ (CD79b) in Normal and Malignant B Cells. Int. J. Mol. Sci. 2024, 25, 10. https://doi.org/10.3390/ijms25010010
Tkachenko A, Kupcova K, Havranek O. B-Cell Receptor Signaling and Beyond: The Role of Igα (CD79a)/Igβ (CD79b) in Normal and Malignant B Cells. International Journal of Molecular Sciences. 2024; 25(1):10. https://doi.org/10.3390/ijms25010010
Chicago/Turabian StyleTkachenko, Anton, Kristyna Kupcova, and Ondrej Havranek. 2024. "B-Cell Receptor Signaling and Beyond: The Role of Igα (CD79a)/Igβ (CD79b) in Normal and Malignant B Cells" International Journal of Molecular Sciences 25, no. 1: 10. https://doi.org/10.3390/ijms25010010
APA StyleTkachenko, A., Kupcova, K., & Havranek, O. (2024). B-Cell Receptor Signaling and Beyond: The Role of Igα (CD79a)/Igβ (CD79b) in Normal and Malignant B Cells. International Journal of Molecular Sciences, 25(1), 10. https://doi.org/10.3390/ijms25010010







