Pyrosequencing Analysis of O-6-Methylguanine-DNA Methyltransferase Methylation at Different Cut-Offs of Positivity Associated with Treatment Response and Disease-Specific Survival in Isocitrate Dehydrogenase-Wildtype Grade 4 Glioblastoma
Abstract
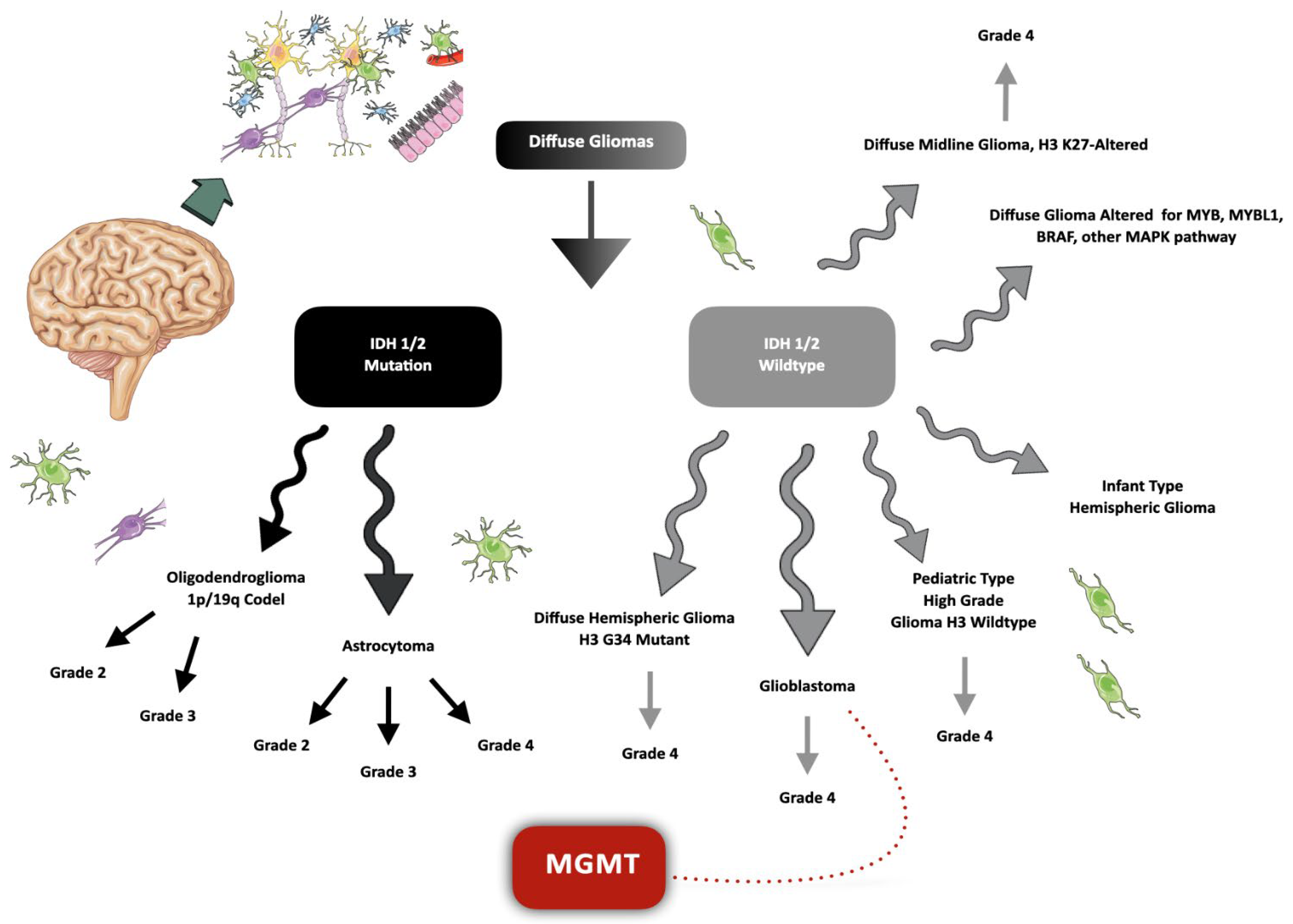
:1. Introduction
2. Results
2.1. Sample Description
2.2. Association of Clinicopathological Parameters with MGMT Methylation Status
2.3. MGMT Methylation Status
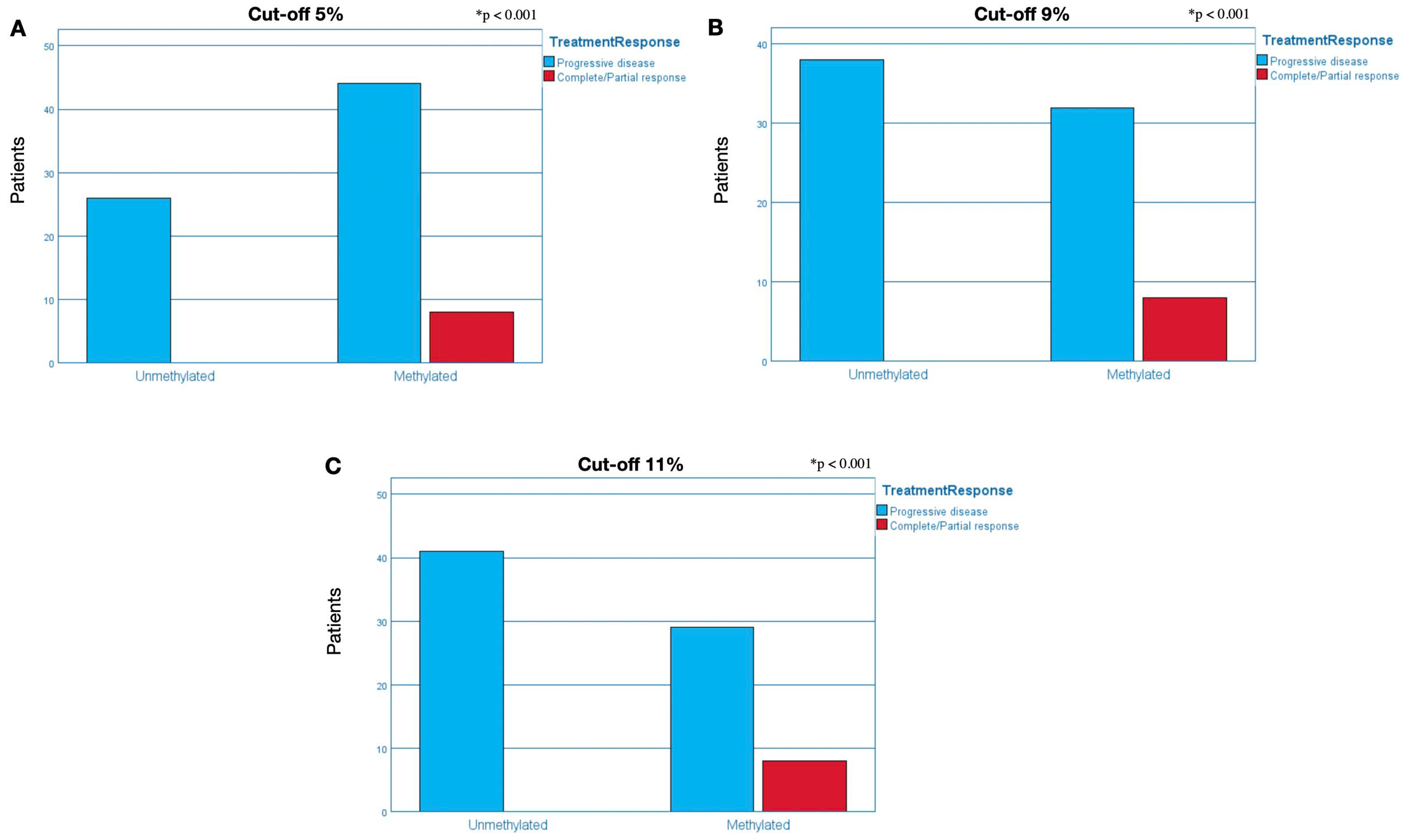
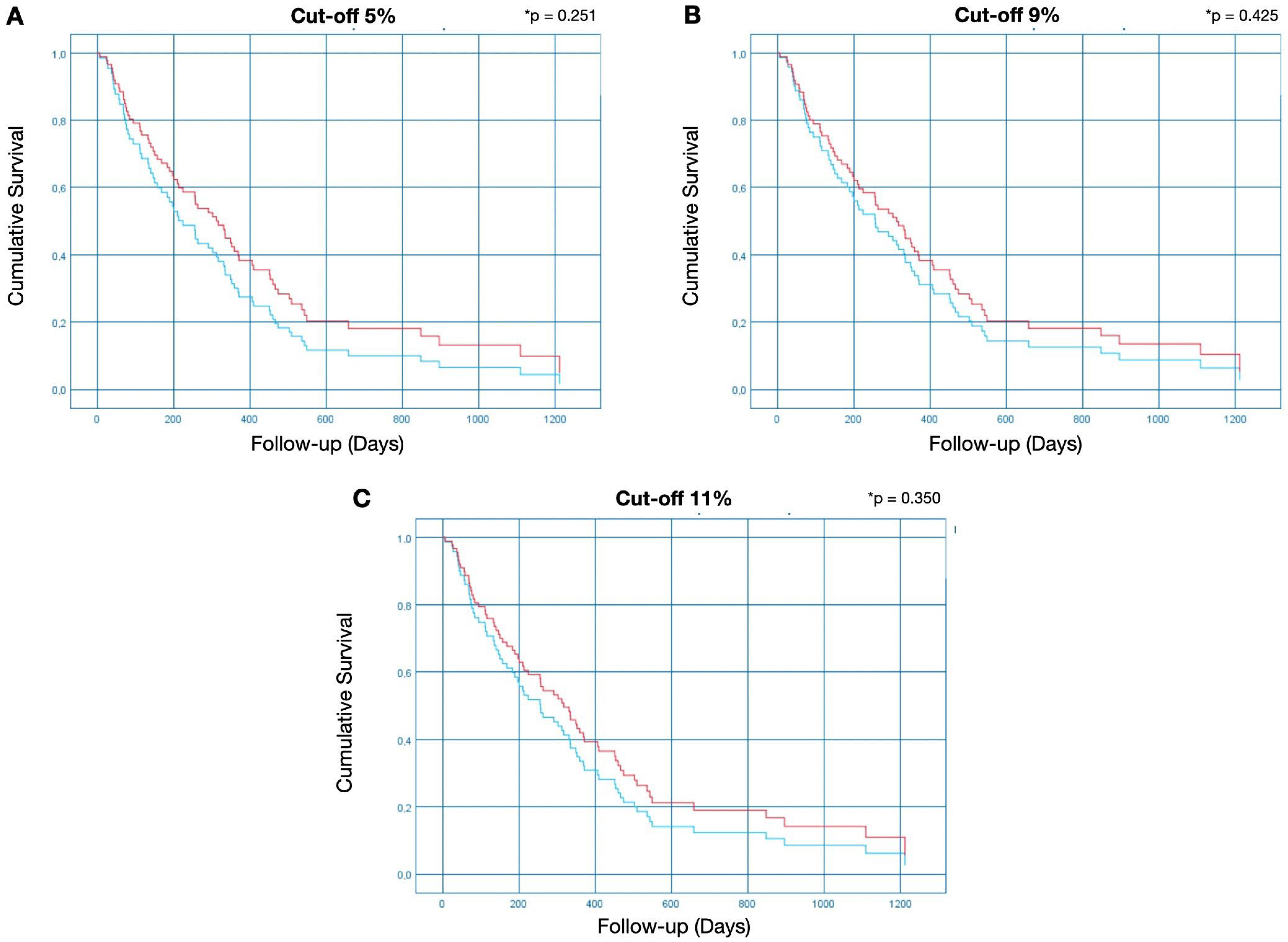
2.3.1. Cut-Off of 5% for Methylation Positivity
2.3.2. Cut-Off of 9% for Methylation Positivity
2.3.3. Cut-Off of 11% for Methylation Positivity
3. Discussion
4. Material and Methods
4.1. Patient Selection
