Genome-Wide Identification and Expression Analysis of the Stearoyl-Acyl Carrier Protein Δ9 Desaturase Gene Family under Abiotic Stress in Barley
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of SAD Proteins in Barley
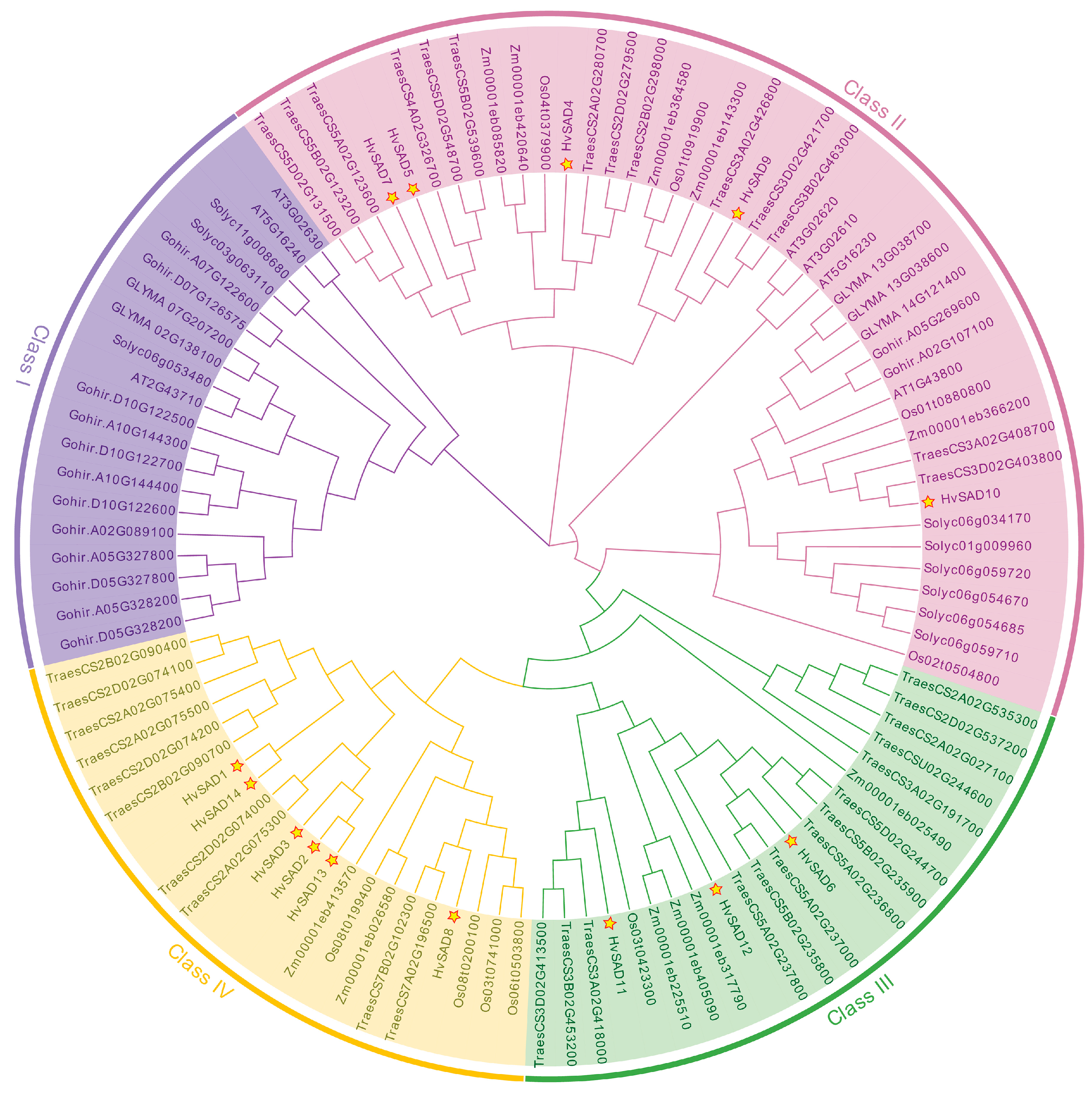
2.2. Sequence Alignment and Phylogenetic Analysis
2.3. Phylogenetic Analysis of SAD Proteins
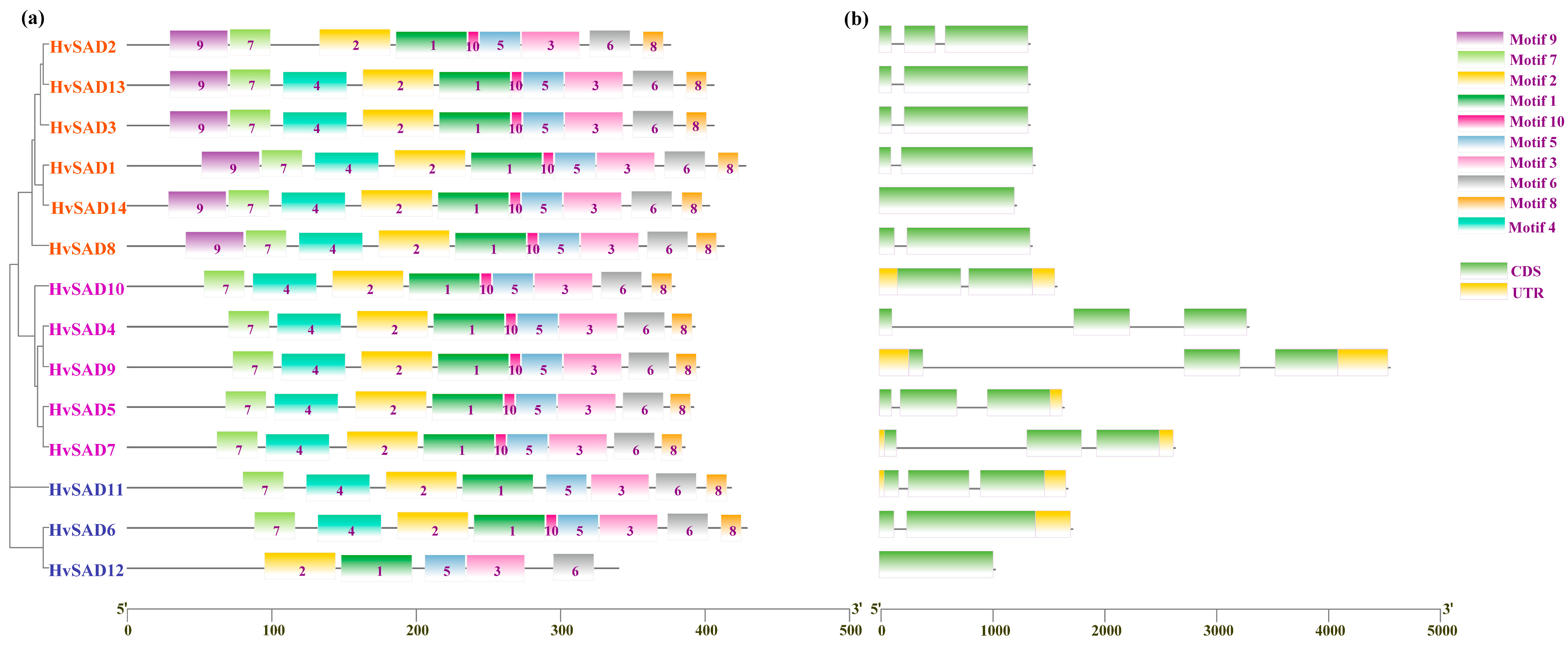
2.4. Gene Structure and Protein Motif Analysis of the HvSADs Family
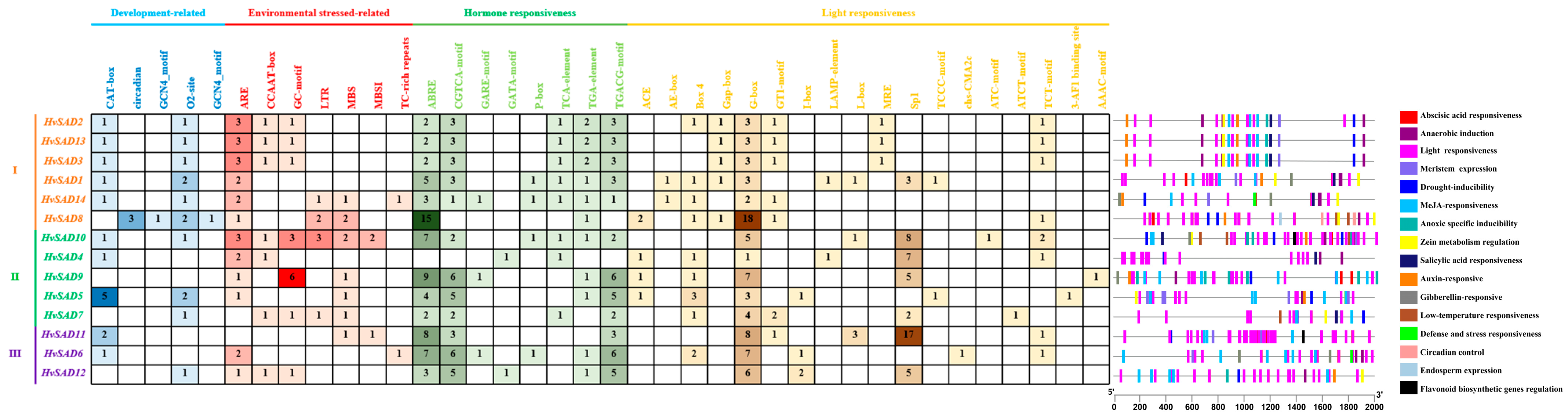
2.5. Cis-Regulatory Elements Analysis in the Promoters of HvSADs
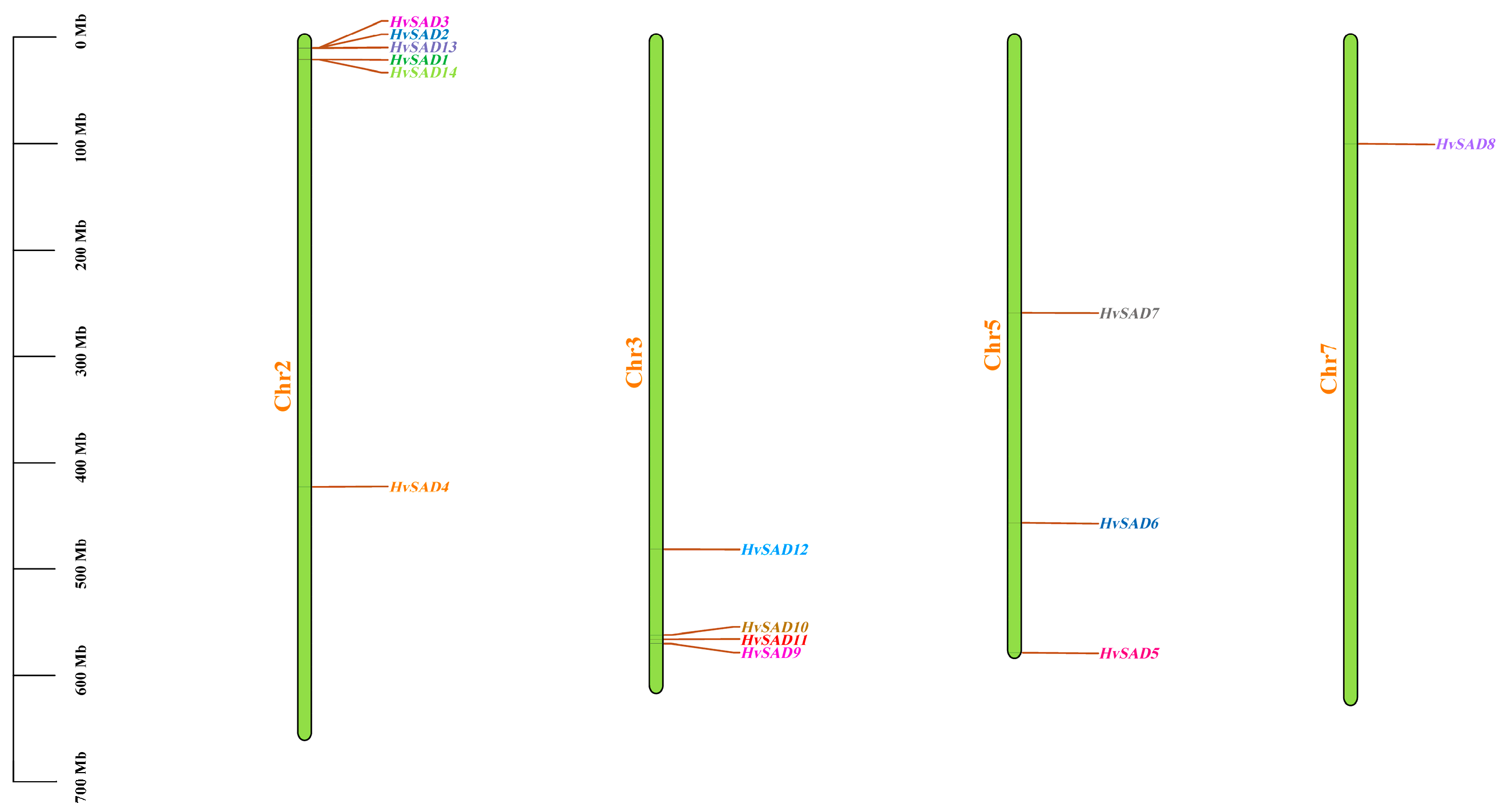
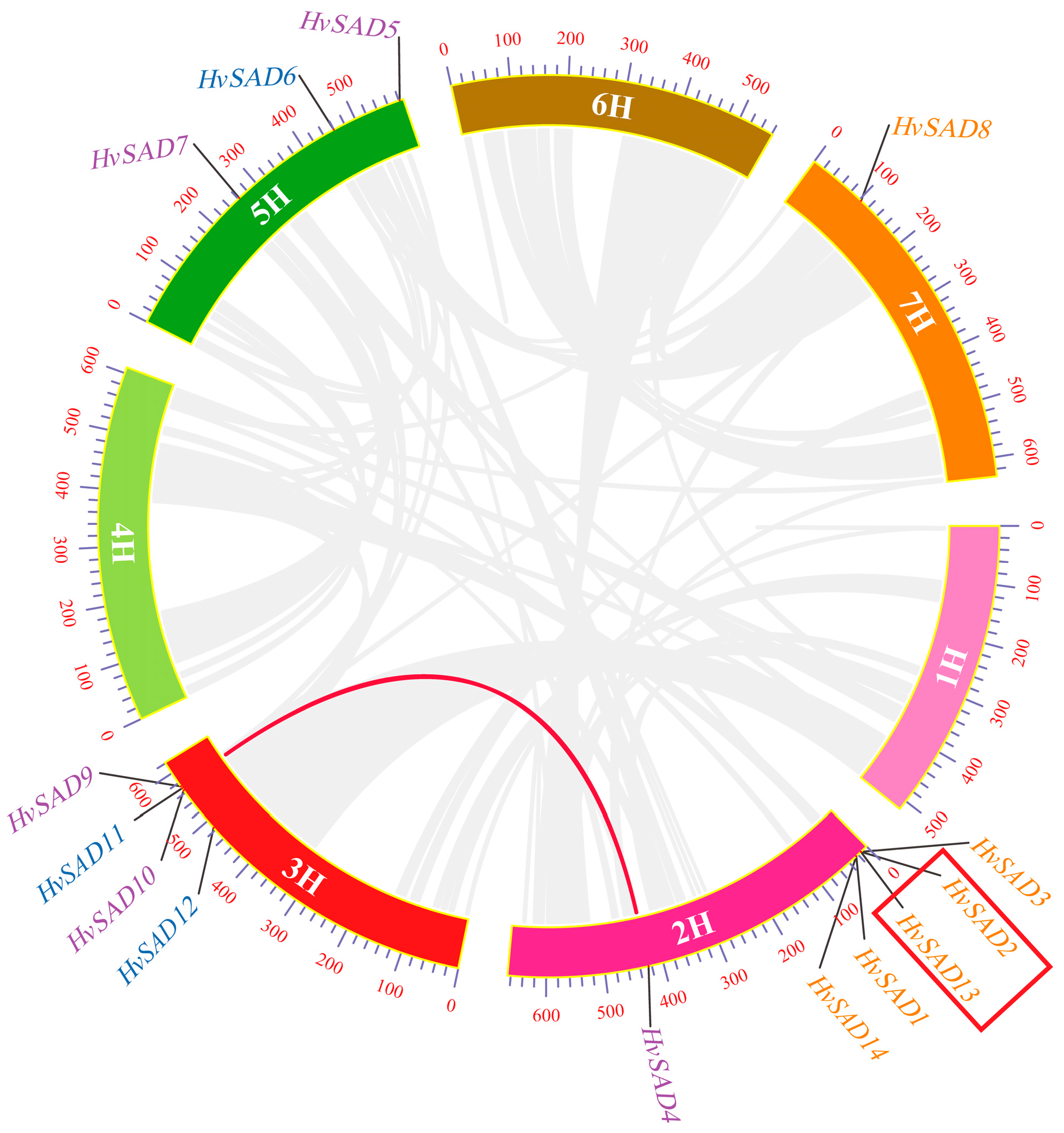
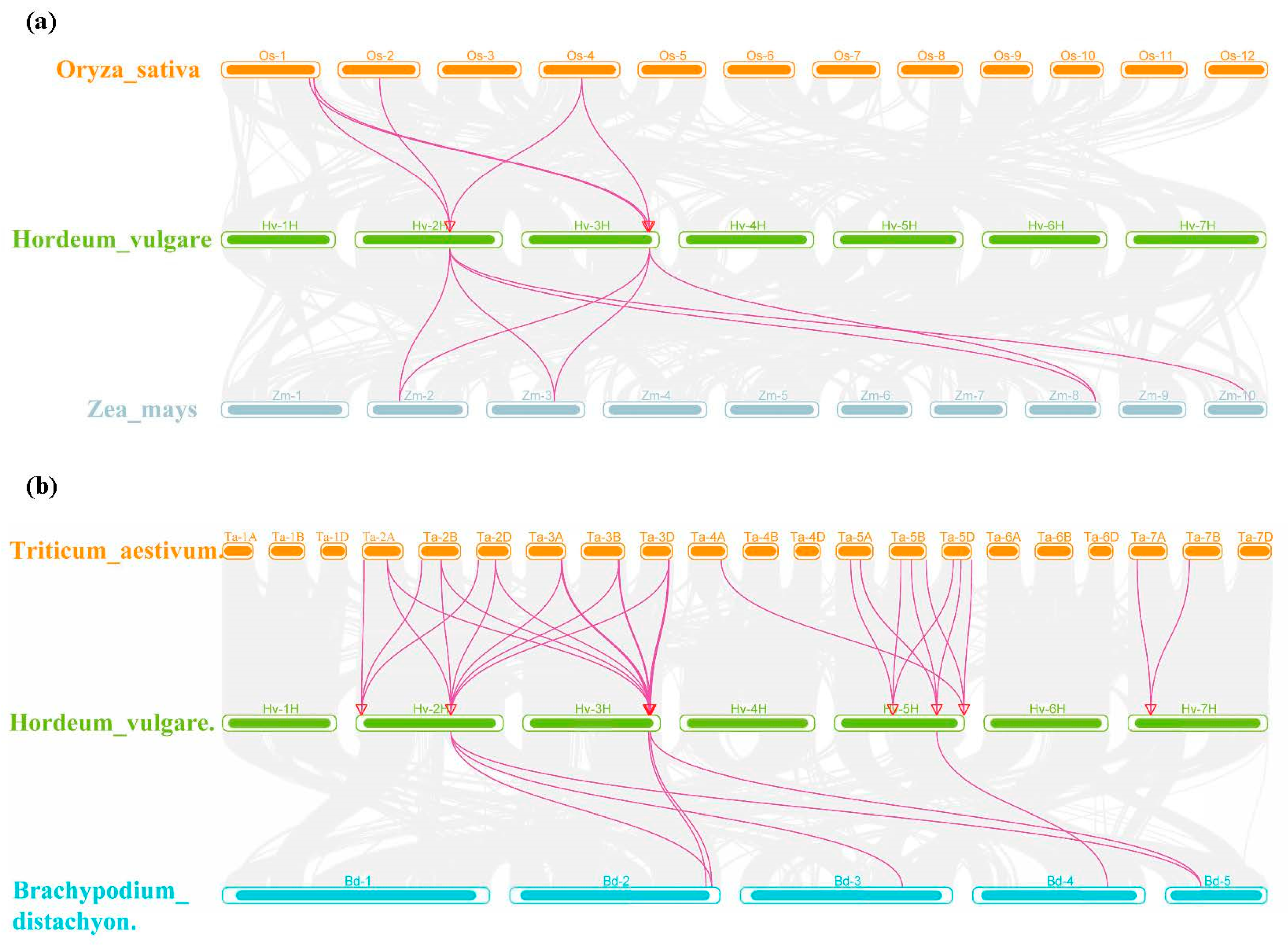
2.6. Chromosome Location and Gene Duplication Analysis of HvSADs
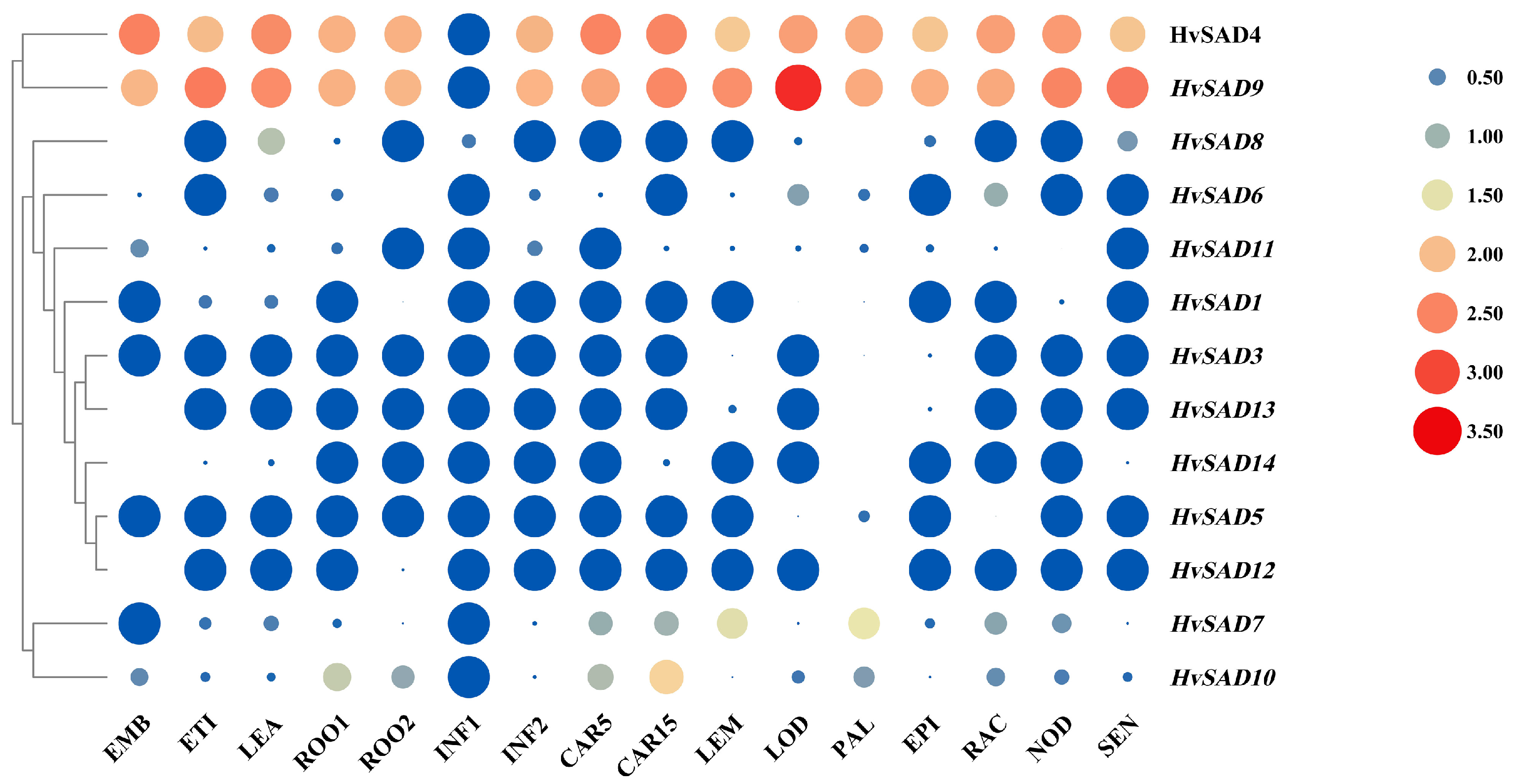
2.7. Expression Profiles of HvSADs in Different Tissues
2.8. Expression of 10 HvSADs under Abiotic Stress by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Identification of SADs in Barley
4.2. Physicochemical Properties Analysis
4.3. Phylogenetic Analysis
4.4. Analysis of Conserved Motif and Gene Structure
4.5. Cis-Regulatory Elements Prediction in the Promoter Regions of HvSADs
4.6. Chromosome Distribution, Gene Duplication, and Selective Pressure Analysis
4.7. Expression Analysis of HvSADs Members
4.8. Plant Material, Stress Treatment, RNA Extraction, and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McKeon, T.A.; Stumpf, P.K. Purification and Characterization of the Stearoyl-Acyl Carrier Protein Desaturase and the Acyl-Acyl Carrier Protein Thioesterase from Maturing Seeds of Safflower. J. Biol. Chem. 1982, 257, 12141–12147. [Google Scholar] [CrossRef] [PubMed]
- Fox, B.G. Stearoyl-Acyl Carrier Protein Δ9 Desaturase from Ricinus Communis Is a Diiron-Oxo Protein. Proc. Natl. Acad. Sci. USA 1993, 90, 2486–2490. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Xu, G.; Du, H.; Hu, J.; Liu, Z.; Li, H.; Li, J.; Yang, X. Natural Variations in Stearoyl-Acp Desaturase Genes Affect the Conversion of Stearic to Oleic Acid in Maize Kernel. Theor. Appl. Genet. 2017, 130, 151–161. [Google Scholar] [CrossRef]
- Ohlrogge, J.; Browse, J. Lipid Biosynthesis. Plant Cell 1995, 7, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Liu, B.; Wang, Z.; Xue, J.; Zhang, H.; Li, R. Identification and Functional Analysis of Soybean Stearoyl-ACP Δ9 Desaturase (GmSAD) Gene Family. Sheng Wu Gong Cheng Xue Bao 2020, 36, 716–731. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, Y.; Xue, J.; Mao, X.; Jia, X.; Li, R. Stearoyl-ACP Δ9 Desaturase 6 and 8 (GhA-SAD6 and GhD-SAD8) Are Responsible for Biosynthesis of Palmitoleic Acid Specifically in Developing Endosperm of Upland Cotton Seeds. Front. Plant Sci. 2019, 10, 703. [Google Scholar] [CrossRef] [PubMed]
