Role of Single-Nucleotide Polymorphisms in Genes Implicated in Capecitabine Pharmacodynamics on the Effectiveness of Adjuvant Therapy in Colorectal Cancer
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
2.2. Genotype Frequency and Distribution
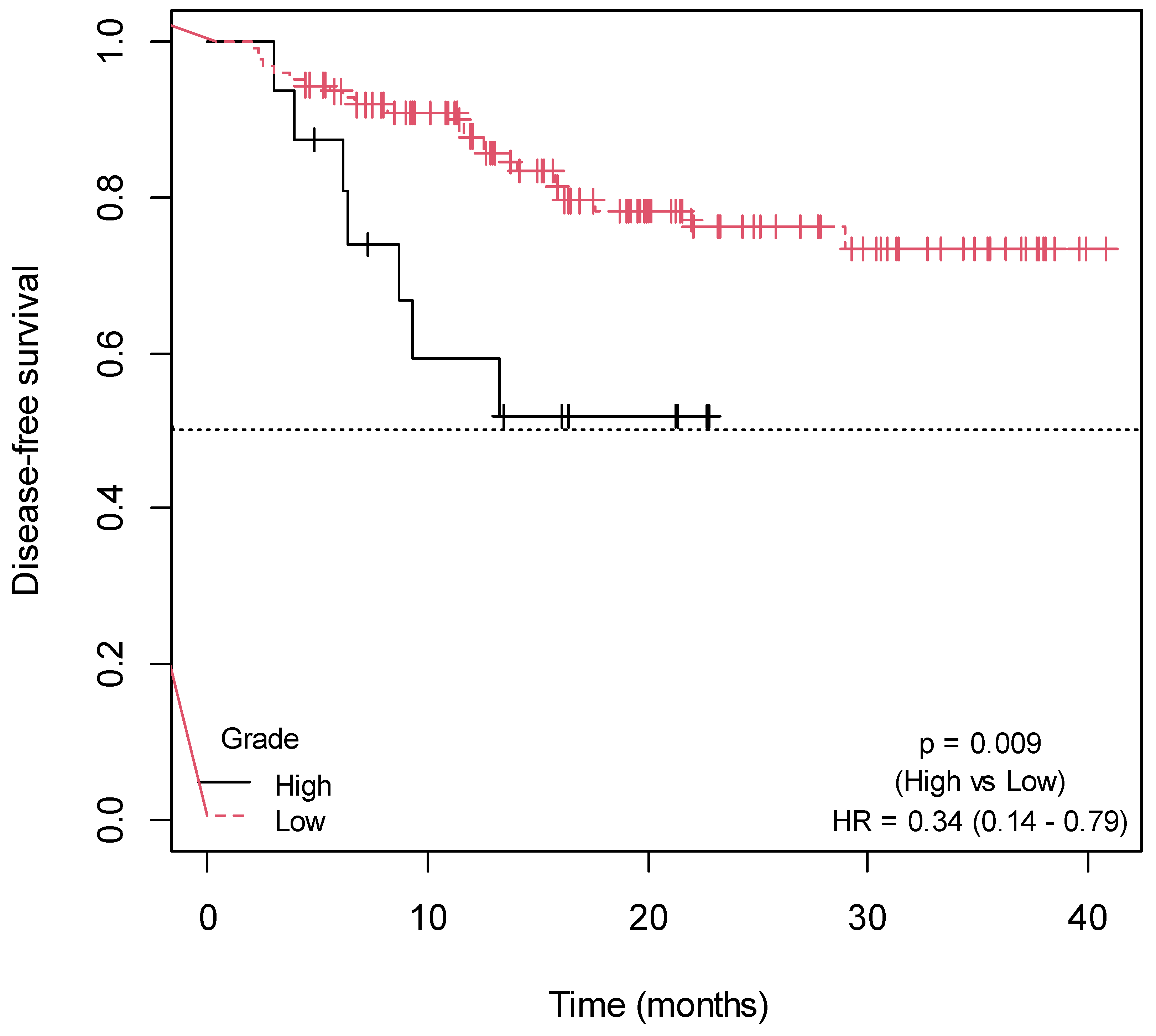
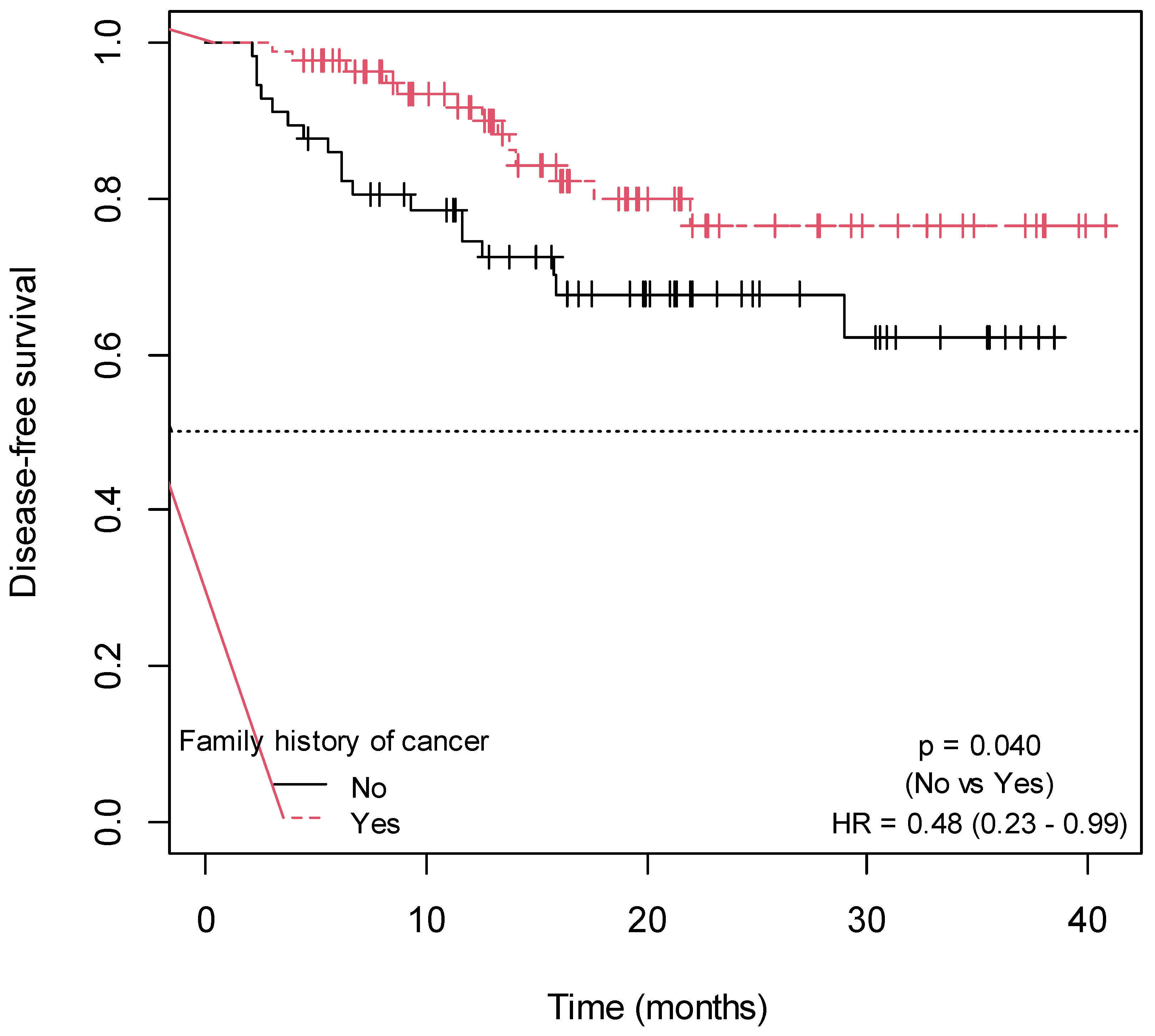
2.3. Influence of Non-Genetic Factors on Disease-Free Survival
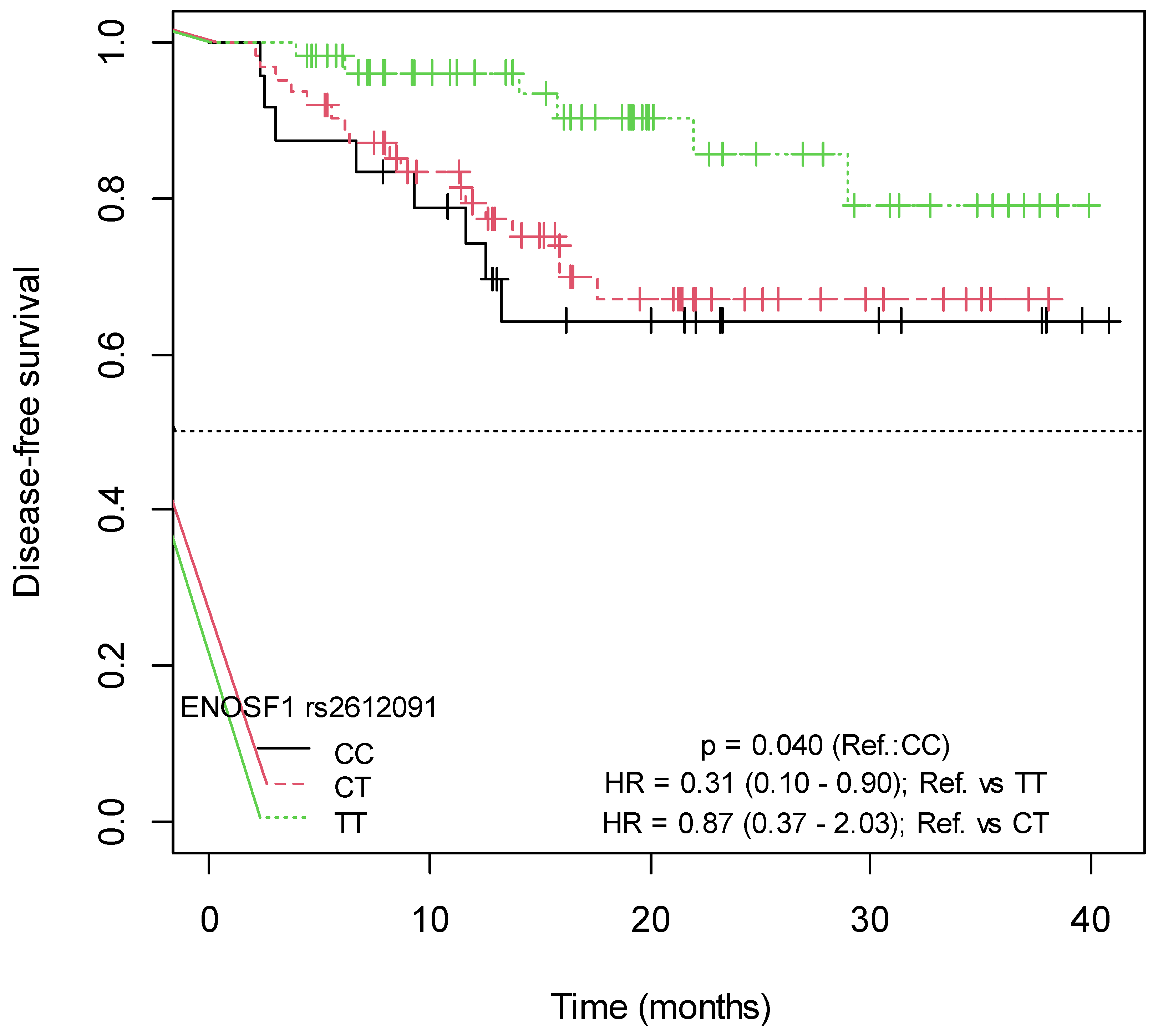
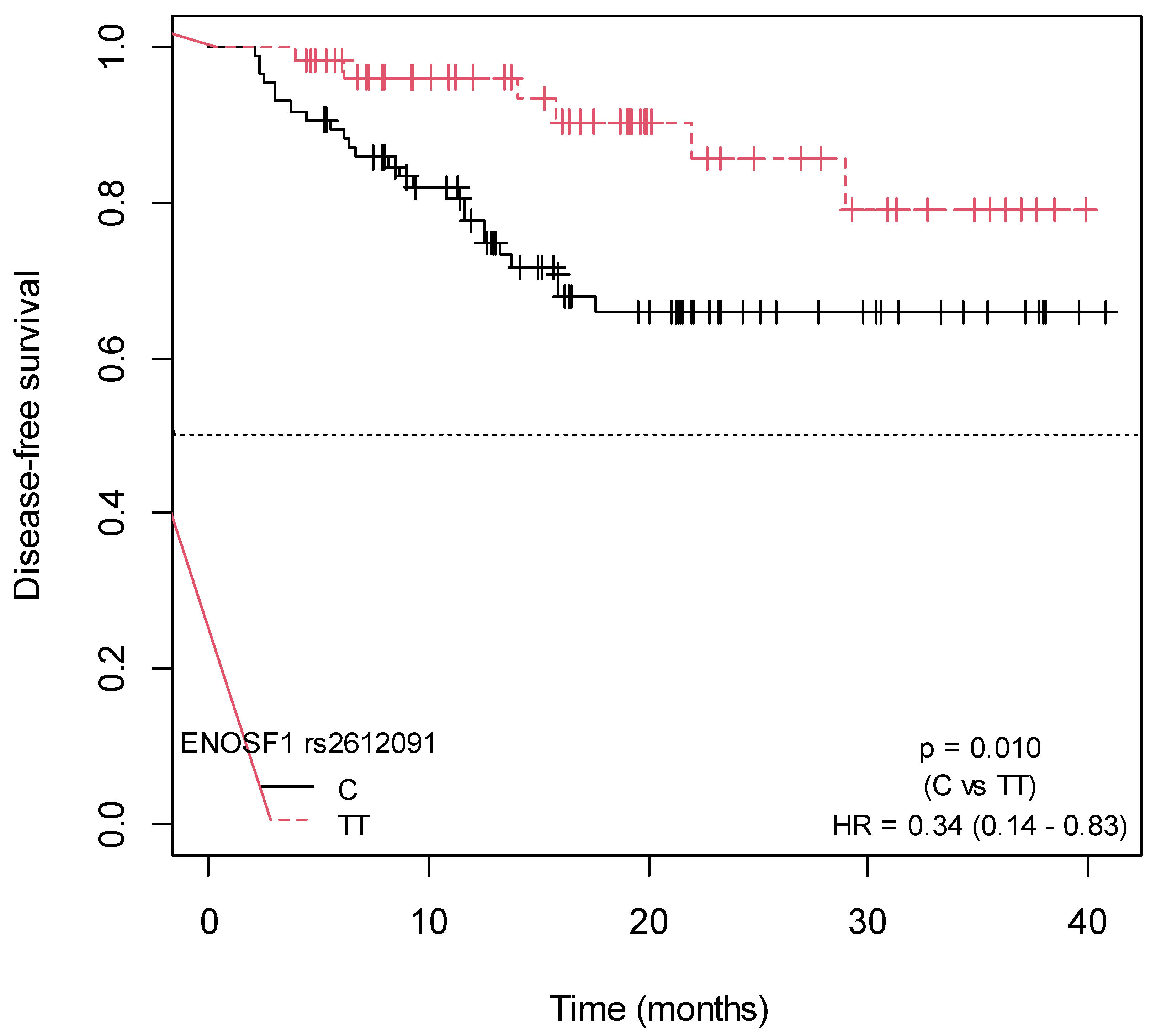
2.4. Influence of SNPs in Capecitabine Pharmacodynamics on Disease-Free Survival
3. Discussion
4. Materials and Methods
4.1. Design and Settings
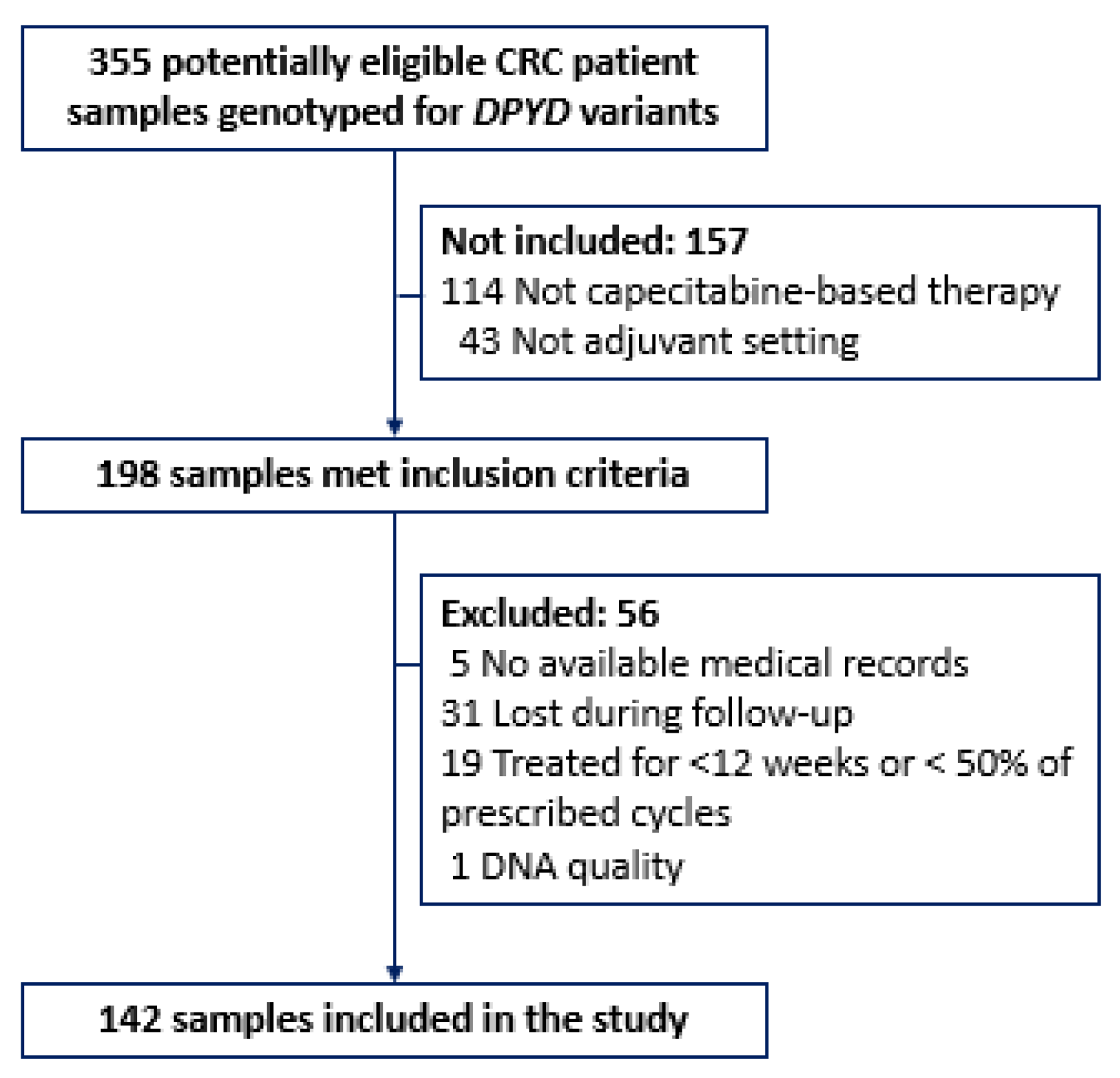
4.2. Study Population
4.3. Variables
4.3.1. Endpoints
4.3.2. Sociodemographic and Clinical Data
4.3.3. Genotyping Data
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Recio-Boiles, A.; Cagir, B. Colon Cancer. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Recio-Boiles, A.; Kashyap, S.; Tsoris, A.; Babiker, H.M. Rectal Cancer. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Gelibter, A.J.; Caponnetto, S.; Urbano, F.; Emiliani, A.; Scagnoli, S.; Sirgiovanni, G.; Napoli, V.M.; Cortesi, E. Adjuvant Chemotherapy in Resected Colon Cancer: When, How and How Long? Surg. Oncol. 2019, 30, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Bertholee, D.; Maring, J.G.; van Kuilenburg, A.B.P. Genotypes Affecting the Pharmacokinetics of Anticancer Drugs. Clin. Pharmacokinet 2017, 56, 317–337. [Google Scholar] [CrossRef] [PubMed]
- Mroziewicz, M.; Tyndale, R.F. Pharmacogenetics: A Tool for Identifying Genetic Factors in Drug Dependence and Response to Treatment. Addict Sci. Clin. Pract. 2010, 5, 17–29. [Google Scholar] [PubMed]
- Cura, Y.; Pérez-Ramírez, C.; Sánchez-Martín, A.; Membrive-Jimenez, C.; Valverde-Merino, M.I.; González-Flores, E.; Morales, A.J. Influence of Single-Nucleotide Polymorphisms on Clinical Outcomes of Capecitabine-Based Chemotherapy in Colorectal Cancer Patients: A Systematic Review. Cancers 2023, 15, 1821. [Google Scholar] [CrossRef] [PubMed]