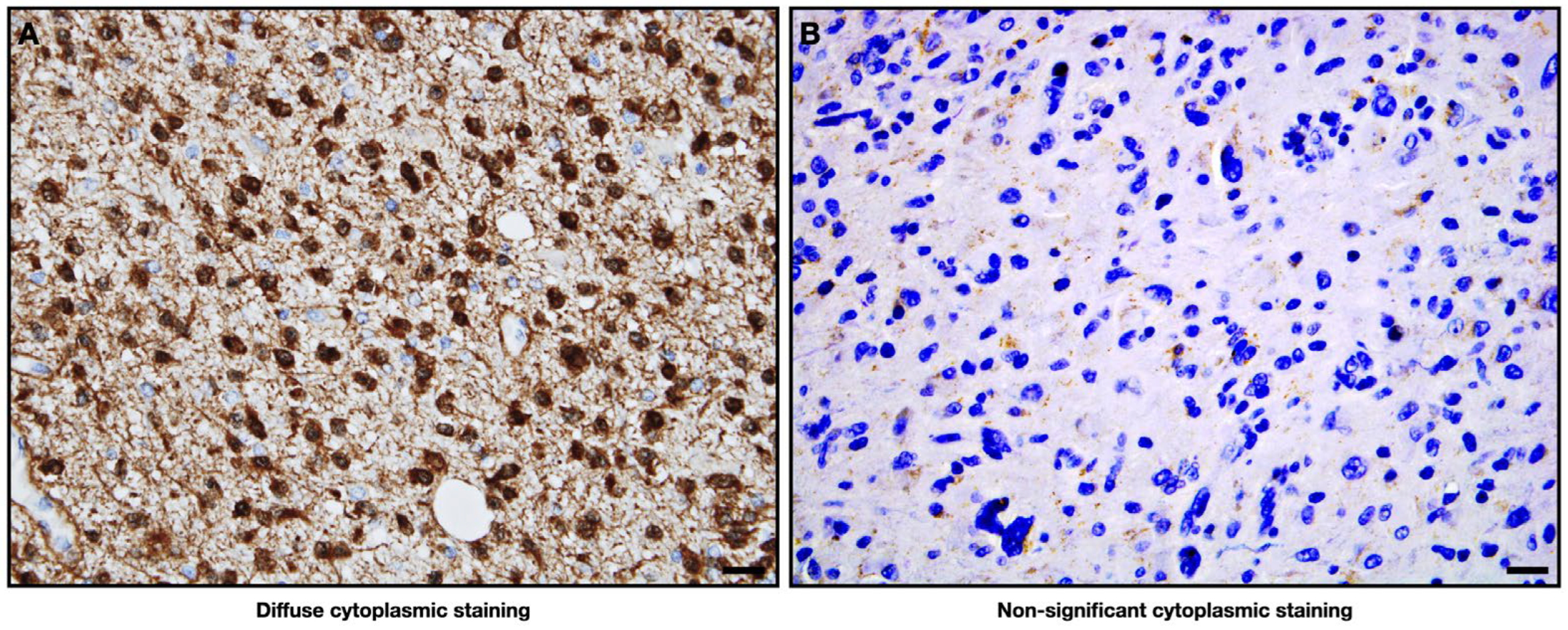
4.2. Immunohistochemistry (IHC)
IHC Evaluation
4.3. Patient Grouping
4.4. Data Extraction
4.5. Analysis of MGMT Methylation
4.5.1. PSQ
4.5.2. Quantification of Methylation
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Girardi, F.; Matz, M.; Stiller, C.; You, H.; Gragera, R.M.; Valkov, M.Y.; Bulliard, J.L.; De, P.; Morrison, D.; Wanner, M.; et al. Global survival trends for brain tumors, by histology: Analysis of individual records for 556,237 adults diagnosed in 59 countries during 2000–2014 (CONCORD-3). Neuro-Oncology 2023, 25, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Gritsch, S.; Batchelor, T.T.; Gonzalez Castro, L.N. Diagnostic, therapeutic, and prognostic implications of the 2021 World Health Organization