- Kazaz, S.; Barthole, G.; Domergue, F.; Ettaki, H.; To, A.; Vasselon, D.; De Vos, D.; Belcram, K.; Lepiniec, L.; Baud, S. Differential Activation of Partially Redundant Δ9 Stearoyl-ACP Desaturase Genes Is Critical for Omega-9 Monounsaturated Fatty Acid Biosynthesis During Seed Development in Arabidopsis. Plant Cell 2020, 32, 3613–3637. [Google Scholar] [CrossRef]
- Zhang, Y.; Maximova, S.N.; Guiltinan, M.J. Characterization of a Stearoyl-Acyl Carrier Protein Desaturase Gene Family from Chocolate Tree, Theobroma cacao L. Front. Plant Sci. 2015, 6, 239. [Google Scholar] [CrossRef][Green Version]
- Tasseva, G.; Davy De Virville, J.; Cantrel, C.; Moreau, F.; Zachowski, A. Changes in the Endoplasmic Reticulum Lipid Properties in Response to Low Temperature in Brassica napus. Plant Physiol. Biochem. 2004, 42, 811–822. [Google Scholar] [CrossRef]
- Li, F.; Bian, C.S.; Xu, J.F.; Pang, W.F.; Liu, J.; Duan, S.G.; Lei, Z.-G.; Jiwan, P.; Jin, L.-P. Cloning and Functional Characterization of SAD Genes in Potato. PLoS ONE 2015, 10, e0122036. [Google Scholar] [CrossRef]
- Byfield, G.E.; Upchurch, R.G. Effect of Temperature on Delta-9 Stearoyl-ACP and Microsomal Omega-6 Desaturase Gene Expression and Fatty Acid Content in Developing Soybean Seeds. Crop Sci. 2007, 47, 1698–1704. [Google Scholar] [CrossRef]
- Sui, N.; Li, M.; Li, K.; Song, J.; Wang, B.S. Increase in Unsaturated Fatty Acids in Membrane Lipids of Suaeda salsa L. Enhances Protection of Photosystem II under High Salinity. Photosynthetica 2010, 48, 623–629. [Google Scholar] [CrossRef]
- Sui, N.; Tian, S.; Wang, W.; Wang, M.; Fan, H. Overexpression of Glycerol-3-Phosphate Acyltransferase from Suaeda salsa Improves Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1337. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Wang, Y.; Liu, S.; Yang, Z.; Wang, F.; Wan, S. Transcriptomic and Physiological Evidence for the Relationship between Unsaturated Fatty Acid and Salt Stress in Peanut. Front. Plant Sci. 2018, 9, 7. [Google Scholar] [CrossRef]
- Mo, S.; Zhang, Y.; Wang, X.; Yang, J.; Sun, Z.; Zhang, D.; Chen, B.; Wang, G.; Ke, H.; Liu, Z.; et al. Cotton GhSSI2 Isoforms from the Stearoyl Acyl Carrier Protein Fatty Acid Desaturase Family Regulate Verticillium Wilt Resistance. Mol. Plant Pathol. 2021, 22, 1041–1056. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Exp. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Farmer, E.E.; Mueller, M.J. ROS-Mediated Lipid Peroxidation and RES-Activated Signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Kumar, A.; Kaur, N. ROS and Oxidative Burst: Roots in Plant Development. Plant Divers. 2020, 42, 33–43. [Google Scholar] [CrossRef]
- Giraldo, P.; Benavente, E.; Manzano-Agugliaro, F.; Gimenez, E. Worldwide Research Trends on Wheat and Barley: A Bibliometric Comparative Analysis. Agronomy 2019, 9, 352. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, S.; Chen, Y.; Dong, M.; Fang, Y.; Zhang, X.; Tong, T.; Zhang, Z.; Zheng, J.; Xue, D.; et al. Genome-Wide Identification of the F-Box Gene Family and Expression Analysis under Drought and Salt Stress in Barley. Phyton 2020, 89, 229–251. [Google Scholar] [CrossRef]
- Zhang, Z.; Tong, T.; Fang, Y.; Zheng, J.; Zhang, X.; Niu, C.; Li, J.; Zhang, X.; Xue, D. Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment. Plants 2020, 9, 1281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Chen, Y.; Wang, S.; Fang, Y.; Zhang, X.; Wu, Y.; Xue, D. Genome-Wide Identification of the SOD Gene Family and Expression Analysis under Drought and Salt Stress in Barley. Plant Growth Regul. 2021, 94, 49–60. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Q.; Zhang, X.; Zhang, X.; Yu, T.; Wu, Y.; Fang, Y.; Xue, D. Genome-Wide Identification of the HMA Gene Family and Expression Analysis under Cd Stress in Barley. Plants 2021, 10, 1849. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Fang, Y.; Zhang, Z.; Zheng, J.; Lu, X.; Zhang, X.; Xue, D. Genome-Wide Identification, Phylogenetic and Expression Analysis of SBP-Box Gene Family in Barley (Hordeum vulgare L.). Plant Growth Regul. 2020, 90, 137–149. [Google Scholar] [CrossRef]
- Tong, T.; Fang, Y.; Zhang, Z.; Zheng, J.; Zhang, X.; Li, J.; Niu, C.; Xue, D.; Zhang, X. Genome-Wide Identification and Expression Pattern Analysis of the KCS Gene Family in Barley. Plant Growth Regul. 2021, 93, 89–103. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, Z.; Tong, T.; Fang, Y.; Zhang, X.; Niu, C.; Li, J.; Wu, Y.; Xue, D.; Zhang, X. Genome-Wide Identification of WRKY Gene Family and Expression Analysis under Abiotic Stress in Barley. Agronomy 2021, 11, 521. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Shanklin, J.; Whittle, E.; Lapchyk, L.; Hildebrand, D.; Kachroo, P. The Arabidopsis Stearoyl-Acyl Carrier Protein-Desaturase Family and the Contribution of Leaf Isoforms to Oleic Acid Synthesis. Plant Mol. Biol. 2006, 63, 257–271. [Google Scholar] [CrossRef]
- Shilman, F.; Brand, Y.; Brand, A.; Hedvat, I.; Hovav, R. Identification and Molecular Characterization of Homeologous Δ9-Stearoyl Acyl Carrier Protein Desaturase 3 Genes from the Allotetraploid Peanut (Arachis hypogaea). Plant Mol. Biol. Rep. 2011, 29, 232–241. [Google Scholar] [CrossRef]
- Shang, X.; Cheng, C.; Ding, J.; Guo, W. Identification of Candidate Genes from the SAD Gene Family in Cotton for Determination of Cottonseed Oil Composition. Mol. Genet. Genom. 2017, 292, 173–186. [Google Scholar] [CrossRef]
- Sperling, P.; Ternes, P.; Zank, T.K.; Heinz, E. The Evolution of Desaturases. Prostag. Leukotr. Ess. 2003, 68, 73–95. [Google Scholar] [CrossRef] [PubMed]
- Guy, J.E.; Whittle, E.; Moche, M.; Lengqvist, J.; Lindqvist, Y.; Shanklin, J. Remote Control of Regioselectivity in Acyl-Acyl Carrier Protein-Desaturases. Proc. Natl. Acad. Sci. USA 2011, 108, 16594–16599. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, V.; Janni, M.; Bellincampi, D.; Giardina, T.; D’Ovidio, R. Intron Retention Regulates the Expression of Pectin Methyl Esterase Inhibitor (Pmei) Genes during Wheat Growth and Development: Isolation and Functional Characterisation of Wheat Pmei Genes. Plant Biol. 2012, 14, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, B.; Manan, S.; Li, P.; Liu, C.; She, G.; Zhao, S.; Zhao, J. Critical Metabolic Pathways and SAD/FADs, WRI1s, and DGATs Cooperate for High-Oleic Acid Oil Production in Developing Oil Tea (Camellia oleifera) Seeds. Hort. Res. 2022, 9, uhac087. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Li, X.; Munir, R.; Khan, A.R.; Azhar, W.; Yasin, M.U.; Jiang, Q.; Bancroft, I.; Gan, Y. The WRKY6 Transcription Factor Affects Seed Oil Accumulation and Alters Fatty Acid Compositions in Arabidopsis thaliana. Physiol. Plant 2020, 169, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Transcriptomic Analysis Reveals the Regulatory Networks and Hub Genes Controlling the Unsaturated Fatty Acid Contents of Developing Seed in Soybean. Front. Plant Sci. 2022, 13, 876371. [Google Scholar] [CrossRef]
- Xi, Y.; Cai, J.; Li, G.; Huang, H.; Peng, X.; Zhu, G. High CO2 Facilitates Fatty Acid Biosynthesis and Mitigates Cellular Oxidative Stress Caused by CAC2 Dysfunction in Arabidopsis. Plant J. 2023, 115, 1316–1330. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Ezoe, A.; Shirai, K.; Hanada, K. Degree of Functional Divergence in Duplicates Is Associated with Distinct Roles in Plant Evolution. Mol. Biol. Evol. 2021, 38, 1447–1459. [Google Scholar] [CrossRef]
- Jia, Y.; Xu, M.; Hu, H.; Chapman, B.; Watt, C.; Buerte, B.; Han, N.; Zhu, M.; Bian, H.; Li, C.; et al. Comparative Gene Retention Analysis in Barley, Wild Emmer, and Bread Wheat Pangenome Lines Reveals Factors Affecting Gene Retention Following Gene Duplication. BMC Biol. 2023, 21, 25. [Google Scholar] [CrossRef]
- To, V.-T.; Shi, Q.; Zhang, Y.; Shi, J.; Shen, C.; Zhang, D.; Cai, W. Genome-Wide Analysis of the GRAS Gene Family in Barley (Hordeum vulgare L.). Genes 2020, 11, 553. [Google Scholar] [CrossRef] [PubMed]
- Cvijovic, I.; Good, B.H.; Desai, M.M. The Effect of Strong Purifying Selection on Genetic Diversity. Genetics 2018, 209, 1235–1278. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A. Arabidopsis Sfd Mutants Affect Plastidic Lipid Composition and Suppress Dwarfing, Cell Death, and the Enhanced Disease Resistance Phenotypes Resulting from the Deficiency of a Fatty Acid Desaturase. Plant Cell 2003, 15, 2383–2398. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C. Jasmonates: An Update on Biosynthesis, Signal Transduction and Action in Plant Stress Response, Growth and Development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Marand, A.P.; Eveland, A.L.; Kaufmann, K.; Springer, N.M. Cis-Regulatory Elements in Plant Development, Adaptation, and Evolution. Annu. Rev. Plant Biol. 2023, 74, 111–137. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Z.; Zhong, Y.; Huang, J.; Hu, Q.; Chen, F. Stearoyl-Acyl Carrier Protein Desaturase Gene from the Oleaginous Microalga Chlorella Zofingiensis: Cloning, Characterization and Transcriptional Analysis. Planta 2012, 236, 1665–1676. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.L.; Sicardo, M.D.; Alfonso, M.; Martínez-Rivas, J.M. Transcriptional Regulation of Stearoyl-Acyl Carrier Protein Desaturase Genes in Response to Abiotic Stresses Leads to Changes in the Unsaturated Fatty Acids Composition of Olive Mesocarp. Front. Plant Sci. 2019, 10, 251. [Google Scholar] [CrossRef]
- Song, Z.; Lai, X.; Chen, H.; Wang, L.; Pang, X.; Hao, Y.; Lu, W.; Chen, W.; Zhu, X.; Li, X. Role of MaABI5-like in Abscisic Acid-Induced Cold Tolerance of ‘Fenjiao’ Banana Fruit. Hortic. Res. 2022, 9, uhac130. [Google Scholar] [CrossRef]
- Cao, J.; Li, M.; Chen, J.; Liu, P.; Li, Z. Effects of MeJA on Arabidopsis Metabolome under Endogenous JA Deficiency. Sci. Rep. 2016, 6, 37674. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The Roles of Methyl Jasmonate to Stress in Plants. Funct. Plant Biol. 2019, 46, 197. [Google Scholar] [CrossRef]
- Contreras, C.; Mariotti, R.; Mousavi, S.; Baldoni, L.; Guerrero, C.; Roka, L.; Cultrera, N.; Pierantozzi, P.; Maestri, D.; Gentili, L.; et al. Characterization and Validation of Olive FAD and SAD Gene Families: Expression Analysis in Different Tissues and during Fruit Development. Mol. Biol. Rep. 2020, 47, 4345–4355. [Google Scholar] [CrossRef]
- Ding, Z.T.; Shen, J.Z.; Pan, L.L.; Wang, Y.U.; Li, Y.S.; Wang, Y.; Sun, H.W. CsSAD: A Fatty Acid Desaturase Gene Involved in Abiotic Resistance in Camellia sinensis (L.). Genet. Mol. Res. 2016, 15, 15017512. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Multiple Roles in Stress Response. Front. Plant Sci. 2020, 11, 562785. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, J.; Zhang, L.; Zhang, L. Tung Tree Stearoyl-acyl Carrier Protein Δ9 Desaturase Improves Oil Content and Cold Resistance of Arabidopsis and Saccharomyces cerevisiae. Front. Plant Sci. 2023, 14, 1144853. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shao, Y.; Yang, X.; Zhang, C.; Guo, Y.; Liu, Z.; Chen, M. Heterogeneous Expression of Stearoyl-Acyl Carrier Protein Desaturase Genes SAD1 and SAD2 from Linum Usitatissimum Enhances Seed Oleic Acid Accumulation and Seedling Cold and Drought Tolerance Capability in Brassica napus. J. Integr. Agric. 2023, S2095311923001387. [Google Scholar] [CrossRef]
- Alexander, R.D.; Wendelboe-Nelson, C.; Morris, P.C. The Barley Transcription Factor HvMYB1 Is a Positive Regulator of Drought Tolerance. Plant Physiol. Biochem. 2019, 142, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.G.; Jang, C.S.; Kim, J.Y.; Kim, D.S.; Park, J.H.; Kim, D.Y.; Seo, Y.W. A Myb Transcription Factor (TaMyb1) from Wheat Roots Is Expressed during Hypoxia: Roles in Response to the Oxygen Concentration in Root Environment and Abiotic Stresses. Physiol. Plant 2007, 129, 375–385. [Google Scholar] [CrossRef]
- Allakhverdiev, S.I.; Kinoshita, M.; Inaba, M.; Suzuki, I.; Murata, N. Unsaturated Fatty Acids in Membrane Lipids Protect the Photosynthetic Machinery against Salt-Induced Damage in Synechococcus. Plant Physiol. 2001, 125, 1842–1853. [Google Scholar] [CrossRef]
- Huang, J.; Xue, C.; Wang, H.; Wang, L.; Schmidt, W.; Shen, R.; Lan, P. Genes of ACYL CARRIER PROTEIN Family Show Different Expression Profiles and Overexpression of ACYL CARRIER PROTEIN 5 Modulates Fatty Acid Composition and Enhances Salt Stress Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 987. [Google Scholar] [CrossRef]
- Kumar, M.; Kumar Patel, M.; Kumar, N.; Bajpai, A.B.; Siddique, K.H.M. Metabolomics and Molecular Approaches Reveal Drought Stress Tolerance in Plants. Int. J. Mol. Sci. 2021, 22, 9108. [Google Scholar] [CrossRef]
- Li, Y.; Si, Y.-T.; He, Y.-X.; Li, J.-X. Comparative Analysis of Drought-Responsive and -Adaptive Genes in Chinese Wingnut (Pterocarya stenoptera C. DC). BMC Genom. 2021, 22, 155. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Wu, D.; Zhang, X.; Xu, Y.; Kuang, L.; Cai, S.; Zhang, G.; Shen, Q. Vacuolar H+-Pyrophosphatase HVP10 Enhances Salt Tolerance via Promoting Na+ Translocation into Root Vacuoles. Plant Physiol. 2022, 188, 1248–1263. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, G.; Zeng, F.; Han, Z.; Qiu, C.; Zeng, M.; Yang, Z.; Xu, F.; Wu, D.; Deng, F.; et al. Molecular Evidence for Adaptive Evolution of Drought Tolerance in Wild Cereals. New Phytol. 2023, 237, 497–514. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.J.; Eddy, S.R. Nhmmer: DNA Homology Search with Profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The Protein Families Database. Nucl. Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Colmsee, C.; Beier, S.; Himmelbach, A.; Schmutzer, T.; Stein, N.; Scholz, U.; Mascher, M. BARLEX—The Barley Draft Genome Explorer. Mol. Plant 2015, 8, 964–966. [Google Scholar] [CrossRef]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomic Data. In Plant Genomics Databases; Van Dijk, A.D.J., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1533, pp. 1–31. ISBN 978-1-4939-6656-1. [Google Scholar]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucl. Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucl. Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucl. Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Bannai, H.; Tamada, Y.; Maruyama, O.; Nakai, K.; Miyano, S. Extensive Feature Detection of N-Terminal Protein Sorting Signals. Bioinformatics 2002, 18, 298–305. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An Online Visualization and Management Tool for Customized and Annotated Phylogenetic Trees. Nucl. Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucl. Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | Protein | Subcellular Localization | Domain | Chromosome | Genomic Location | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Size (aa) | MW (Da) | pI | Aliphatic Index | GRAVY | ||||||
| HvSAD1 | HORVU.MOREX.r3.2HG0106740 | 427 | 47,347 | 8.47 | 85.55 | −0.267 | Chloroplast | FA_desaturase_2 | Chr2 | 23,954,883−23,956,257 |
| HvSAD2 | HORVU.MOREX.r3.2HG0101470 | 375 | 41,677.74 | 8.68 | 87.33 | −0.257 | Mitochondrial | FA_desaturase_2 | Chr2 | 13,191,866−13,193,197 |
| HvSAD3 | HORVU.MOREX.r3.2HG0101440 | 405 | 44,883.21 | 7.19 | 84.96 | −0.241 | Mitochondrial | FA_desaturase_2 | Chr2 | 13,111,139−13,112,470 |
| HvSAD4 | HORVU.MOREX.r3.2HG0161410 | 392 | 44,559.57 | 6.04 | 73.44 | −0.490 | Chloroplast | FA_desaturase_2 | Chr2 | 425,614,894−425,618,180 |
| HvSAD5 | HORVU.MOREX.r3.5HG0535350 | 391 | 44,667.93 | 6.05 | 80.36 | −0.405 | Chloroplast | FA_desaturase_2 | Chr5 | 581,725,723−581,727,357 |
| HvSAD6 | HORVU.MOREX.r3.5HG0486420 | 428 | 47,617.36 | 7.59 | 73.93 | −0.306 | Chloroplast | FA_desaturase_2 | Chr5 | 459,736,400−459,738,111 |
