Exploring the Multiple Roles of Notch1 in Biological Development: An Analysis and Study Based on Phylogenetics and Transcriptomics
Abstract
1. Introduction
2. Results
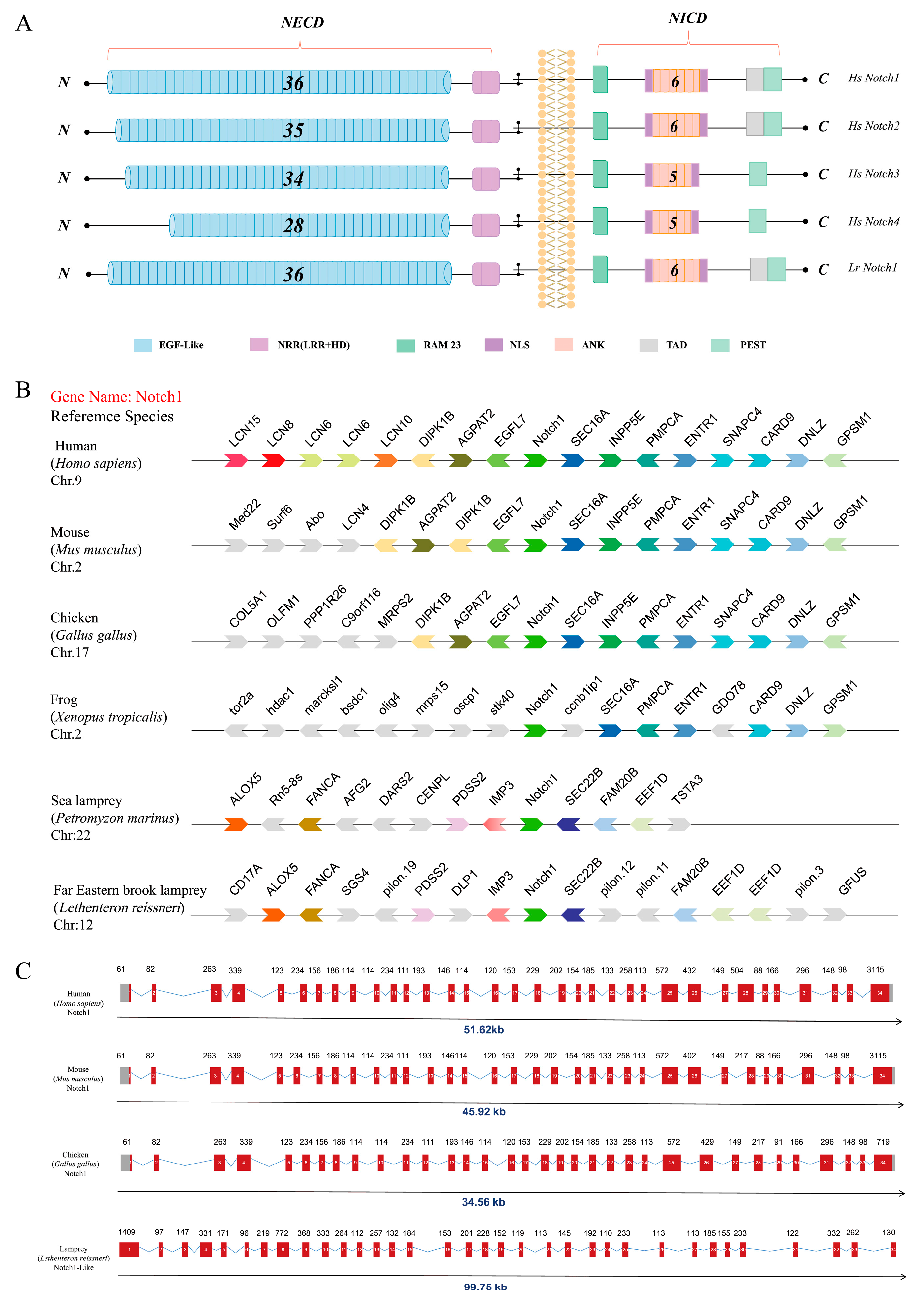
2.1. Phylogenetic Analysis of Notch1
2.1.1. Identification and Evolutionary Analysis of the Notch Members in the Genome Database of the Notch Gene Family
2.1.2. Analysis of Collinearity and Gene Structure of the Notch1 Gene in the Lamprey
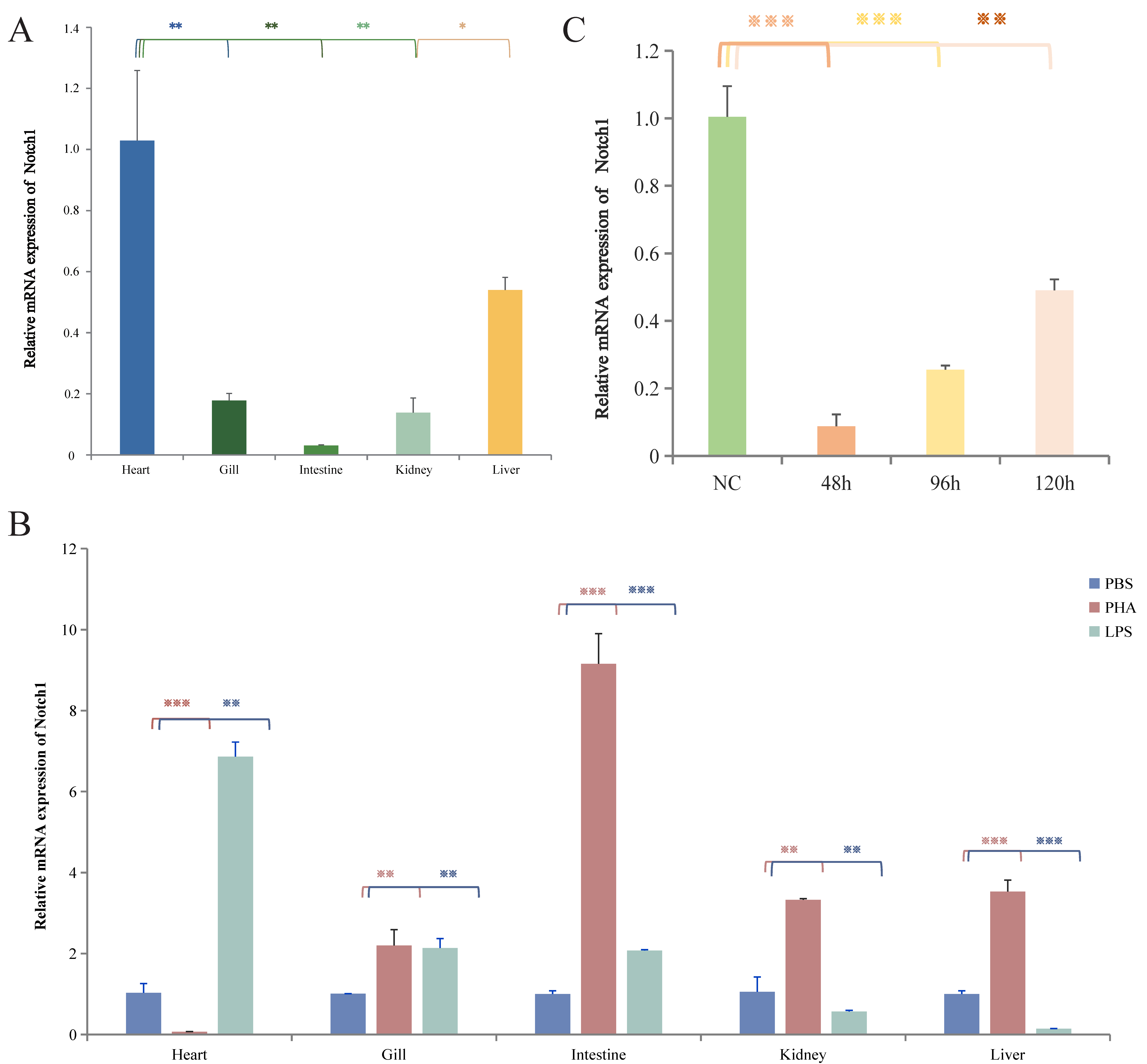
2.2. Tissue Distribution, Immune Response, and siRNA-Mediated Specific Gene Silencing
2.2.1. Distribution of Lr. Notch1 in Various Tissues of L. reissneri and siRNA-Mediated Specific Gene Silencing
2.2.2. The Immune Response to Various Stimuli Mediated by Lr. Notch1
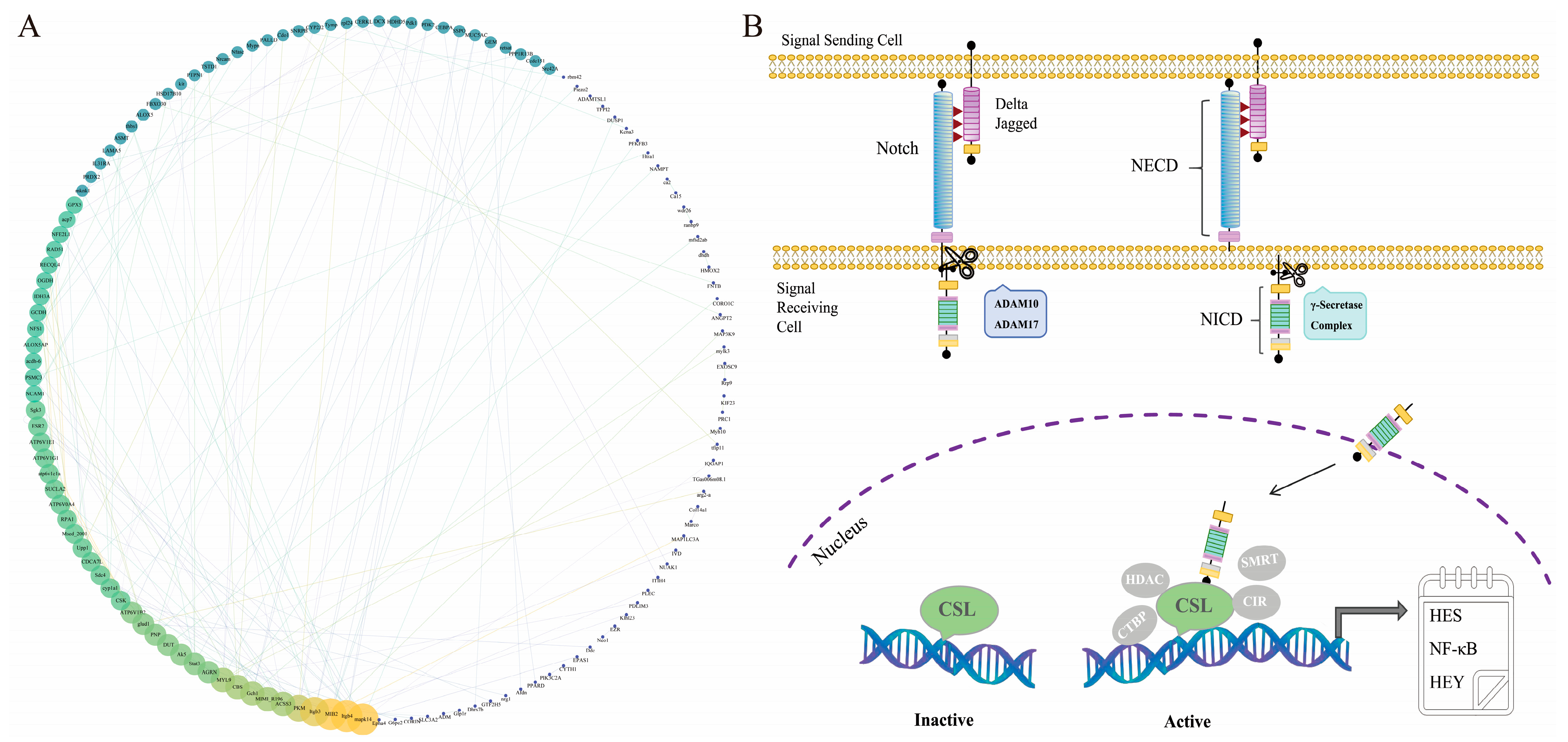
2.3. Analysis of Transcriptional Data from Lr. Notch1 Gene Silencing
2.3.1. Enrichment Analysis of Differentially Expressed Genes
2.3.2. Validation of RNA-Seq Results by qPCR
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. Multiple Sequence Alignment and Phylogenetic Analysis
4.3. Conservation Domain Prediction, Tertiary Structure Prediction
4.4. Gene Structure, Collinearity, and Motif Analysis
4.5. Real-Time Quantitative Polymerase Chain Reaction (qPCR) and Tissue Distribution Analysis, as Well as Determination of Immune Response to Different Stimuli
4.6. siRNA Target Design, Injection, and Silencing Validation
4.7. Transcriptome Sequencing and Data Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Penton, A.L.; Leonard, L.D.; Spinner, N.B. Notch signaling in human development and disease. Semin. Cell Dev. Biol. 2012, 23, 450–457. [Google Scholar] [CrossRef]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef]
- Iso, T.; Kedes, L.; Hamamori, Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 2003, 194, 237–255. [Google Scholar] [CrossRef]
- Williams, C.K.; Li, J.L.; Murga, M.; Harris, A.L.; Tosato, G. Up-regulation of the Notch ligand Delta-like 4 inhibits VEGF-induced endothelial cell function. Blood 2006, 107, 931–939. [Google Scholar] [CrossRef]
- Hirose, H.; Ishii, H.; Mimori, K.; Ohta, D.; Ohkuma, M.; Tsujii, H.; Saito, T.; Sekimoto, M.; Doki, Y.; Mori, M. Notch pathway as candidate therapeutic target in Her2/Neu/ErbB2 receptor-negative breast tumors. Oncol. Rep. 2010, 23, 35–43. [Google Scholar]
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef]
- Borggrefe, T.; Oswald, F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 2009, 66, 1631–1646. [Google Scholar] [CrossRef]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef]
- Pibiri, M.; Simbula, G. Role of the Hippo pathway in liver regeneration and repair: Recent advances. Inflamm. Regen. 2022, 42, 59. [Google Scholar] [CrossRef]
- Li, L.; Tang, P.; Li, S.; Qin, X.; Yang, H.; Wu, C.; Liu, Y. Notch signaling pathway networks in cancer metastasis: A new target for cancer therapy. Med. Oncol. 2017, 34, 180. [Google Scholar] [CrossRef]
- Abby, E.; Dentro, S.C.; Hall, M.W.J.; Fowler, J.C.; Ong, S.H.; Sood, R.; Herms, A.; Piedrafita, G.; Abnizova, I.; Siebel, C.W.; et al. Notch1 mutations drive clonal expansion in normal esophageal epithelium but impair tumor growth. Nat. Genet. 2023, 55, 232–245. [Google Scholar] [CrossRef]
- Yokoyama, A.; Kakiuchi, N.; Yoshizato, T.; Nannya, Y.; Suzuki, H.; Takeuchi, Y.; Shiozawa, Y.; Sato, Y.; Aoki, K.; Kim, S.K.; et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 2019, 565, 312–317. [Google Scholar] [CrossRef]
- Martincorena, I.; Fowler, J.C.; Wabik, A.; Lawson, A.R.; Abascal, F.; Hall, M.W.; Cagan, A.; Murai, K.; Mahbubani, K.; Stratton, M.R.; et al. Somatic mutant clones colonize the human esophagus with age. Science 2018, 362, 911–917. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Chen, X. NOTCH and Esophageal Squamous Cell Carcinoma. Adv. Exp. Med. Biol. 2021, 1287, 59–68. [Google Scholar]
- Xiu, M.X.; Liu, Y.M. The role of oncogenic Notch2 signaling in cancer: A novel therapeutic target. Am. J. Cancer Res. 2019, 9, 837–854. [Google Scholar]
- Motooka, Y.; Fujino, K.; Sato, Y.; Kudoh, S.; Suzuki, M.; Ito, T. Pathobiology of Notch2 in lung cancer. Pathology 2017, 49, 486–493. [Google Scholar] [CrossRef]
- Deng, J.; Liu, A.D.; Hou, G.Q.; Zhang, X.; Ren, K.; Chen, X.-Z.; Li, S.S.C.; Wu, Y.-S.; Cao, X. N-acetylcysteine decreases malignant characteristics of glioblastoma cells by inhibiting Notch2 signaling. J. Exp. Clin. Cancer Res. 2019, 38, 2. [Google Scholar] [CrossRef]
- Hu, Z.I.; Bendell, J.C.; Bullock, A.; LoConte, N.K.; Hatoum, H.; Ritch, P.; Hool, H.; Leach, J.W.; Sanchez, J.; Sohal, D.P.S.; et al. A randomized phase II trial of nab-paclitaxel and gemcitabine with tarextumab or placebo in patients with untreated metastatic pancreatic cancer. Cancer Med. 2019, 8, 5148–5157. [Google Scholar] [CrossRef]
- Xiu, M.; Zeng, X.; Shan, R.; Wen, W.; Li, J.; Wan, R. Targeting Notch4 in Cancer: Molecular Mechanisms and Therapeutic Perspectives. Cancer Manag. Res. 2021, 13, 7033–7045. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Li, J.; Pang, Y.; Li, Q.W. Lamprey: An important animal model of evolution and disease research. Yi Chuan 2020, 42, 847–857. [Google Scholar]
- Kasamatsu, J.; Sutoh, Y.; Fugo, K.; Otsuka, N.; Iwabuchi, K.; Kasahara, M. Identification of a third variable lymphocyte receptor in the lamprey. Proc. Natl. Acad. Sci. USA 2010, 107, 14304–14308. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Iyer, L.M.; Liang, L.; Glazko, G.V.; Liston, V.G.; Pavlov, Y.I.; Aravind, L.; Pancer, Z. Evolution and diversification of lamprey antigen receptors: Evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat. Immunol. 2007, 8, 647–656. [Google Scholar] [CrossRef]
- Pancer, Z.; Amemiya, C.T.; Ehrhardt, G.R.; Ceitlin, J.; Gartland, G.L.; Cooper, M.D. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 2004, 430, 174–180. [Google Scholar] [CrossRef]
- Han, B.W.; Herrin, B.R.; Cooper, M.D.; Wilson, I.A. Antigen recognition by variable lymphocyte receptors. Science 2008, 321, 1834–1837. [Google Scholar] [CrossRef]
- Hirano, M.; Das, S.; Guo, P.; Cooper, M.D. The evolution of adaptive immunity in vertebrates. Adv. Immunol. 2011, 109, 125–157. [Google Scholar]
- Boehm, T.; McCurley, N.; Sutoh, Y.; Schorpp, M.; Kasahara, M.; Cooper, M.D. VLR-based adaptive immunology. Annu. Rev. Immunol. 2012, 30, 203–220. [Google Scholar] [CrossRef]
- Das, S.; Hirano, M.; Aghaallaei, N.; Bajoghli, B.; Boehm, T.; Cooper, M.D. Organization of lamprey variable lymphocyte receptor C locus and repertoire development. Proc. Natl. Acad. Sci. USA 2013, 110, 6043–6048. [Google Scholar] [CrossRef]
- Das, S.; Boehm, T.; Holland, S.J.; Rast, J.P.; Fontenla-Iglesias, F.; Morimoto, R.; Valadez, J.G.; Heimroth, R.D.; Hirano, M.; Cooper, M.D. Evolution of two distinct variable lymphocyte receptors in lampreys: VLRD and VLRE. Cell Rep. 2023, 42, 112933. [Google Scholar] [CrossRef]
- Boehm, T.; Hirano, M.; Holland, S.J.; Das, S.; Schorpp, M.; Cooper, M.D. Evolution of Alternative Adaptive Immune Systems in Vertebrates. Annu. Rev. Immunol. 2018, 36, 19–42. [Google Scholar] [CrossRef]
- Zhu, T.; Li, Y.; Pang, Y.; Han, Y.; Li, J.; Wang, Z.; Liu, X.; Li, H.; Hua, Y.; Jiang, H.; et al. Chromosome-level genome assembly of L. reissneri provides insights into lamprey evolution. Mol. Ecol. Resour. 2021, 21, 448–463. [Google Scholar] [CrossRef]
- Beatus, P.; Lundkvist, J.; Oberg, C.; Pedersen, K.; Lendahl, U. The origin of the ankyrin repeat region in Notch intracellular domains is critical for regulation of HES promoter activity. Mech. Dev. 2001, 104, 3–20. [Google Scholar] [CrossRef]
- Simón, R.; Díaz-Rosales, P.; Tafalla, C. The Ancient Cytokine BAFF- and APRIL-like Molecule Regulates the Functionality of Teleost IgM+ B Cells Similarly to BAFF and APRIL. J. Immunol. 2021, 206, 1765–1775. [Google Scholar] [CrossRef]
- Ye, S.; Wang, C.; Xu, Z.; Lin, H.; Wan, X.; Yu, Y.; Adhicary, S.; Zhang, J.Z.; Zhou, Y.; Liu, C.; et al. Impaired Human Cardiac Cell Development due to NOTCH1 Deficiency. Circ. Res. 2023, 132, 187–204. [Google Scholar] [CrossRef]
- Kokubo, H.; Miyagawa-Tomita, S.; Johnson, R.L. Hesr, a mediator of the Notch signaling, functions in heart and vessel development. Trends Cardiovasc. Med. 2005, 15, 190–194. [Google Scholar] [CrossRef]
- Zhang, B.; Elmabsout, A.A.; Khalaf, H.; Basic, V.T.; Jayaprakash, K.; Kruse, R.; Bengtsson, T.; Sirsjö, A. The periodontal pathogen Porphyromonas gingivalis changes the gene expression in vascular smooth muscle cells involving the TGFbeta/Notch signalling pathway and increased cell proliferation. BMC Genom. 2013, 14, 770. [Google Scholar] [CrossRef]
- Yi, L.; Zhou, X.; Li, T.; Liu, P.; Hai, L.; Tong, L.; Ma, H.; Tao, Z.; Xie, Y.; Zhang, C.; et al. Notch1 signaling pathway promotes invasion, self-renewal and growth of glioma initiating cells via modulating chemokine system CXCL12/CXCR4. J. Exp. Clin. Cancer Res. 2019, 38, 339. [Google Scholar] [CrossRef]
- Kleinjan, A.; Tindemans, I.; Montgomery, J.E.; Lukkes, M.; de Bruijn, M.J.; van Nimwegen, M.; Bergen, I.; Moellering, R.E.; Hoogsteden, H.C.; Boon, L.; et al. The Notch pathway inhibitor stapled alpha-helical peptide derived from mastermind-like 1 (SAHM1) abrogates the hallmarks of allergic asthma. J. Allergy Clin. Immunol. 2018, 142, 76–85.e8. [Google Scholar] [CrossRef]
- Palaga, T.; Buranaruk, C.; Rengpipat, S.; Fauq, A.H.; Golde, T.E.; Kaufmann, S.H.E.; Osborne, B.A. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur. J. Immunol. 2008, 38, 174–183. [Google Scholar] [CrossRef]
- Atanga, R.; Romero, A.S.; Hernandez, A.J.; Peralta-Herrera, E.; Merkley, S.D.; In, J.G.; Castillo, E.F. Inflammatory macrophages prevent colonic goblet and enteroendocrine cell differentiation through Notch signaling. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.; Chi, C.; Xu, Y. Cross-talk between Notch and LPS-TLR4-NF-κB inflammatory signaling pathways in LPS-activated HepG2 cells. J. Prac. Hepatol. 2019, 22, 470–473. [Google Scholar]
- Guo, P.; Hirano, M.; Herrin, B.R.; Li, J.; Yu, C.; Sadlonova, A.; Cooper, M.D. Dual nature of the adaptive immune system in lampreys. Nature 2009, 459, 796–801, Erratum in Nature 2009, 460, 1044. [Google Scholar] [CrossRef]
- Zhang, C.; Berndt-Paetz, M.; Neuhaus, J. A Comprehensive Bioinformatics Analysis of Notch Pathways in Bladder Cancer. Cancers 2021, 13, 3089. [Google Scholar] [CrossRef]
- Jelloul, F.Z.; Yang, R.; Garces, S.; Kanagal-Shamanna, R.; Ok, C.Y.; Loghavi, S.; Routbort, M.J.; Zuo, Z.; Yin, C.C.; Floyd, K.; et al. Landscape of NOTCH1 mutations and co-occurring biomarker alterations in chronic lymphocytic leukemia. Leuk. Res. 2022, 116, 106827. [Google Scholar] [CrossRef]
- Brechbiel, J.; Miller-Moslin, K.; Adjei, A.A. Crosstalk between hedgehog and other signaling pathways as a basis for combination therapies in cancer. Cancer Treat. Rev. 2014, 40, 750–759. [Google Scholar] [CrossRef]
- Zhou, W.W.; Dai, C.; Liu, W.Z.; Zhang, C.; Zhang, Y.; Yang, G.S.; Guo, Q.H.; Li, S.; Yang, H.X.; Li, A.Y. Gentianella acuta improves TAC-induced cardiac remodelling by regulating the Notch and PI3K/Akt/FOXO1/3 pathways. Biomed. Pharmacother. 2022, 154, 113564. [Google Scholar] [CrossRef]
- Garcia-Lezana, T.; Lopez-Canovas, J.L.; Villanueva, A. Signaling pathways in hepatocellular carcinoma. Adv. Cancer Res. 2021, 149, 63–101. [Google Scholar]
- Morimoto, R.; O’Meara, C.P.; Holland, S.J.; Trancoso, I.; Souissi, A.; Schorpp, M.; Vassaux, D.; Iwanami, N.; Giorgetti, O.B.; Evanno, G.; et al. Cytidine deaminase 2 is required for VLRB antibody gene assembly in lampreys. Sci. Immunol. 2020, 5, eaba0925. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Yan, Z.; Pang, Y.; Jiang, Y.; Zhuang, R.; Zhang, S.; Nurmamat, A.; Xiu, M.; Li, D.; Zhao, L.; et al. Exploring the Multiple Roles of Notch1 in Biological Development: An Analysis and Study Based on Phylogenetics and Transcriptomics. Int. J. Mol. Sci. 2024, 25, 611. https://doi.org/10.3390/ijms25010611
Zhou Y, Yan Z, Pang Y, Jiang Y, Zhuang R, Zhang S, Nurmamat A, Xiu M, Li D, Zhao L, et al. Exploring the Multiple Roles of Notch1 in Biological Development: An Analysis and Study Based on Phylogenetics and Transcriptomics. International Journal of Molecular Sciences. 2024; 25(1):611. https://doi.org/10.3390/ijms25010611
Chicago/Turabian StyleZhou, Yuesi, Zihao Yan, Ya Pang, Yao Jiang, Ruyu Zhuang, Shuyuan Zhang, Ayqeqan Nurmamat, Min Xiu, Ding Li, Liang Zhao, and et al. 2024. "Exploring the Multiple Roles of Notch1 in Biological Development: An Analysis and Study Based on Phylogenetics and Transcriptomics" International Journal of Molecular Sciences 25, no. 1: 611. https://doi.org/10.3390/ijms25010611
APA StyleZhou, Y., Yan, Z., Pang, Y., Jiang, Y., Zhuang, R., Zhang, S., Nurmamat, A., Xiu, M., Li, D., Zhao, L., Liu, X., Li, Q., & Han, Y. (2024). Exploring the Multiple Roles of Notch1 in Biological Development: An Analysis and Study Based on Phylogenetics and Transcriptomics. International Journal of Molecular Sciences, 25(1), 611. https://doi.org/10.3390/ijms25010611