- Thorn, C.F.; Marsh, S.; Carrillo, M.W.; McLeod, H.L.; Klein, T.E.; Altman, R.B. PharmGKB Summary: Fluoropyrimidine Pathways. Pharmacogenet. Genom. 2011, 21, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Ruzzo, A.; Graziano, F.; Loupakis, F.; Santini, D.; Catalano, V.; Bisonni, R.; Ficarelli, R.; Fontana, A.; Andreoni, F.; Falcone, A.; et al. Pharmacogenetic Profiling in Patients with Advanced Colorectal Cancer Treated with First-Line FOLFIRI Chemotherapy. Pharmacogenomics J. 2008, 8, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Grimaldi, M.-C.; Milano, G.; Maindrault-Goebel, F.; Chibaudel, B.; Formento, J.-L.; Francoual, M.; Lledo, G.; André, T.; Mabro, M.; Mineur, L.; et al. Methylenetetrahydrofolate Reductase (MTHFR) Gene Polymorphisms and FOLFOX Response in Colorectal Cancer Patients. Br. J. Clin. Pharmacol. 2010, 69, 58–66. [Google Scholar] [CrossRef]
- Huang, M.-Y.; Huang, M.-L.; Chen, M.-J.; Lu, C.-Y.; Chen, C.-F.; Tsai, P.-C.; Chuang, S.-C.; Hou, M.-F.; Lin, S.-R.; Wang, J.-Y. Multiple Genetic Polymorphisms in the Prediction of Clinical Outcome of Metastatic Colorectal Cancer Patients Treated with First-Line FOLFOX-4 Chemotherapy. Pharmacogenet Genom. 2011, 21, 18–25. [Google Scholar] [CrossRef]
- Li, M.; Zhao, Y.; Zhao, E.; Wang, K.; Lu, W.; Yuan, L. Predictive Value of Two Polymorphisms of ERCC2, Rs13181 and Rs1799793, in Clinical Outcomes of Chemotherapy in Gastric Cancer Patients: A Meta-Analysis. Disease Markers 2018, 2018, 3947626. [Google Scholar] [CrossRef]
- Yousef, A.-M.; Zawiah, M.; Al-Yacoub, S.; Kadi, T.; Tantawi, D.A.; Al-Ramadhani, H. The Association of Polymorphisms in Folate-Metabolizing Genes with Response to Adjuvant Chemotherapy of Colorectal Cancer. Cancer Chemother. Pharmacol. 2018, 82, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Boldrin, E.; Malacrida, S.; Rumiato, E.; Battaglia, G.; Ruol, A.; Amadori, A.; Saggioro, D. Association Between ERCC1 Rs3212986 and ERCC2/XPD Rs1799793 and OS in Patients with Advanced Esophageal Cancer. Front. Oncol. 2019, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Boige, V.; Mollevi, C.; Gourgou, S.; Azria, D.; Seitz, J.-F.; Vincent, M.; Bigot, L.; Juzyna, B.; Miran, I.; Gerard, J.-P.; et al. Impact of Single-Nucleotide Polymorphisms in DNA Repair Pathway Genes on Response to Chemoradiotherapy in Rectal Cancer Patients: Results from ACCORD-12/PRODIGE-2 Phase III Trial. Int. J. Cancer 2019, 145, 3163–3172. [Google Scholar] [CrossRef] [PubMed]
- Salimzadeh, H.; Lindskog, E.B.; Gustavsson, B.; Wettergren, Y.; Ljungman, D. Association of DNA Repair Gene Variants with Colorectal Cancer: Risk, Toxicity, and Survival. BMC Cancer 2020, 20, 409. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.-J.; Cho, S.-H.; Kim, E.-J.; Ryu, C.-S.; Park, H.-S.; Kim, J.-W.; Lee, J.-Y.; An, H.-J.; Kim, N.-K. 3’-UTR Polymorphisms in Thymidylate Synthase with Colorectal Cancer Prevalence and Prognosis. J. Pers. Med. 2021, 11, 537. [Google Scholar] [CrossRef] [PubMed]
- Arjmandi, K.; Ameli, F.; Salahshourifar, I.; Esfandbod, M.; Irani, S. Evaluation of TS and ENOSF1 Variants as a Biomarker in Response to Neoadjuvant Chemotherapy Based on 5FU in Gastric Cancer Patients. Asian Pac. J. Cancer Prev. 2022, 23, 2983–2989. [Google Scholar] [CrossRef]
- Pesola, F.; Eloranta, S.; Martling, A.; Saraste, D.; Smedby, K.E. Family History of Colorectal Cancer and Survival: A Swedish Population-Based Study. J. Intern. Med. 2020, 287, 723–733. [Google Scholar] [CrossRef]
- Oh, M.G.; Kim, J.H.; Han, M.A.; Park, J.; Ryu, S.Y.; Choi, S.W. Family History and Survival of Patients with Gastric Cancer: A Meta-Analysis. Asian Pac. J. Cancer Prev. 2014, 15, 3465–3470. [Google Scholar] [CrossRef]
- Brook, M.N.; Ní Raghallaigh, H.; Govindasami, K.; Dadaev, T.; Rageevakumar, R.; Keating, D.; Hussain, N.; Osborne, A.; Lophatananon, A.; UKGPCS Collaborators; et al. Family History of Prostate Cancer and Survival Outcomes in the UK Genetic Prostate Cancer Study. Eur. Urol. 2023, 83, 257–266. [Google Scholar] [CrossRef]
- Barresi, V.; Reggiani Bonetti, L.; Ieni, A.; Caruso, R.A.; Tuccari, G. Histological Grading in Colorectal Cancer: New Insights and Perspectives. Histol. Histopathol. 2015, 30, 1059–1067. [Google Scholar] [CrossRef]
- Sagaert, X.; Vanstapel, A.; Verbeek, S. Tumor Heterogeneity in Colorectal Cancer: What Do We Know So Far? Pathobiology 2018, 85, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Johncilla, M.; Yantiss, R.K. Histology of Colorectal Carcinoma. Surg. Pathol. Clin. 2020, 13, 503–520. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhang, G.; Bao, H.; Ren, J.; Wang, Z.; Zhang, J.; Zhao, Z.; Yan, X.; Chai, Y.; Zhou, J. Development of a Dual-Energy Spectral Computed Tomography-Based Nomogram for the Preoperative Discrimination of Histological Grade in Colorectal Adenocarcinoma Patients. J. Gastrointest. Oncol. 2021, 12, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Zou, G.; Wang, R.; Wang, M. Clinical Response and Prognostic Significance of Serum miR-497 Expression in Colorectal Cancer. Cancer Biomark 2019, 25, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Cheng, Z.; Huang, Y.; Liang, C.; He, L.; Ma, Z.; Chen, X.; Wu, X.; Li, Y.; Liang, C.; et al. CT-Based Radiomics Signature to Discriminate High-Grade from Low-Grade Colorectal Adenocarcinoma. Acad Radiol. 2018, 25, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Mehrkhani, F.; Nasiri, S.; Donboli, K.; Meysamie, A.; Hedayat, A. Prognostic Factors in Survival of Colorectal Cancer Patients after Surgery. Color. Dis. 2009, 11, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.B.; Chun, H.-K.; Yun, H.R.; Kim, H.C.; Yun, S.H.; Lee, W.Y. Histological Grade Predicts Survival Time Associated with Recurrence after Resection for Colorectal Cancer. Hepatogastroenterology 2009, 56, 1335–1340. [Google Scholar]
- van den Berg, I.; Coebergh van den Braak, R.R.J.; van Vugt, J.L.A.; Ijzermans, J.N.M.; Buettner, S. Actual Survival after Resection of Primary Colorectal Cancer: Results from a Prospective Multicenter Study. World J. Surg. Oncol. 2021, 19, 96. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, Y.; Xu, G.; Baklaushev, V.P.; Chekhonin, V.P.; Peltzer, K.; Ma, W.; Wang, X.; Wang, G.; Zhang, C. Nomogram for Predicting Overall Survival in Colorectal Cancer with Distant Metastasis. BMC Gastroenterol. 2021, 21, 103. [Google Scholar] [CrossRef]
- Hamzic, S.; Kummer, D.; Froehlich, T.K.; Joerger, M.; Aebi, S.; Palles, C.; Thomlinson, I.; Meulendijks, D.; Schellens, J.H.M.; García-González, X.; et al. Evaluating the Role of ENOSF1 and TYMS Variants as Predictors in Fluoropyrimidine-Related Toxicities: An IPD Meta-Analysis. Pharmacol. Res. 2020, 152, 104594. [Google Scholar] [CrossRef]
- Wichelecki, D.J.; Froese, D.S.; Kopec, J.; Muniz, J.R.C.; Yue, W.W.; Gerlt, J.A. Enzymatic and Structural Characterization of rTSγ Provides Insights into the Function of rTSβ. Biochemistry 2014, 53, 2732–2738. [Google Scholar] [CrossRef] [PubMed]
- Rosmarin, D.; Palles, C.; Pagnamenta, A.; Kaur, K.; Pita, G.; Martin, M.; Domingo, E.; Jones, A.; Howarth, K.; Freeman-Mills, L.; et al. A Candidate Gene Study of Capecitabine-Related Toxicity in Colorectal Cancer Identifies New Toxicity Variants at DPYD and a Putative Role for ENOSF1 Rather than TYMS. Gut 2015, 64, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Meulendijks, D.; Rozeman, E.A.; Cats, A.; Sikorska, K.; Joerger, M.; Deenen, M.J.; Beijnen, J.H.; Schellens, J.H.M. Pharmacogenetic Variants Associated with Outcome in Patients with Advanced Gastric Cancer Treated with Fluoropyrimidine and Platinum-Based Triplet Combinations: A Pooled Analysis of Three Prospective Studies. Pharmacogenomics J. 2017, 17, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Dolnick, B.J.; Angelino, N.J.; Dolnick, R.; Sufrin, J.R. A Novel Function for the rTS Gene. Cancer Biol. Ther. 2003, 2, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Dolnick, B.J. Detection of Thymidylate Synthase Modulators by a Novel Screening Assay. Mol. Pharmacol. 2003, 63, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Matsusaka, S.; Lenz, H.-J. Pharmacogenomics of Fluorouracil -Based Chemotherapy Toxicity. Expert Opin Drug Metab. Toxicol. 2015, 11, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and Predicted Protein-Protein Associations, Integrated and Transferred across Organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef]
- Lautner-Csorba, O.; Gézsi, A.; Erdélyi, D.J.; Hullám, G.; Antal, P.; Semsei, Á.F.; Kutszegi, N.; Kovács, G.; Falus, A.; Szalai, C. Roles of Genetic Polymorphisms in the Folate Pathway in Childhood Acute Lymphoblastic Leukemia Evaluated by Bayesian Relevance and Effect Size Analysis. PLoS ONE 2013, 8, e69843. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Toolset for Whole-Genome Association and Population-Based Linkage Analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Tregouet, D.A.; Garelle, V. A New JAVA Interface Implementation of THESIAS: Testing Haplotype Effects in Association Studies. Bioinformatics 2007, 23, 1038–1039. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]

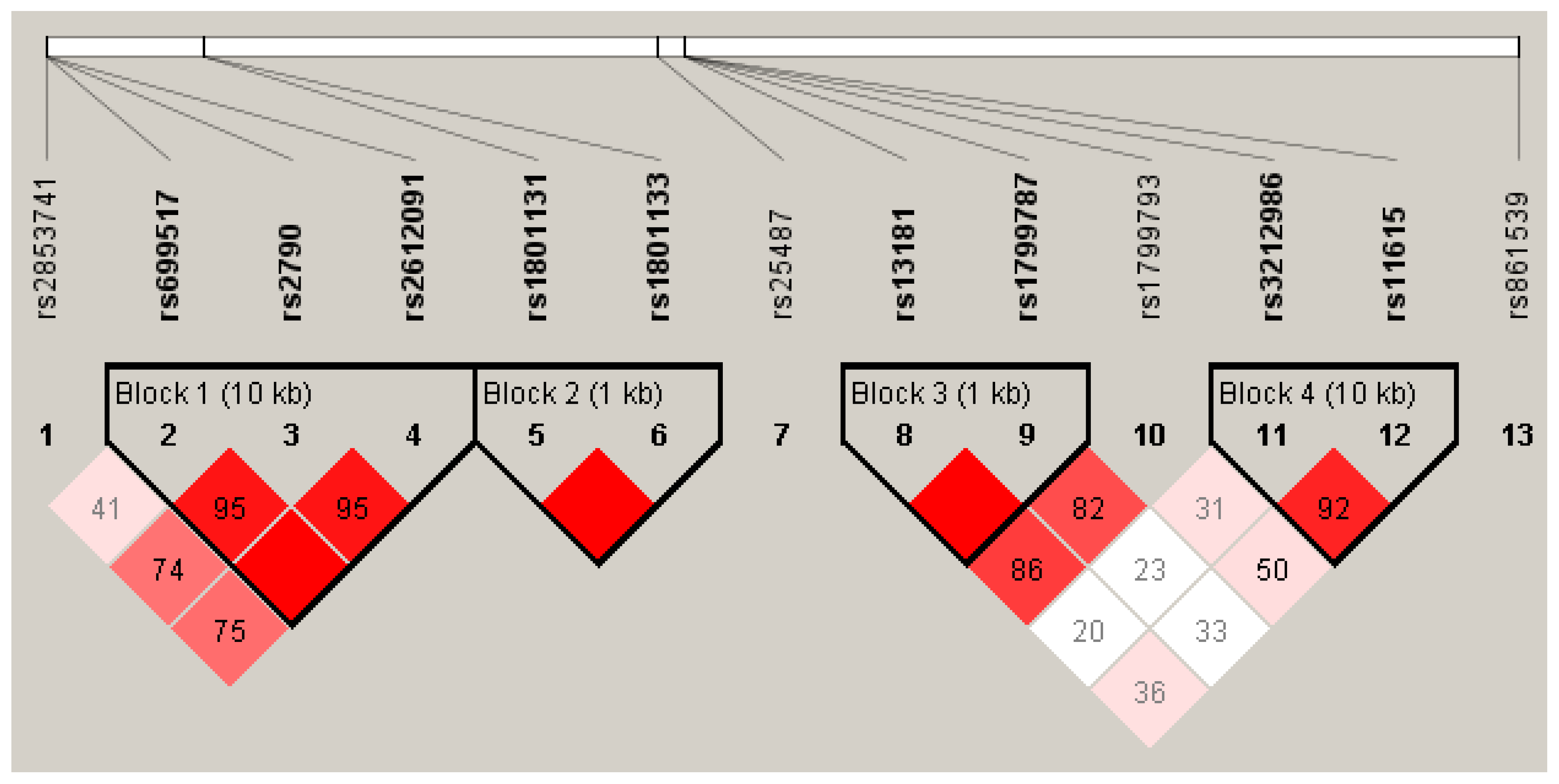
| Characteristic | n (%) | Median [p25–p75] |
|---|---|---|
| Sex | ||
| Female | 53 (37.32) | - |
| Male | 89 (62.68) | - |
| Age at CRC diagnosis (years) | - | 65.00 [57.00–73.00] |
| Family history of cancer | ||
| Yes | 85 (59.86) | - |
| No | 57 (40.14) | - |
| Performance status | ||
| 0 | 107 (75.35) | - |
| 1 | 28 (19.72) | - |
| 2 | 7 (4.93) | - |
| Primary tumor site | ||
| Colon | 81 (57.04) | - |
| Rectum | 61 (42.96) | - |
| Primary tumor size (cm) | - | 4.30 [3.00–6.00] |
| Stage | ||
| 0-IIC | 17 (11.97) | - |
| IIIA-IV | 125 (88.03) | - |
| Grade | ||
| Low | 126 (88.73) | - |
| High | 16 (11.27) | - |
| Capecitabine-based regimen | ||
| Monotherapy | 65 (45.77) | - |
| Combination | 77 (54.23) | - |
| H | TYMS/ENOSF1 rs2790 | TYMS/ENOSF1 rs699517 | ENOSF1 rs2612091 | Frequency | HR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| 0 | A | C | C | 0.3830 | 1.00 | - |
| 1 | G | T | T | 0.2670 | 0.58 (0.31–1.07) | 0.083 |
| 2 | A | C | T | 0.2150 | 0.37 (0.17–0.80) | 0.012 |
| 3 | A | T | T | 0.1270 | 0.89 (0.36–2.15) | 0.796 |
| 4 | G | C | C | 0.0040 | - | - |
| 5 | G | C | T | 0.0040 | - | - |
| Goodness-of-fit: −2 × Log-likelihood (with covariates) = 278.098421 | ||||||
| Goodness-of-fit: −2 × Log-likelihood (without covariates) = 285.795061 | ||||||
| df = 3; model p-value: 0.052 | ||||||
| ENOSF1 rs2612091 (TT) Family history of cancer (Yes) Grade (low) | Disease-Free Survival | ||
| HR (95% CI) | p-value | p-BH * | |
| 0.30 (0.12–0.74) 0.41 (0.20–0.85) 0.29 (0.12–0.68) | 0.009 0.017 0.004 | 0.013 0.017 0.012 | |
| Model p-value < 0.001 | |||
| Gene | Location * | SNP rs | SNP Type | TaqMan Assay ID |
|---|---|---|---|---|
| TYMS | Chr. 18, g.657352T>C | rs2853741 | 2KB upstream variant | C__26612342_10 |
| TYMS-ENOSF1 | Chr. 18, g.673086A>G | rs2790 | 3 primer UTR variant/ Intron variant | C___7486263_10 |
| TYMS-ENOSF1 | Chr. 18, g.673016C>T | rs699517 | 3 primer UTR variant/ Non-coding transcript variant | C___7486269_10 |
| ENOSF1 | Chr. 18, g.683607C>T | rs2612091 | Intron variant | C__15908768_10 |
| MTHFR | Chr. 1, g.11794419T>G | rs1801131 | Missense variant (p.Glu469Ala) | C____850486_20 |
| MTHFR | Chr. 1, g.11796321G>A | rs1801133 | Missense variant (p.Ala262Val) | C_1202883_20 |
| ERCC1 | Chr. 19, g.45420395A>G | rs11615 | Synonymous variant (p.Asn118=) | C__2532959_10 |
| ERCC1 | Chr. 19, g.45409478C>A | rs3212986 | 3 primer UTR variant | C__2532948_10 |
| ERCC2 | Chr. 19, g.45352886G>A | rs1799787 | Intron variant | C__11973901_10 |
| ERCC2 | Chr. 19, g.45351661T>G | rs13181 | Missense variant (p.Lys751Gln) | C__3145033_10 |
| ERCC2 | Chr. 19, g.45364001C>T | rs1799793 | Missense variant (p.Asp312Asn) | C__3145050_10 |
| XRCC1 | Chr. 19, g.43551574T>C | rs25487 | Missense variant (p.Gln399Arg) | C__622564_10 |
| XRCC3 | Chr. 14, g.103699416G>A | rs861539 | Missense variant (p.Thr241Met) | C__8901525_10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cura, Y.; Sánchez-Martín, A.; Márquez-Pete, N.; González-Flores, E.; Martínez-Martínez, F.; Pérez-Ramírez, C.; Jiménez-Morales, A. Role of Single-Nucleotide Polymorphisms in Genes Implicated in Capecitabine Pharmacodynamics on the Effectiveness of Adjuvant Therapy in Colorectal Cancer. Int. J. Mol. Sci. 2024, 25, 104. https://doi.org/10.3390/ijms25010104
Cura Y, Sánchez-Martín A, Márquez-Pete N, González-Flores E, Martínez-Martínez F, Pérez-Ramírez C, Jiménez-Morales A. Role of Single-Nucleotide Polymorphisms in Genes Implicated in Capecitabine Pharmacodynamics on the Effectiveness of Adjuvant Therapy in Colorectal Cancer. International Journal of Molecular Sciences. 2024; 25(1):104. https://doi.org/10.3390/ijms25010104
Chicago/Turabian StyleCura, Yasmin, Almudena Sánchez-Martín, Noelia Márquez-Pete, Encarnación González-Flores, Fernando Martínez-Martínez, Cristina Pérez-Ramírez, and Alberto Jiménez-Morales. 2024. "Role of Single-Nucleotide Polymorphisms in Genes Implicated in Capecitabine Pharmacodynamics on the Effectiveness of Adjuvant Therapy in Colorectal Cancer" International Journal of Molecular Sciences 25, no. 1: 104. https://doi.org/10.3390/ijms25010104