| HvSAD7 | HORVU.MOREX.r3.5HG0457600 | 385 | 43,660.82 | 5.97 | 74.75 | −0.381 | Chloroplast | FA_desaturase_2 | Chr5 | 262,136,160−262,138,788 |
| HvSAD8 | HORVU.MOREX.r3.7HG0668240 | 412 | 45,390.46 | 6.87 | 72.91 | −0.344 | Chloroplast | FA_desaturase_2 | Chr7 | 103,262,598−103,263,947 |
| HvSAD9 | HORVU.MOREX.r3.3HG0310210 | 395 | 44,530.92 | 6.65 | 78.61 | −0.409 | Chloroplast | FA_desaturase_2 | Chr3 | 573,217,384−573,221,932 |
| HvSAD10 | HORVU.MOREX.r3.3HG0307490 | 378 | 42,389.33 | 6.24 | 79.29 | −0.324 | Chloroplast | FA_desaturase_2 | Chr3 | 564,864,759−564,866,328 |
| HvSAD11 | HORVU.MOREX.r3.3HG0309010 | 417 | 45,352.59 | 6.87 | 78.01 | −0.177 | Chloroplast | FA_desaturase_2 | Chr3 | 569,309,951−569,311,619 |
| HvSAD12 | HORVU.MOREX.r3.3HG0288610 | 339 | 38,752.42 | 9.03 | 76.31 | −0.297 | Cytoplasmic | FA_desaturase_2 | Chr3 | 484,477,447−484,478,466 |
| HvSAD13 | HORVU.MOREX.r3.2HG0101500 | 405 | 44,883.21 | 7.19 | 84.96 | −0.241 | Mitochondrial | FA_desaturase_2 | Chr2 | 13,265,580−13,266,911 |
| HvSAD14 | HORVU.MOREX.r3.2HG0106840 | 402 | 44,563 | 8.4 | 85.55 | −0.236 | Chloroplast | FA_desaturase_2 | Chr2 | 24,197,160−24,198,368 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, M.; Zhou, D.; Ye, Y.; Wen, S.; Zhang, X.; Tian, Q.; Zhang, X.; Mou, W.; Dang, C.; Fang, Y.; et al. Genome-Wide Identification and Expression Analysis of the Stearoyl-Acyl Carrier Protein Δ9 Desaturase Gene Family under Abiotic Stress in Barley. Int. J. Mol. Sci. 2024, 25, 113. https://doi.org/10.3390/ijms25010113
Ding M, Zhou D, Ye Y, Wen S, Zhang X, Tian Q, Zhang X, Mou W, Dang C, Fang Y, et al. Genome-Wide Identification and Expression Analysis of the Stearoyl-Acyl Carrier Protein Δ9 Desaturase Gene Family under Abiotic Stress in Barley. International Journal of Molecular Sciences. 2024; 25(1):113. https://doi.org/10.3390/ijms25010113
Chicago/Turabian StyleDing, Mingyu, Danni Zhou, Yichen Ye, Shuting Wen, Xian Zhang, Quanxiang Tian, Xiaoqin Zhang, Wangshu Mou, Cong Dang, Yunxia Fang, and et al. 2024. "Genome-Wide Identification and Expression Analysis of the Stearoyl-Acyl Carrier Protein Δ9 Desaturase Gene Family under Abiotic Stress in Barley" International Journal of Molecular Sciences 25, no. 1: 113. https://doi.org/10.3390/ijms25010113
APA StyleDing, M., Zhou, D., Ye, Y., Wen, S., Zhang, X., Tian, Q., Zhang, X., Mou, W., Dang, C., Fang, Y., & Xue, D. (2024). Genome-Wide Identification and Expression Analysis of the Stearoyl-Acyl Carrier Protein Δ9 Desaturase Gene Family under Abiotic Stress in Barley. International Journal of Molecular Sciences, 25(1), 113. https://doi.org/10.3390/ijms25010113