classification of tumors of the central nervous system. Cancer 2022, 128, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Horbinski, C.; Berger, T.; Packer, R.J.; Wen, P.Y. Clinical implications of the 2021 edition of the WHO classification of central nervous system tumours. Nat. Rev. Neurol. 2022, 18, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.L.; Wadhwani, N.; Horbinski, C. Major Features of the 2021 WHO Classification of CNS Tumors. Neurother. J. Am. Soc. Exp. NeuroTherapeutics 2022, 19, 1691–1704. [Google Scholar] [CrossRef] [PubMed]
- Wong, Q.H.W.; Li, K.K.W.; Wang, W.W.; Malta, T.M.; Noushmehr, H.; Grabovska, Y.; Jones, C.; Chan, A.K.Y.; Kwan, J.S.H.; Huang, Q.J.Q.; et al. Molecular landscape of IDH-mutant primary astrocytoma Grade IV/glioblastomas. Mod. Pathol. 2021, 34, 1245–1260. [Google Scholar] [CrossRef]
- Hertler, C.; Felsberg, J.; Gramatzki, D.; Rhun, E.L.; Clarke, J.; Soffietti, R.; Wick, W.; Chinot, O.; Ducray, F.; Roth, P.; et al. Long-term survival with IDH wildtype glioblastoma: First results from the ETERNITY Brain Tumor Funders’ Collaborative Consortium (EORTC 1419). Eur. J. Cancer 2023, 189, 112913. [Google Scholar] [CrossRef]
- Guo, X.; Gu, L.; Li, Y.; Zheng, Z.; Chen, W.; Wang, Y.; Wang, Y.; Xing, H.; Shi, Y.; Liu, D.; et al. Histological and molecular glioblastoma, IDH-wildtype: A real-world landscape using the 2021 WHO classification of central nervous system tumors. Front. Oncol. 2023, 13, 1200815. [Google Scholar] [CrossRef]
- Chehade, G.; