Specificity Enhancement of Glutenase Bga1903 toward Celiac Disease-Eliciting Pro-Immunogenic Peptides via Active-Site Modification
Abstract
:1. Introduction
2. Results
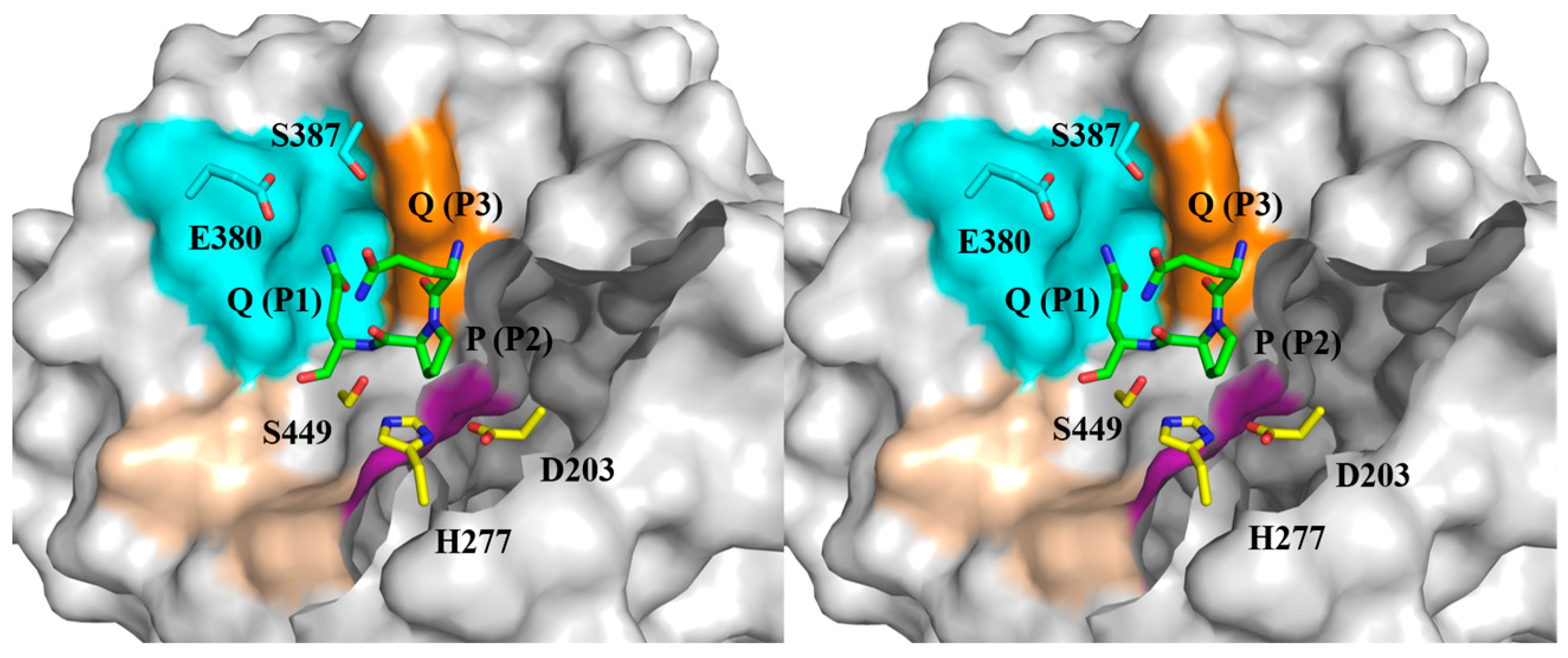
2.1. The Substrate-Binding Subsites of Bga1903
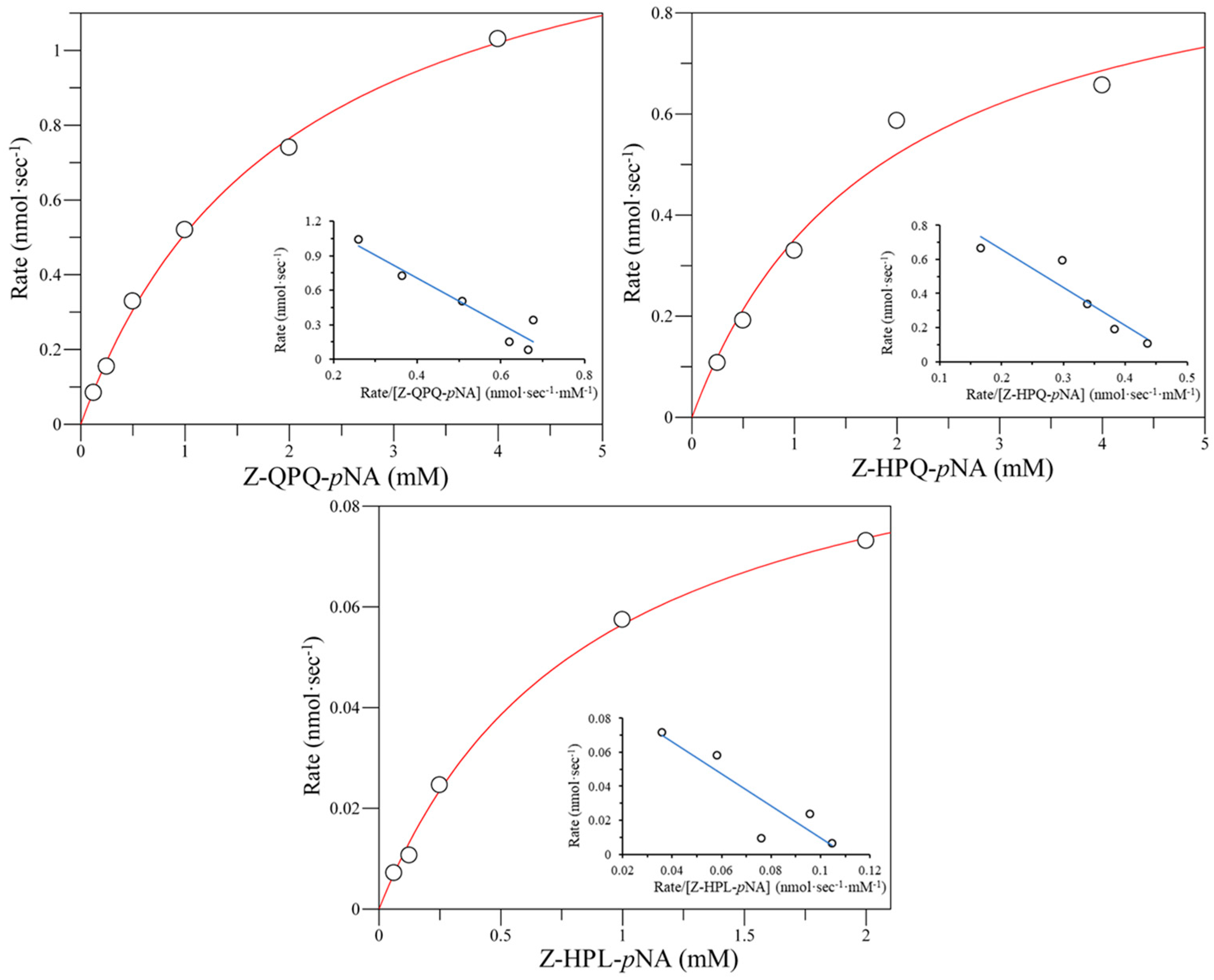
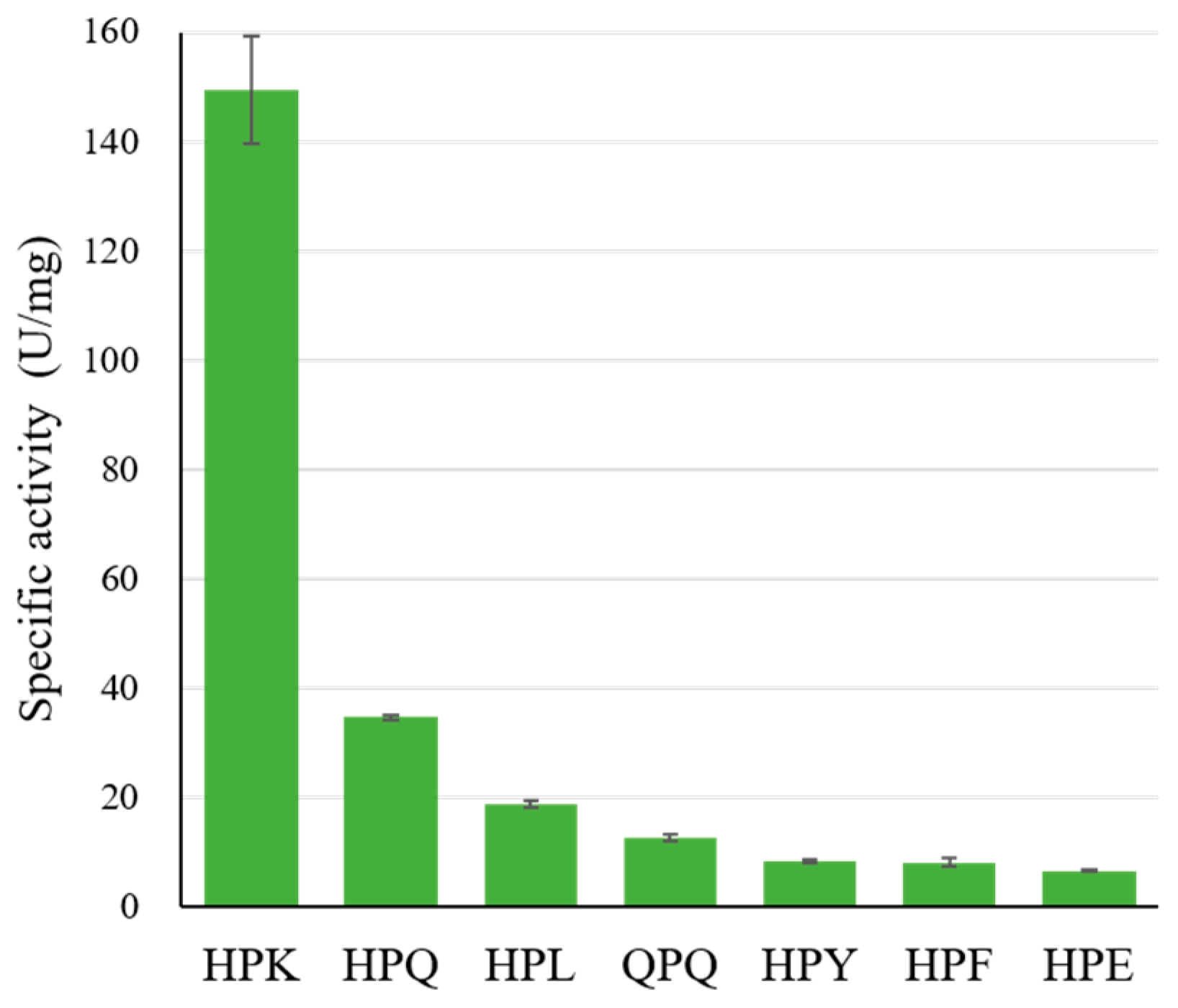
2.2. The Specific Activity of Bga1903 toward Various Chromogenic Tripeptidyl Substrates
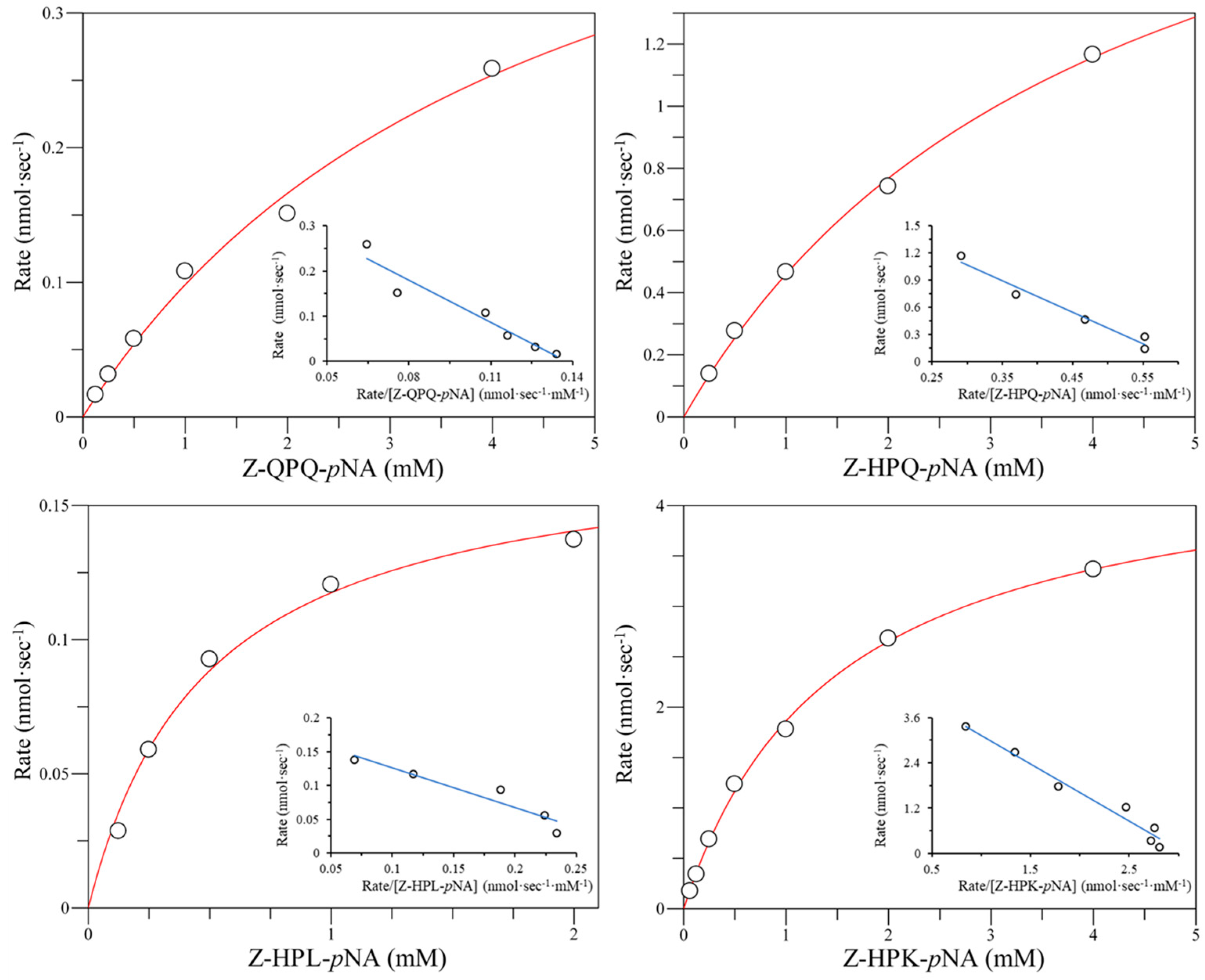
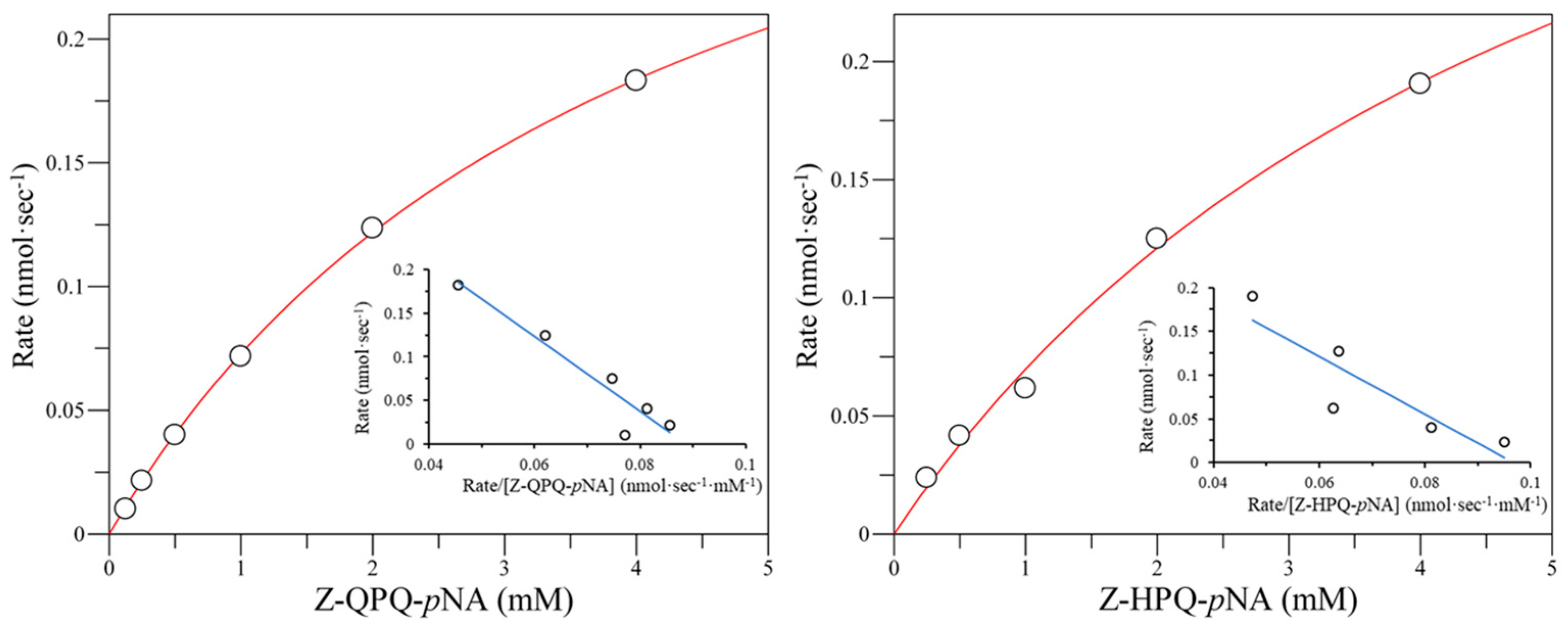
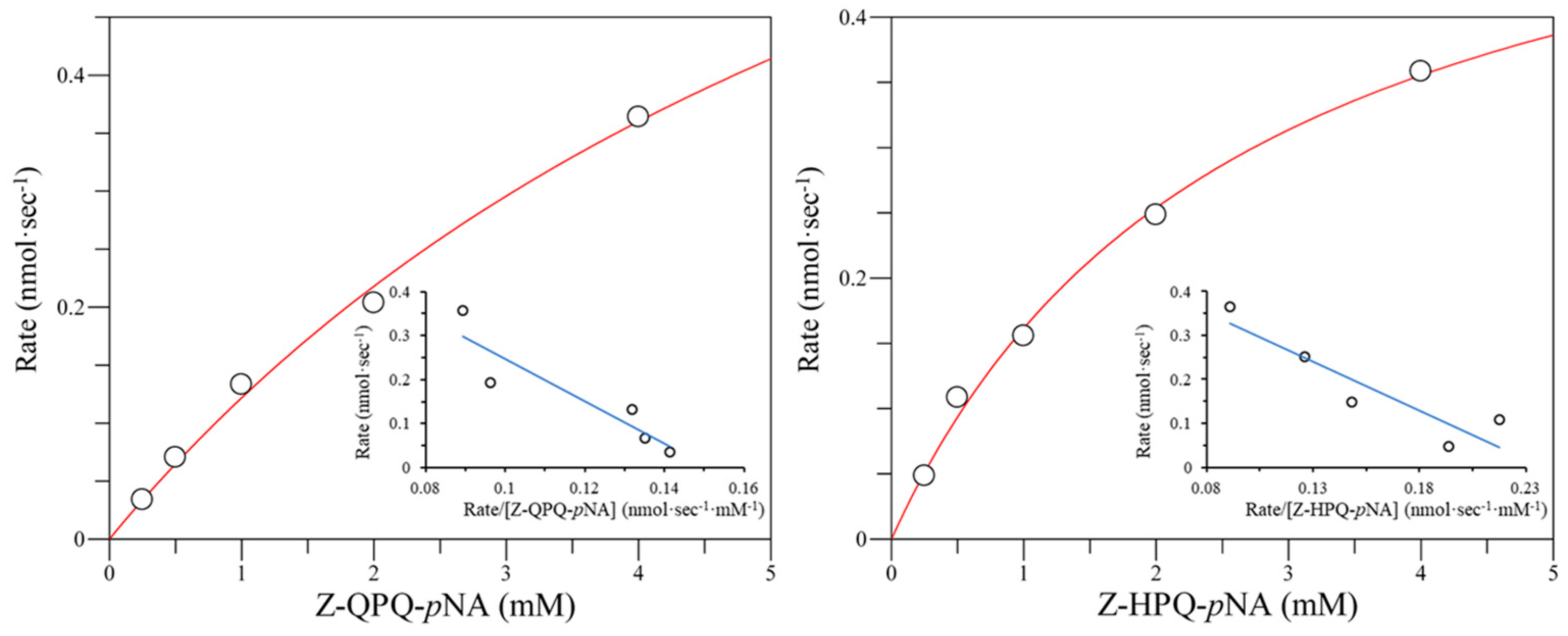
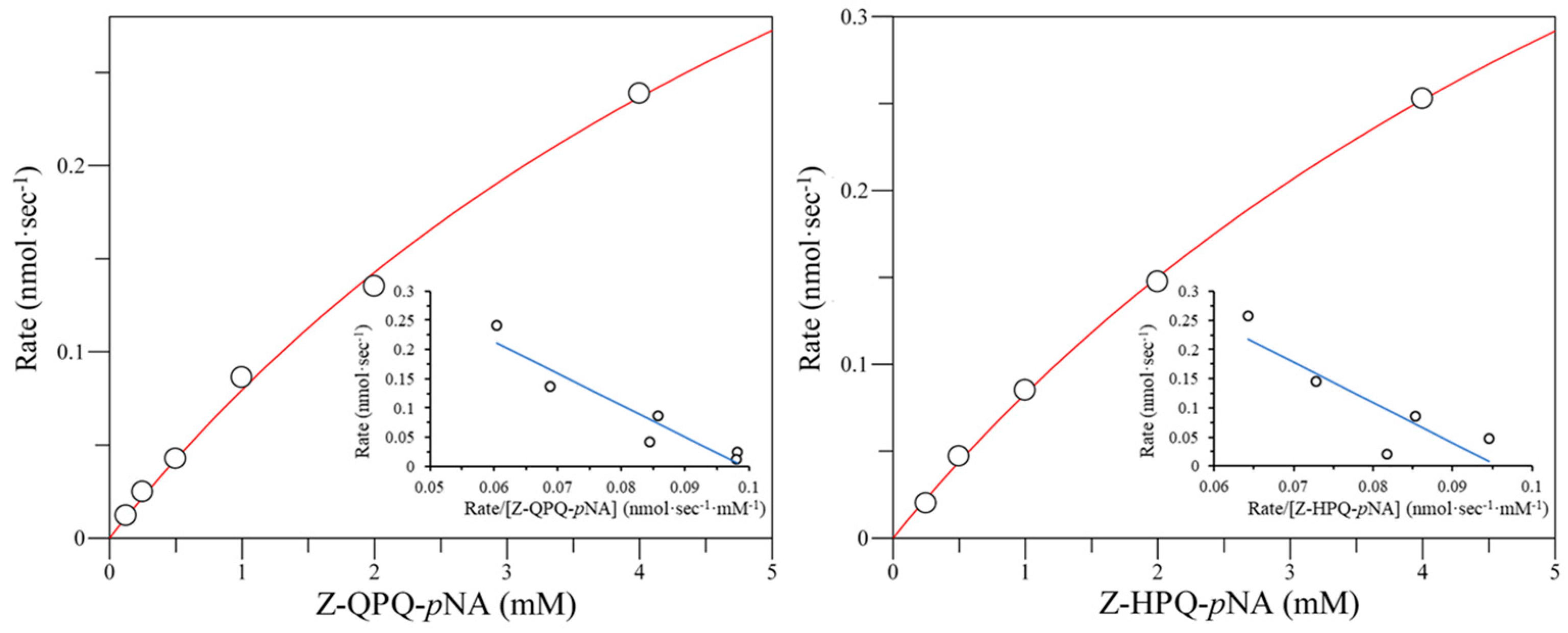
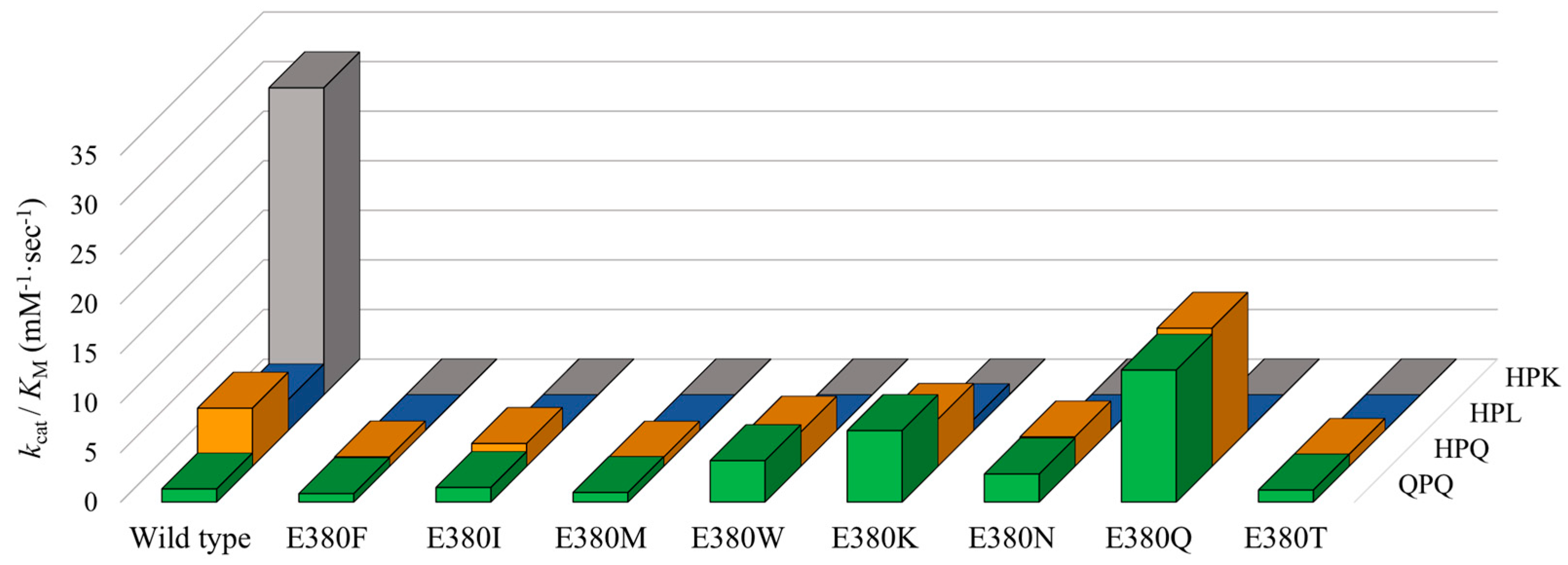
2.3. Effect of E380 Substitutions on the Enzymatic Activity of Bga1903
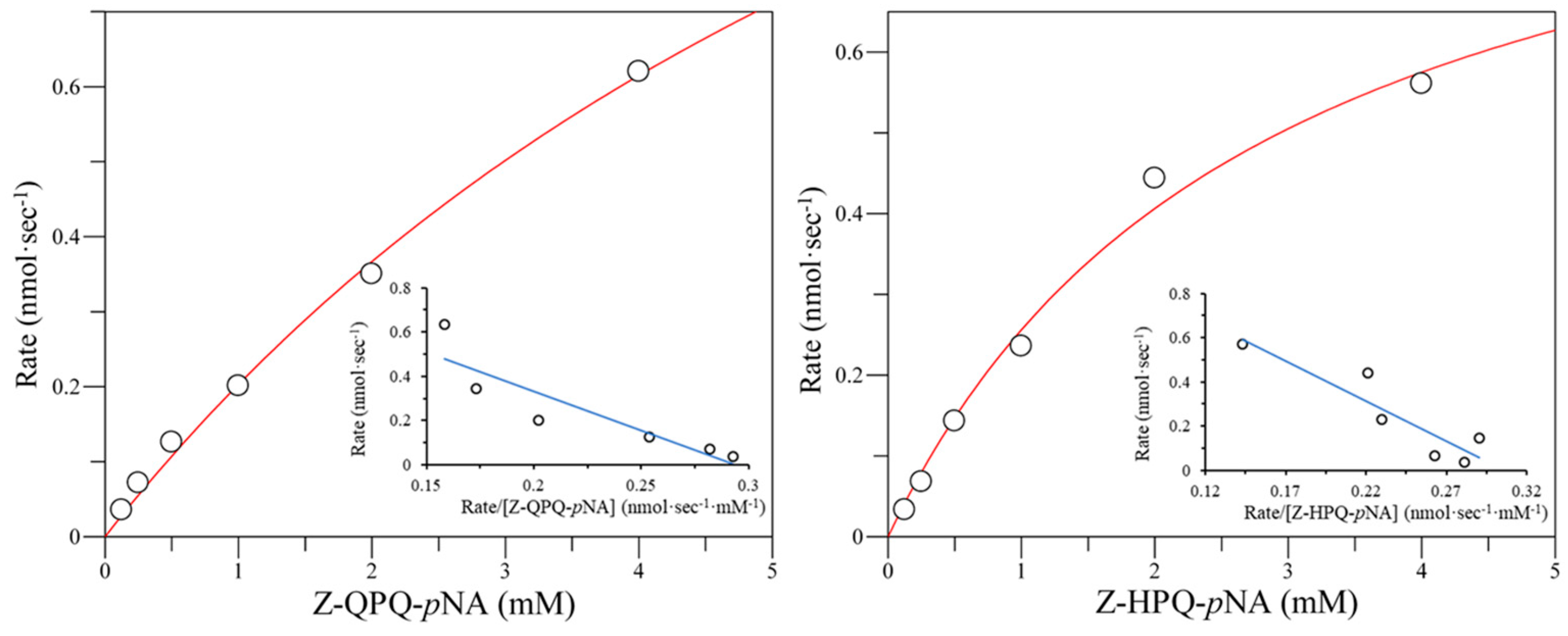
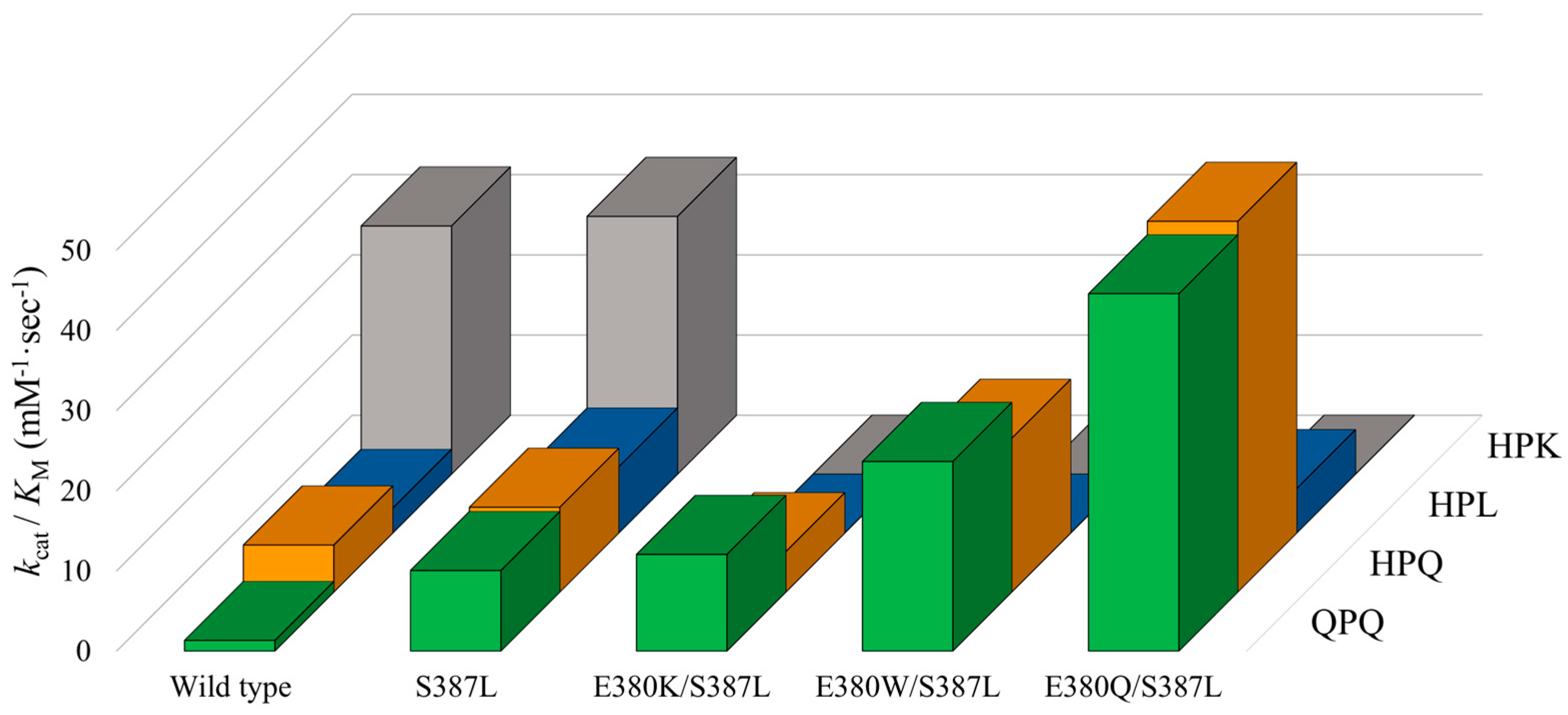
2.4. Cumulative Effect of S387L with Different E380 Mutations on the Enzymatic Activity
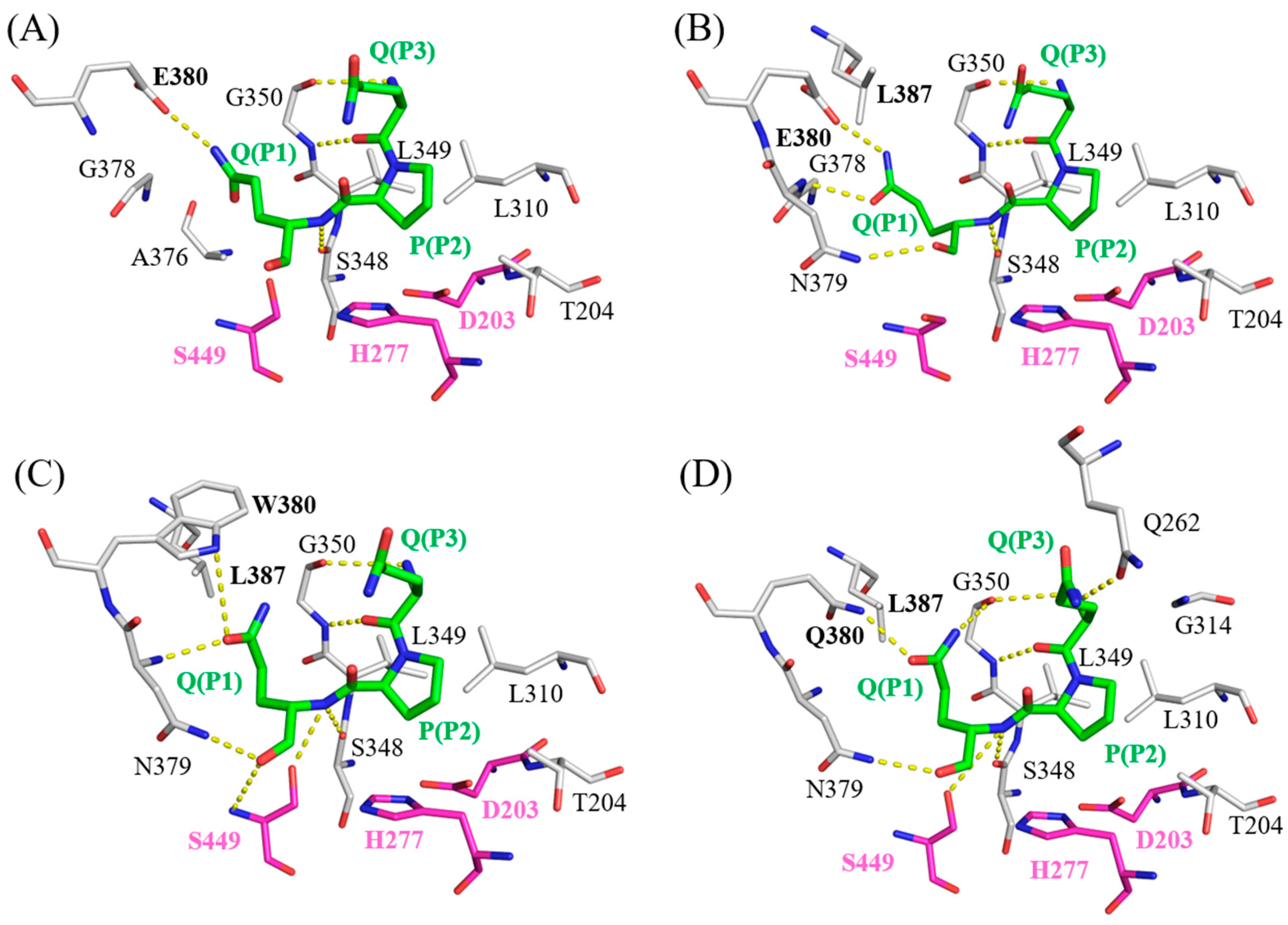
2.5. Molecular Docking Models of Bga1903s and QPQ Peptides
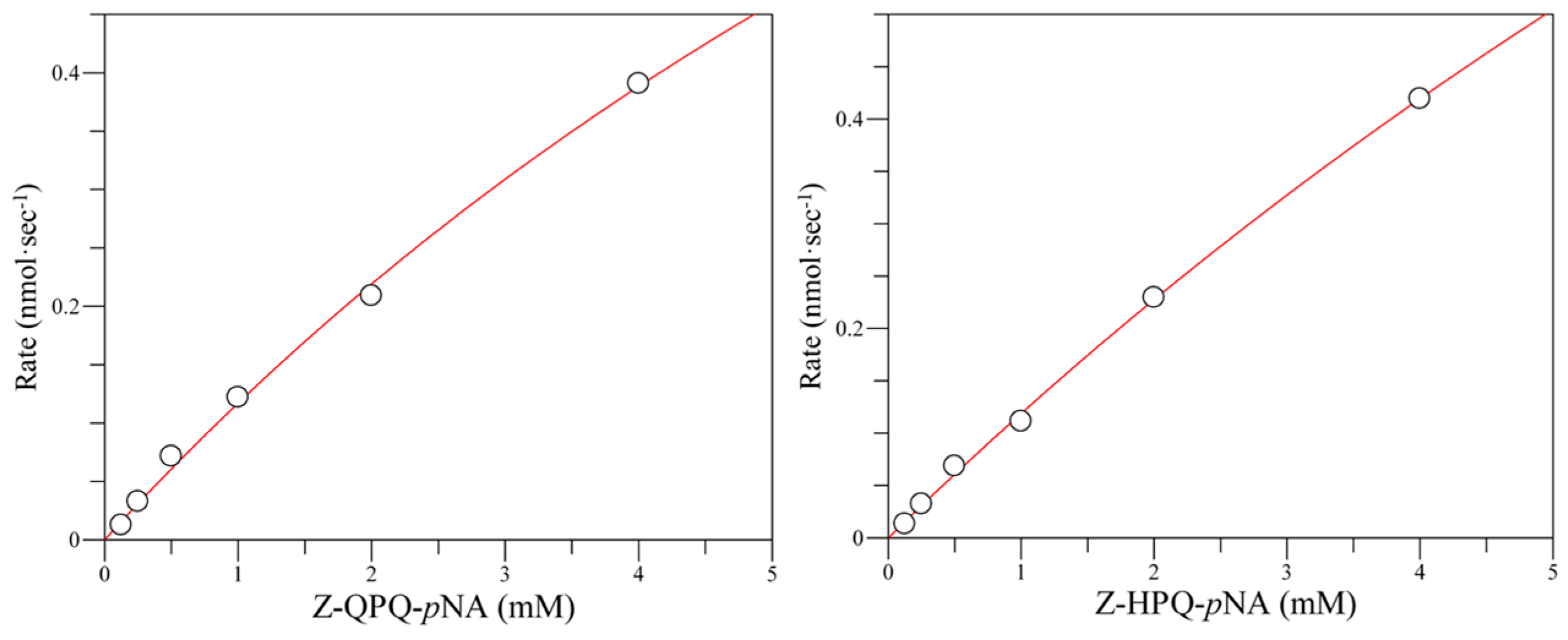
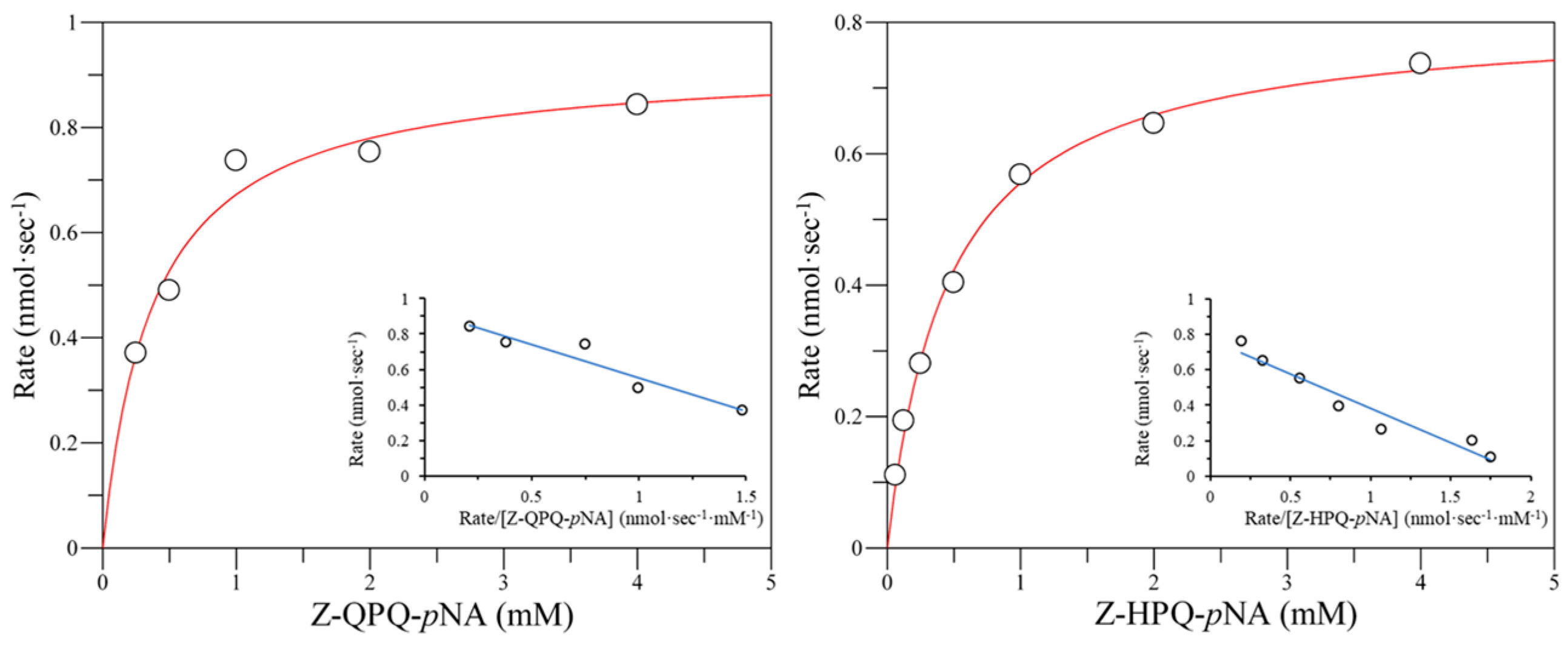
2.6. Hydrolysis of Pro-Immunogenic Peptides by Mutant E380W/S387L and E380Q/S387L
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. E. coli Strains and Site-Directed Mutagenesis
4.3. Protein Expression and Purification
4.4. Enzyme Activity Assay Utilizing Chromogenic Peptidyl Substrates
4.5. Computational Evaluation of the Interaction between Bga1903s and QPQ Peptide
4.6. Hydrolysis of the 26- and 33-mer Pro-Immunogenic Peptides
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A




References
- Dewar, D.H.; Ciclitira, P.J. Clinical features and diagnosis of celiac disease. Gastroenterology 2005, 128, S19–S24. [Google Scholar] [CrossRef] [PubMed]
- Tollefsen, S.; Arentz-Hansen, H.; Fleckenstein, B.; Molberg, Ø.; Ráki, M.; Kwok, W.W.; Jung, G.; Lundin, K.E.A.; Sollid, L.M. HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. J. Clin. Investig. 2006, 116, 2226–2236. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Arora, S.; Lal, S.; Strand, T.A.; Makharia, G.K. Risk of Celiac Disease in the First- and Second-Degree Relatives of Patients with Celiac Disease: A Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2015, 110, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Gasbarrini, A.; Cammarota, G. Endoscopic tools for the diagnosis and evaluation of celiac disease. World J. Gastroenterol. 2013, 19, 8562–8570. [Google Scholar] [CrossRef] [PubMed]
- Schwertz, E.; Kahlenberg, F.; Sack, U.; Richter, T.; Stern, M.; Conrad, K.; Zimmer, K.P.; Mothes, T. Serologic assay based on gliadin-related nonapeptides as a highly sensitive and specific diagnostic aid in celiac disease. Clin. Chem. 2004, 50, 2370–2375. [Google Scholar] [CrossRef]
- Shewry, P.R.; Tatham, A.S.; Forde, J.; Kreis, M.; Miflin, B.J. The classification and nomenclature of wheat gluten proteins: A reassessment. J. Cereal Sci. 1986, 4, 97–106. [Google Scholar] [CrossRef]
- Shan, L.; Molberg, Ø.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef]
- Shan, L.; Qiao, S.W.; Arentz-Hansen, H.; Molberg, Ø.; Gray, G.M.; Sollid, L.M.; Khosla, C. Identification and analysis of multivalent proteolytically resistant peptides from gluten: Implications for celiac sprue. J. Proteome Res. 2005, 4, 1732–1741. [Google Scholar] [CrossRef]
- Molberg, Ø.; McAdam, S.N.; Körner, R.; Quarsten, H.; Kristiansen, C.; Madsen, L.; Fugger, L.; Scott, H.; Norén, O.; Roepstorff, P.; et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 1998, 4, 713–717. [Google Scholar] [CrossRef]
- Unsworth, D.J.; Kieffer, M.; Holborow, E.J.; Coombs, R.R.; Walker-Smith, J.A. IgA anti-gliadin antibodies in coeliac disease. Clin. Exp. Immunol. 1981, 46, 286–293. [Google Scholar]
- Mankai, A.; Sakly, W.; Landolsi, H.; Gueddah, L.; Sriha, B.; Ayadi, A.; Sfar, M.T.; Skandrani, K.; Harbi, A.; Essoussi, A.S.; et al. Tissue transglutaminase antibodies in celiac disease, comparison of an enzyme linked immunosorbent assay and a dot blot assay. Pathol. Biol. 2005, 53, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Nurminskaya, M.V.; Belkin, A.M. Cellular Functions of Tissue Transglutaminase. Int. Rev. Cell Mol. Biol. 2012, 294, 1–97. [Google Scholar] [PubMed]
- Uhde, M.; Ajamian, M.; Caio, G.; De Giorgio, R.; Indart, A.; Green, P.H.; Verna, E.C.; Volta, U.; Alaedini, A. Intestinal cell damage and systemic immune activation in individuals reporting sensitivity to wheat in the absence of coeliac disease. Gut 2016, 65, 1930–1937. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Whelan, K. Limited availability and higher cost of gluten-free foods. J. Hum. Nutr. Diet. 2011, 24, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Lerner, B.A.; Phan Vo, L.T.; Yates, S.; Rundle, A.G.; Green, P.H.R.; Lebwohl, B. Detection of gluten in gluten-free labeled restaurant food: Analysis of crowd-sourced data. Am. J. Gastroenterol. 2019, 114, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Christophersen, A.; Risnes, L.F.; Dahal-Koirala, S.; Sollid, L.M. Therapeutic and Diagnostic Implications of T Cell Scarring in Celiac Disease and Beyond. Trends Mol. Med. 2019, 25, 836–852. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A. New therapeutic strategies for celiac disease. Autoimmun. Rev. 2010, 9, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Bethune, M.T.; Khosla, C. Oral enzyme therapy for celiac sprue. Methods Enzymol. 2012, 502, 241–271. [Google Scholar]
- Scherf, K.A.; Wieser, H.; Koehler, P. Novel approaches for enzymatic gluten degradation to create high-quality gluten-free products. Food Res. Int. 2018, 110, 62–72. [Google Scholar] [CrossRef]
- Caputo, I.; Lepretti, M.; Martucciello, S.; Esposito, C. Enzymatic strategies to detoxify gluten: Implications for celiac disease. Enzym. Res. 2010, 2010, 174354. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Lee, C.C.; Hsu, J.H.; Leu, W.M.; Meng, M. Efficient hydrolysis of gluten-derived celiac disease-triggering immunogenic peptides by a bacterial serine protease from Burkholderia gladioli. Biomolecules 2021, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Lin, I.C.; Chen, P.-C.; Lee, C.-C.; Meng, M. Crystal structure of a Burkholderia peptidase and modification of the substrate-binding site for enhanced hydrolytic activity toward gluten-derived pro-immunogenic peptides. Int. J. Biol. Macromol. 2022, 222, 2258–2269. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Helmerhorst, E.J.; Darwish, G.; Blumenkranz, G.; Schuppan, D. Gluten degrading enzymes for treatment of celiac disease. Nutrients 2020, 12, 2095. [Google Scholar] [CrossRef] [PubMed]
- Rey, M.; Yang, M.; Lee, L.; Zhang, Y.; Sheff, J.G.; Sensen, C.W.; Mrazek, H.; Halada, P.; Man, P.; McCarville, J.L.; et al. Addressing proteolytic efficiency in enzymatic degradation therapy for celiac disease. Sci. Rep. 2016, 6, 30980. [Google Scholar] [CrossRef] [PubMed]
- Ting, T.-Y.; Baharin, A.; Ramzi, A.B.; Ng, C.-L.; Goh, H.-H. Neprosin belongs to a new family of glutamic peptidase based on in silico evidence. Plant Physiol. Biochem. 2022, 183, 23–35. [Google Scholar] [CrossRef] [PubMed]
- del Amo-Maestro, L.; Mendes, S.R.; Rodríguez-Banqueri, A.; Garzon-Flores, L.; Girbal, M.; Rodríguez-Lagunas, M.J.; Guevara, T.; Franch, À.; Pérez-Cano, F.J.; Eckhard, U.; et al. Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy. Nat. Commun. 2022, 13, 4446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Leu, W.M.; Meng, M. Hydrolysis of Gluten-Derived Celiac Disease-Triggering Peptides across a Broad pH Range by RmuAP1: A Novel Aspartic Peptidase Isolated from Rhodotorula mucilaginosa. J. Agric. Food Chem. 2023, 71, 17202–17213. [Google Scholar] [CrossRef] [PubMed]
- Akeroyd, M.; van Zandycke, S.; den Hartog, J.; Mutsaers, J.; Edens, L.; van den Berg, M.; Christis, C. AN-PEP, Proline-Specific Endopeptidase, Degrades All Known Immunostimulatory Gluten Peptides in Beer Made from Barley Malt. J. Am. Soc. Brew. Chem. 2018, 74, 91–99. [Google Scholar] [CrossRef]
- Sestak, K.; Thwin, H.; Dufour, J.; Liu, D.X.; Alvarez, X.; Laine, D.; Clarke, A.; Doyle, A.; Aye, P.P.; Blanchard, J.; et al. Supplementation of Reduced Gluten Barley Diet with Oral Prolyl Endopeptidase Effectively Abrogates Enteropathy-Associated Changes in Gluten-Sensitive Macaques. Nutrients 2016, 8, 401. [Google Scholar] [CrossRef]
- Knorr, V.; Wieser, H.; Koehler, P. Production of gluten-free beer by peptidase treatment. Eur. Food Res. Technol. 2015, 242, 1129–1140. [Google Scholar] [CrossRef]
- Wei, G.; Tian, N.; Siezen, R.; Schuppan, D.; Helmerhorst, E.J. Identification of food-grade subtilisins as gluten-degrading enzymes to treat celiac disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, G571–G580. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.; Eliasson, M.; Uhlen, M.; Flock, J.I. Cloning sequencing and expression of subtilisin Carlsberg from Bacillus licheniformis. Nucleic Acids Res. 1985, 13, 8913–8926. [Google Scholar] [CrossRef] [PubMed]
- Darwish, G.; Helmerhorst, E.J.; Schuppan, D.; Oppenheim, F.G.; Wei, G. Pharmaceutically modified subtilisins withstand acidic conditions and effectively degrade gluten in vivo. Sci. Rep. 2019, 9, 7505. [Google Scholar] [CrossRef] [PubMed]
- Bethune, M.T.; Strop, P.; Tang, Y.; Sollid, L.M.; Khosla, C. Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem. Biol. 2006, 13, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Gass, J.; Vora, H.; Bethune, M.T.; Gray, G.M.; Khosla, C. Effect of barley endoprotease EP-B2 on gluten digestion in the intact rat. J. Pharmacol. Exp. Ther. 2006, 318, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Marti, T.; Sollid, L.M.; Gray, G.M.; Khosla, C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: Implications for coeliac sprue. Biochem. J. 2004, 383, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Siegel, M.; Garber, M.E.; Spencer, A.G.; Botwick, W.; Kumar, P.; Williams, R.N.; Kozuka, K.; Shreeniwas, R.; Pratha, V.; Adelman, D.C. Safety, tolerability, and activity of ALV003: Results from two phase 1 single, escalating-dose clinical trials. Dig. Dis. Sci. 2012, 57, 440–450. [Google Scholar] [CrossRef]
- Lähdeaho, M.L.; Kaukinen, K.; Laurila, K.; Vuotikka, P.; Koivurova, O.P.; Kärjä-Lahdensuu, T.; Marcantonio, A.; Adelman, D.C.; Mäki, M. Glutenase ALV003 attenuates gluten-induced mucosal injury in patients with celiac disease. Gastroenterology 2014, 146, 1649–1658. [Google Scholar] [CrossRef]
- Syage, J.A.; Murray, J.A.; Green, P.H.R.; Khosla, C. Latiglutenase Improves Symptoms in Seropositive Celiac Disease Patients While on a Gluten-Free Diet. Dig. Dis. Sci. 2017, 62, 2428–2432. [Google Scholar] [CrossRef]
- Murray, J.A.; Kelly, C.P.; Green, P.H.R.; Marcantonio, A.; Wu, T.-T.; Mäki, M.; Adelman, D.C.; Ansari, S.; Ayub, K.; Basile, A.; et al. No Difference Between Latiglutenase and Placebo in Reducing Villous Atrophy or Improving Symptoms in Patients with Symptomatic Celiac Disease. Gastroenterology 2017, 152, 787–798. [Google Scholar] [CrossRef]
- Murray, J.A.; Syage, J.A.; Wu, T.T.; Dickason, M.A.; Ramos, A.G.; Van Dyke, C.; Horwath, I.; Lavin, P.T.; Mäki, M.; Hujoel, I.; et al. Latiglutenase Protects the Mucosa and Attenuates Symptom Severity in Patients with Celiac Disease Exposed to a Gluten Challenge. Gastroenterology 2022, 163, 1510–1521. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.R.; Stanley, E.J.; Wolf, S.; Toland, A.; Wu, S.J.; Hadidi, D.; Mills, J.H.; Baker, D.; Pultz, I.S.; Siegel, J.B. Computational design of an alpha-gliadin peptidase. J. Am. Chem. Soc. 2012, 134, 20513–20520. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.; Siegel, J.B.; Tinberg, C.; Camarca, A.; Gianfrani, C.; Paski, S.; Guan, R.; Montelione, G.; Baker, D.; Pultz, I.S. Engineering of Kuma030: A Gliadin Peptidase That Rapidly Degrades Immunogenic Gliadin Peptides in Gastric Conditions. J. Am. Chem. Soc. 2015, 137, 13106–13113. [Google Scholar] [CrossRef] [PubMed]
- Pultz, I.S.; Hill, M.; Vitanza, J.M.; Wolf, C.; Saaby, L.; Liu, T.; Winkle, P.; Leffler, D.A. Gluten Degradation, Pharmacokinetics, Safety, and Tolerability of TAK-062, an Engineered Enzyme to Treat Celiac Disease. Gastroenterology 2021, 161, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Honorato, R.V.; Koukos, P.I.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A.M.J.J. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]

| Variants | QPQ | HPQ | HPL | HPK | ||||
|---|---|---|---|---|---|---|---|---|
| kcat a | KM | kcat | KM | kcat | KM | kcat | KM | |
| (s−1) | (mM) | (s−1) | (mM) | (s−1) | (mM) | (s−1) | (mM) | |
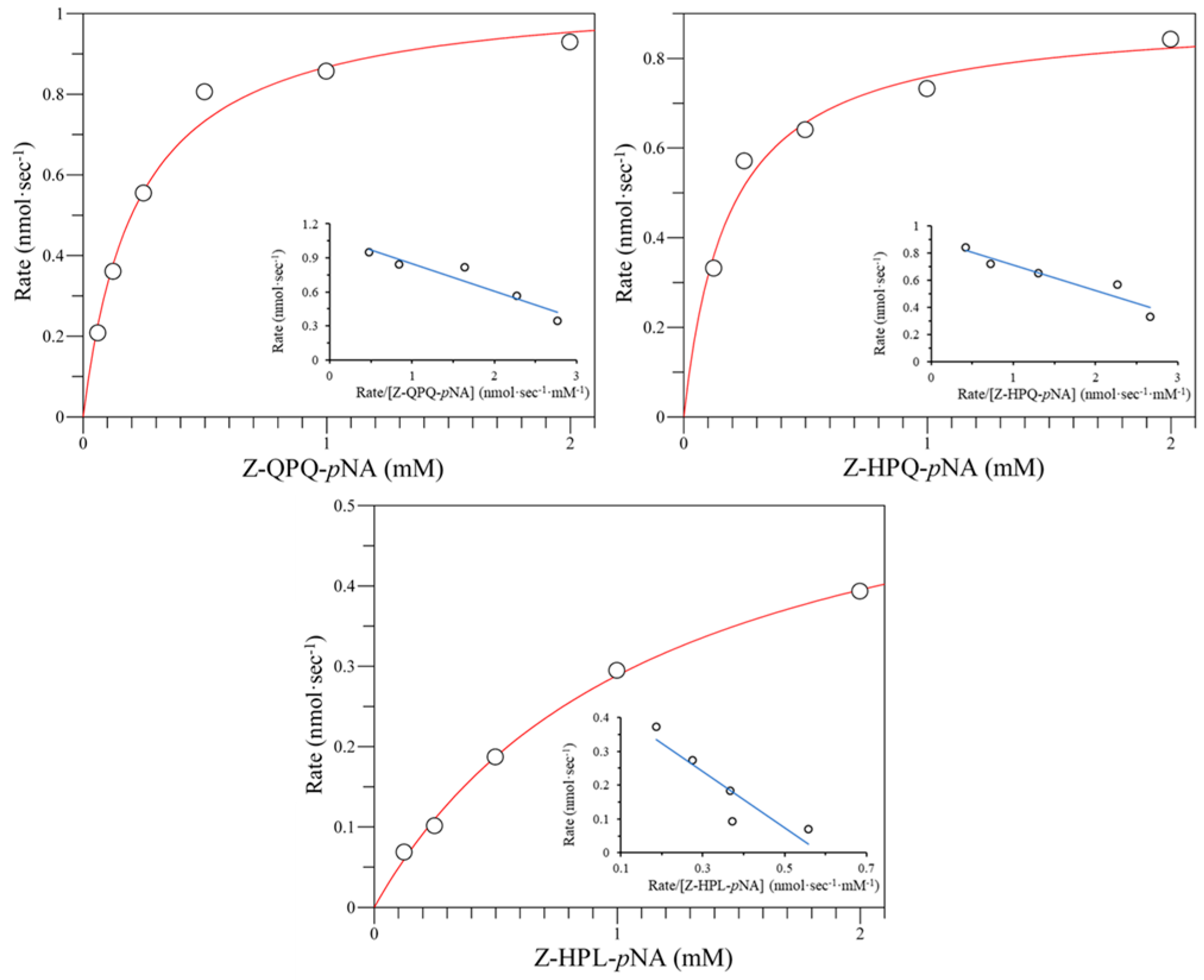
| Wild type | 4.17 ± 0.06 | 3.16 ± 0.08 | 20.43 ± 0.37 | 3.49 ± 0.09 | 1.77 ± 0.12 | 0.57 ± 0.07 | 49.90 ± 7.44 | 1.61 ± 0.40 |
| E380F | 3.69 ± 0.05 | 4.34 ± 0.06 | 2.99 ± 0.09 | 3.17 ± 0.21 | BDL b | BDL | BDL | BDL |
| E380I | 7.54 ± 0.45 | 5.16 ± 0.30 | 5.09 ± 0.14 | 2.20 ± 0.10 | BDL | BDL | BDL | BDL |
| E380M | 4.97 ± 0.38 | 5.18 ± 0.30 | 6.19 ± 0.44 | 6.72 ± 0.58 | BDL | BDL | BDL | BDL |
| E380W | 10.49 ± 0.09 | 2.51 ± 0.01 | 9.13 ± 0.40 | 2.65 ± 0.20 | BDL | BDL | BDL | BDL |
| E380K | 14.86 ± 0.28 | 2.07 ± 0.07 | 10.42 ± 0.21 | 2.21 ± 0.02 | 1.00 ± 0.01 | 0.90 ± 0.07 | BDL | BDL |
| E380N | 9.93 ± 0.12 | 3.52 ± 0.01 | 10.50 ± 0.46 | 3.55 ± 0.11 | BDL | BDL | BDL | BDL |
| E380Q | 11.75 ± 0.41 | 0.88 ± 0.02 | 9.60 ± 0.30 | 0.69 ± 0.03 | BDL | BDL | BDL | BDL |
| E380T | UD c | UD | UD | UD | BDL | BDL | BDL | BDL |
| Variants | QPQ | HPQ | HPL | HPK | ||||
|---|---|---|---|---|---|---|---|---|
| kcat a | KM | kcat | KM | kcat | KM | kcat | KM | |
| (s−1) | (mM) | (s−1) | (mM) | (s−1) | (mM) | (s−1) | (mM) | |
| Wild type | 4.17 ± 0.06 | 3.16 ± 0.08 | 20.43 ± 0.37 | 3.49 ± 0.09 | 1.77 ± 0.12 | 0.57 ± 0.07 | 49.90 ± 7.44 | 1.61 ± 0.40 |
| S387L | 10.17 ± 0.15 | 1.01 ± 0.05 | 9.68 ± 0.42 | 0.92 ± 0.07 | 7.39 ± 0.14 | 0.90 ± 0.05 | 22.50 ± 1.04 | 0.70 ± 0.02 |
| E380K/S387L | 7.20 ± 0.09 | 0.60 ± 0.03 | 5.83 ± 0.33 | 1.16 ± 0.11 | BDL b | BDL | BDL | BDL |
| E380W/S387L | 8.92 ± 0.06 | 0.38 ± 0.01 | 7.42 ± 0.10 | 0.39 ± 0.03 | BDL | BDL | BDL | BDL |
| E380Q/S387L | 10.36 ± 0.16 | 0.23 ± 0.01 | 8.67 ± 0.11 | 0.19 ± 0.02 | 5.47 ± 0.79 | 0.99 ± 0.19 | BDL | BDL |
| Primer Name | Sequence (5′→3′) |
|---|---|
| 1903+ E380-R | GGCCACAACCACGGTGGCACCTTTGGC |
| 1903+ E380I-F | GCCGGTAATATTGCGAGCCCGGTTAGC |
| 1903+ E380K-F | GCCGGTAATAAAGCGAGCCCGGTTAGC |
| 1903+ E380M-F | GCCGGTAATATGGCGAGCCCGGTTAGC |
| 1903+ E380N-F | GCCGGTAATAACGCGAGCCCGGTTAGC |
| 1903+ E380Q-F | GCCGGTAATCAGGCGAGCCCGGTTAGC |
| 1903+ E380T-F | GCCGGTAATACCGCGAGCCCGGTTAGC |
| 1903+ E380W-F | GCCGGTAATTGGGCGAGCCCGGTTAGC |
| 1903+ E380Y-F | GCCGGTAATTATGCGAGCCCGGTTAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.-Y.; Ye, R.-L.; Meng, M. Specificity Enhancement of Glutenase Bga1903 toward Celiac Disease-Eliciting Pro-Immunogenic Peptides via Active-Site Modification. Int. J. Mol. Sci. 2024, 25, 505. https://doi.org/10.3390/ijms25010505
Liu Y-Y, Ye R-L, Meng M. Specificity Enhancement of Glutenase Bga1903 toward Celiac Disease-Eliciting Pro-Immunogenic Peptides via Active-Site Modification. International Journal of Molecular Sciences. 2024; 25(1):505. https://doi.org/10.3390/ijms25010505
Chicago/Turabian StyleLiu, Yu-You, Rui-Ling Ye, and Menghsiao Meng. 2024. "Specificity Enhancement of Glutenase Bga1903 toward Celiac Disease-Eliciting Pro-Immunogenic Peptides via Active-Site Modification" International Journal of Molecular Sciences 25, no. 1: 505. https://doi.org/10.3390/ijms25010505
APA StyleLiu, Y.-Y., Ye, R.-L., & Meng, M. (2024). Specificity Enhancement of Glutenase Bga1903 toward Celiac Disease-Eliciting Pro-Immunogenic Peptides via Active-Site Modification. International Journal of Molecular Sciences, 25(1), 505. https://doi.org/10.3390/ijms25010505








