Unique tRNA Fragment Upregulation with SARS-CoV-2 but Not with SARS-CoV Infection
Abstract
:1. Introduction
2. Results
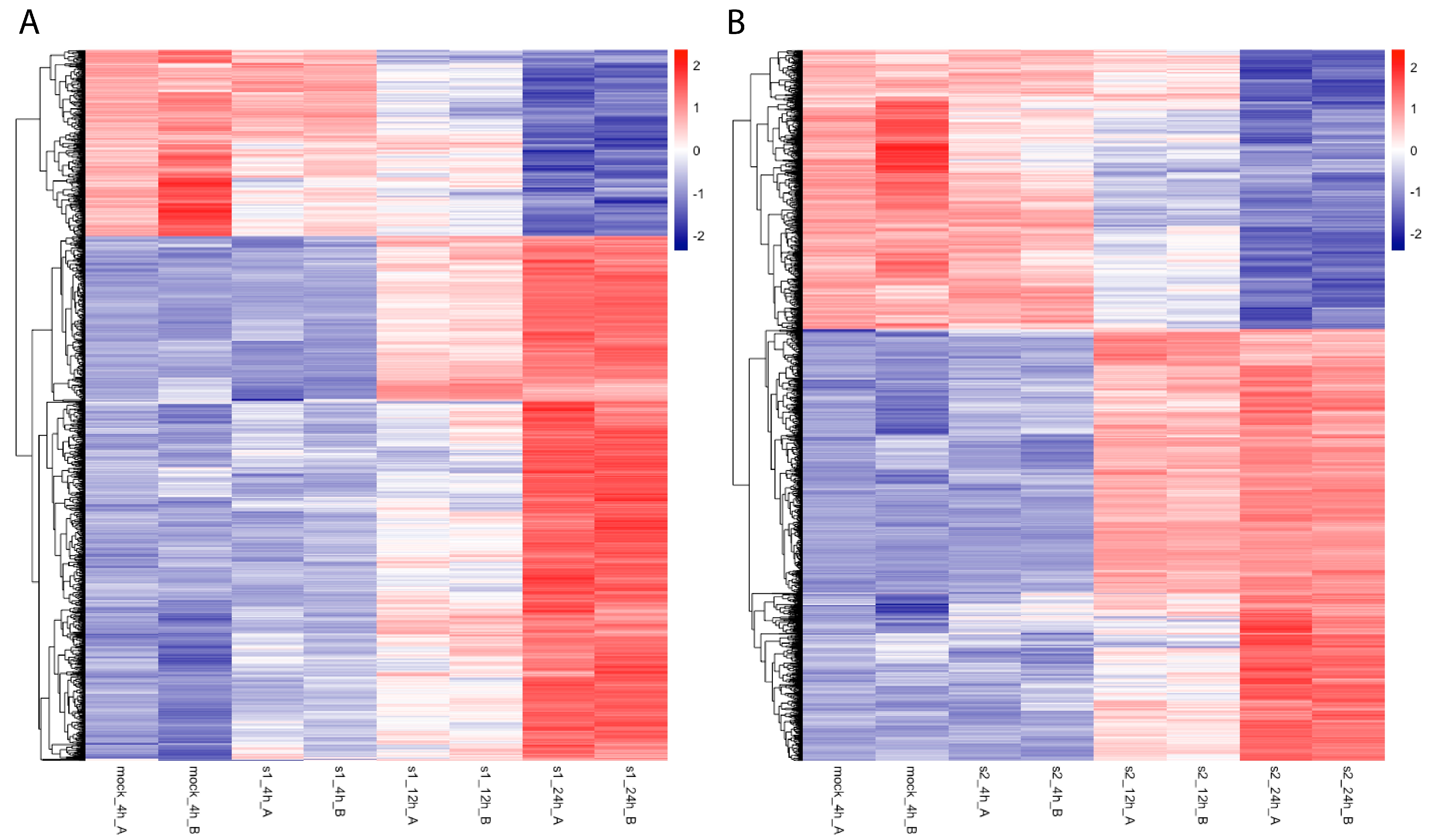
2.1. Uniquely Upregulated tRF5s with CoV2 Infection
2.2. More Downregulated Genes with CoV2 Infection Than with CoV Infection
2.3. Enriched Neural Functions of Theoretical Targets of Upregulated tRF5s
3. Discussion
4. Materials and Methods
4.1. Downloading Fastq Files from a GEO Dataset
4.2. Aligning Sequencing to Transcripts
4.3. Differential Expression Analysis
4.4. Involving Student Scientists
4.5. tRNA Methyltransferase Collection
4.6. Candidate Target Prediction
4.7. Enrichment Testing of tRF5 Targets
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Masre, S.F.; Jufri, N.F.; Ibrahim, F.W.; Abdul Raub, S.H. Classical and alternative receptors for SARS-CoV-2 therapeutic strategy. Rev. Med. Virol. 2021, 31, e2207. [Google Scholar] [CrossRef] [PubMed]
- Zalpoor, H.; Akbari, A.; Samei, A.; Forghaniesfidvajani, R.; Kamali, M.; Afzalnia, A.; Manshouri, S.; Heidari, F.; Pornour, M.; Khoshmirsafa, M.; et al. The roles of Eph receptors, neuropilin-1, P2X7, and CD147 in COVID-19-associated neurodegenerative diseases: Inflammasome and JaK inhibitors as potential promising therapies. Cell Mol. Biol. Lett. 2022, 27, 10. [Google Scholar] [CrossRef]
- Moldofsky, H.; Patcai, J. Chronic widespread musculoskeletal pain, fatigue, depression and disordered sleep in chronic post-SARS syndrome; a case-controlled study. BMC Neurol. 2011, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.U.; Hanif, M.; Ali, M.J.; Haider, M.A.; Kherani, D.; Memon, G.M.; Karim, A.H.; Sattar, A. Neurological Manifestations of COVID-19 (SARS-CoV-2): A Review. Front. Neurol. 2020, 11, 518. [Google Scholar] [CrossRef]
- Troyer, E.A.; Kohn, J.N.; Hong, S. Are we facing a crashing wave of neuropsychiatric sequelae of COVID-19? Neuropsychiatric symptoms and potential immunologic mechanisms. Brain Behav. Immun. 2020, 87, 34–39. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Choi, E.J.; Wu, W.; Zhang, K.; Lee, I.; Kim, I.H.; Lee, Y.S.; Bao, X. ELAC2, an Enzyme for tRNA Maturation, Plays a Role in the Cleavage of a Mature tRNA to Produce a tRNA-Derived RNA Fragment During Respiratory Syncytial Virus Infection. Front. Mol. Biosci. 2020, 7, 609732. [Google Scholar] [CrossRef]
- Wu, W.; Choi, E.J.; Wang, B.; Zhang, K.; Adam, A.; Huang, G.; Tunkle, L.; Huang, P.; Goru, R.; Imirowicz, I.; et al. Changes of Small Non-coding RNAs by Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Front. Mol. Biosci. 2022, 9, 821137. [Google Scholar] [CrossRef] [PubMed]
- Wyler, E.; Mosbauer, K.; Franke, V.; Diag, A.; Gottula, L.T.; Arsie, R.; Klironomos, F.; Koppstein, D.; Honzke, K.; Ayoub, S.; et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 2021, 24, 102151. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Zhang, W.; Foo, M.; Eren, A.M.; Pan, T. tRNA modification dynamics from individual organisms to metaepitranscriptomics of microbiomes. Mol. Cell 2022, 82, 891–906. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Ptashkin, R.N.; Chen, Y.; Cheng, Z.; Liu, G.; Phan, T.; Deng, X.; Zhou, J.; Lee, I.; Lee, Y.S.; et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol. Ther. 2015, 23, 1622–1629. [Google Scholar] [CrossRef]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5’-UTR and 3’-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef]
- Yin, L.; Li, J.; Wang, J.; Pu, T.; Wei, J.; Li, Q.; Wu, B.J. MAOA promotes prostate cancer cell perineural invasion through SEMA3C/PlexinA2/NRP1-cMET signaling. Oncogene 2021, 40, 1362–1374. [Google Scholar] [CrossRef]
- Patton, M.J.; Orihuela, C.J.; Harrod, K.S.; Bhuiyan, M.A.N.; Dominic, P.; Kevil, C.G.; Fort, D.; Liu, V.X.; Farhat, M.; Koff, J.L.; et al. COVID-19 bacteremic co-infection is a major risk factor for mortality, ICU admission, and mechanical ventilation. Crit. Care 2023, 27, 34. [Google Scholar] [CrossRef]
- Mautner, L.; Hoyos, M.; Dangel, A.; Berger, C.; Ehrhardt, A.; Baiker, A. Replication kinetics and infectivity of SARS-CoV-2 variants of concern in common cell culture models. Virol. J. 2022, 19, 76. [Google Scholar] [CrossRef]
- Chuckran, C.A.; Liu, C.; Bruno, T.C.; Workman, C.J.; Vignali, D.A. Neuropilin-1: A checkpoint target with unique implications for cancer immunology and immunotherapy. J. Immunother. Cancer 2020, 8, e000967. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.; Chang, C.J.; Hung, P.H. Possible Pathogenesis and Prevention of Long COVID: SARS-CoV-2-Induced Mitochondrial Disorder. Int. J. Mol. Sci. 2023, 24, 8034. [Google Scholar] [CrossRef] [PubMed]
- Bowe, B.; Xie, Y.; Al-Aly, Z. Postacute sequelae of COVID-19 at 2 years. Nat. Med. 2023, 29, 2347–2357. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Lee, I.; Spratt, H.; Fang, X.; Bao, X. tRNA-Derived Fragments in Alzheimer’s Disease: Implications for New Disease Biomarkers and Neuropathological Mechanisms. J. Alzheimers Dis. 2021, 79, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Kodama, Y.; Shumway, M.; Leinonen, R.; International Nucleotide Sequence Database, C. The Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 2012, 40, D54–D56. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database, C. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]


| Gene Symbol | SARS-CoV Infection | SARS-CoV-2 Infection | ||
|---|---|---|---|---|
| Log2FC | p-Value | Log2FC | p-Value | |
| tRNA Methyltransferase | ||||
| FTSJ1 | −0.41 | 0.058 | −0.70 | 2.3 × 10−4 |
| METTL1 | −0.54 | 0.026 | −0.17 | 0.53 |
| METTL8 | −0.53 | 0.052 | −0.81 | 7.7 × 10−4 |
| THUMPD3 | 0.24 | 0.17 | 0.36 | 0.022 |
| TRDMT1 | −0.30 | 0.41 | −1.43 | 3.2 × 10−5 |
| TRMT2B | −0.57 | 0.026 | −0.37 | 0.13 |
| TRMT44 | 0.59 | 0.036 | 0.67 | 0.60 |
| TRMT61A | −0.56 | 0.0039 | −0.15 | 0.49 |
| TRMT9B | 0.07 | 0.89 | 0.63 | 0.049 |
| TYW3 | −0.22 | 0.36 | −0.42 | 0.033 |
| Demethylase | ||||
| ALKBH3 | −0.77 | 0.0022 | −0.56 | 0.019 |
| Ribonuclease 1 | ||||
| DICER1 | 1.18 | 3.0 × 10−29 | 1.75 | 2.8 × 10−69 |
| KEGG ID | KEGG Description | Observed Gene Count | Background Gene Count | False Discovery Rate |
|---|---|---|---|---|
| Commonly downregulated genes | ||||
| hsa03010 | Ribosome | 83 | 130 | 9.43 × 10−36 |
| hsa01100 | Metabolic pathways | 209 | 1447 | 5.57× 10−12 |
| hsa00190 | Oxidative phosphorylation | 45 | 130 | 1.83 × 10−11 |
| hsa04714 | Thermogenesis | 61 | 229 | 1.83 × 10−11 |
| hsa05012 | Parkinson disease | 57 | 240 | 3.47 × 10−9 |
| hsa05014 | Amyotrophic lateral sclerosis | 68 | 352 | 9.68 × 10−8 |
| hsa05010 | Alzheimer disease | 68 | 355 | 1.12 × 10−7 |
| hsa05016 | Huntington disease | 60 | 298 | 1.75 × 10−7 |
| hsa05020 | Prion disease | 52 | 265 | 3.40 × 10−6 |
| hsa04142 | Lysosome | 29 | 126 | 0.00015 |
| hsa04510 | Focal adhesion | 36 | 198 | 0.0011 |
| hsa01212 | Fatty acid metabolism | 16 | 54 | 0.0015 |
| hsa04932 | Non-alcoholic fatty liver disease | 29 | 148 | 0.0016 |
| hsa03060 | Protein export | 10 | 23 | 0.0026 |
| hsa04260 | Cardiac muscle contraction | 20 | 87 | 0.003 |
| hsa00062 | Fatty acid elongation | 10 | 25 | 0.0039 |
| hsa05110 | Vibrio cholerae infection | 13 | 48 | 0.0105 |
| hsa05100 | Bacterial invasion of epithelial cells | 16 | 70 | 0.012 |
| hsa01200 | Carbon metabolism | 22 | 117 | 0.013 |
| hsa04810 | Regulation of actin cytoskeleton | 32 | 209 | 0.0219 |
| hsa03008 | Ribosome biogenesis in eukaryotes | 16 | 77 | 0.0246 |
| hsa05205 | Proteoglycans in cancer | 30 | 196 | 0.0279 |
| hsa04146 | Peroxisome | 16 | 79 | 0.0282 |
| hsa00010 | Glycolysis/Gluconeogenesis | 14 | 65 | 0.031 |
| hsa04910 | Insulin signaling pathway | 22 | 133 | 0.0415 |
| Downregulated only in SARS-CoV | ||||
| hsa03010 | Ribosome | 19 | 130 | 0.00021 |
| hsa05012 | Parkinson disease | 26 | 240 | 0.00021 |
| hsa05016 | Huntington disease | 30 | 298 | 0.00021 |
| hsa05020 | Prion disease | 28 | 265 | 0.00021 |
| hsa05014 | Amyotrophic lateral sclerosis | 30 | 352 | 0.0011 |
| hsa01100 | Metabolic pathways | 81 | 1447 | 0.0015 |
| hsa05010 | Alzheimer disease | 28 | 355 | 0.0051 |
| hsa03050 | Proteasome | 8 | 43 | 0.0116 |
| hsa00190 | Oxidative phosphorylation | 14 | 130 | 0.0134 |
| hsa04932 | Non-alcoholic fatty liver disease | 15 | 148 | 0.0137 |
| Downregulated only in SARS-CoV-2 | ||||
| hsa01100 | Metabolic pathways | 149 | 1447 | 4.78 × 10−14 |
| hsa00280 | Valine, leucine, and isoleucine degradation | 12 | 46 | 0.0021 |
| hsa05132 | Salmonella infection | 27 | 209 | 0.0021 |
| hsa00100 | Steroid biosynthesis | 8 | 20 | 0.0025 |
| hsa00640 | Propanoate metabolism | 9 | 34 | 0.0094 |
| hsa01200 | Carbon metabolism | 17 | 117 | 0.0099 |
| hsa04723 | Retrograde endocannabinoid signaling | 19 | 145 | 0.012 |
| hsa05031 | Amphetamine addiction | 12 | 66 | 0.012 |
| hsa05110 | Vibrio cholerae infection | 10 | 48 | 0.0121 |
| hsa04071 | Sphingolipid signaling pathway | 16 | 116 | 0.0153 |
| hsa00020 | Citrate cycle (TCA cycle) | 7 | 29 | 0.036 |
| hsa00190 | Oxidative phosphorylation | 16 | 130 | 0.036 |
| hsa00340 | Histidine metabolism | 6 | 21 | 0.036 |
| hsa00620 | Pyruvate metabolism | 8 | 38 | 0.036 |
| hsa04728 | Dopaminergic synapse | 16 | 128 | 0.036 |
| hsa04730 | Long-term depression | 10 | 59 | 0.036 |
| hsa00564 | Glycerophospholipid metabolism | 13 | 97 | 0.0375 |
| hsa04015 | Rap1 signaling pathway | 21 | 202 | 0.0375 |
| hsa04962 | Vasopressin-regulated water reabsorption | 8 | 44 | 0.0478 |
| Term ID | Term Description | tRF5 Name |
|---|---|---|
| GO:0001764 | Neuron migration | tRF5-Glu-CTC-2-1 |
| GO:0007399 | Nervous system development | tRF5-Gln-CTG-2-1 |
| tRF5-Glu-CTC-2-1 | ||
| tRF5-Leu-AAG-3-1 | ||
| GO:0007417 | Central nervous system development | tRF5-Glu-CTC-2-1 |
| tRF5-Leu-AAG-3-1 | ||
| GO:0010975 | Regulation of neuron projection development | tRF5-SeC-TCA-2-1 |
| GO:0010976 | Positive regulation of neuron projection development | tRF5-Glu-CTC-2-1 |
| GO:0022008 | Neurogenesis | tRF5-Gln-CTG-2-1 |
| tRF5-Glu-CTC-2-1 | ||
| GO:0030182 | Neuron differentiation | tRF5-Gln-CTG-2-1 |
| tRF5-Glu-CTC-2-1 | ||
| tRF5-Leu-AAG-3-1 | ||
| GO:0031175 | Neuron projection development | tRF5-Leu-AAG-3-1 |
| GO:0045664 | Regulation of neuron differentiation | tRF5-Glu-CTC-2-1 |
| tRF5-SeC-TCA-2-1 | ||
| GO:0045665 | Negative regulation of neuron differentiation | tRF5-Glu-CTC-2-1 |
| GO:0045666 | Positive regulation of neuron differentiation | tRF5-Glu-CTC-2-1 |
| GO:0048666 | Neuron development | tRF5-Gln-CTG-2-1 |
| tRF5-Glu-CTC-2-1 | ||
| tRF5-Leu-AAG-3-1 | ||
| GO:0048699 | Generation of neurons | tRF5-Gln-CTG-2-1 |
| tRF5-Glu-CTC-2-1 | ||
| tRF5-Leu-AAG-3-1 | ||
| GO:0048812 | Neuron projection morphogenesis | tRF5-Leu-AAG-3-1 |
| GO:0050767 | Regulation of neurogenesis | tRF5-Glu-CTC-2-1 |
| tRF5-Leu-AAG-3-1 | ||
| GO:0050769 | Positive regulation of neurogenesis | tRF5-Glu-CTC-2-1 |
| GO:0051960 | Regulation of nervous system development | tRF5-Gln-CTG-2-1 |
| tRF5-Glu-CTC-2-1 | ||
| tRF5-Leu-AAG-3-1 | ||
| GO:0051962 | Positive regulation of nervous system development | tRF5-Glu-CTC-2-1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imirowicz, I.; Saifee, A.; Henry, L.; Tunkle, L.; Popescu, A.; Huang, P.; Jakpor, J.; Barbano, A.; Goru, R.; Gunawan, A.; et al. Unique tRNA Fragment Upregulation with SARS-CoV-2 but Not with SARS-CoV Infection. Int. J. Mol. Sci. 2024, 25, 399. https://doi.org/10.3390/ijms25010399
Imirowicz I, Saifee A, Henry L, Tunkle L, Popescu A, Huang P, Jakpor J, Barbano A, Goru R, Gunawan A, et al. Unique tRNA Fragment Upregulation with SARS-CoV-2 but Not with SARS-CoV Infection. International Journal of Molecular Sciences. 2024; 25(1):399. https://doi.org/10.3390/ijms25010399
Chicago/Turabian StyleImirowicz, Isabella, Azeem Saifee, Leanne Henry, Leo Tunkle, Alexander Popescu, Philip Huang, Jibiana Jakpor, Ava Barbano, Rohit Goru, Audrey Gunawan, and et al. 2024. "Unique tRNA Fragment Upregulation with SARS-CoV-2 but Not with SARS-CoV Infection" International Journal of Molecular Sciences 25, no. 1: 399. https://doi.org/10.3390/ijms25010399
APA StyleImirowicz, I., Saifee, A., Henry, L., Tunkle, L., Popescu, A., Huang, P., Jakpor, J., Barbano, A., Goru, R., Gunawan, A., Sicilia, M., Ono, M., Bao, X., & Lee, I. (2024). Unique tRNA Fragment Upregulation with SARS-CoV-2 but Not with SARS-CoV Infection. International Journal of Molecular Sciences, 25(1), 399. https://doi.org/10.3390/ijms25010399







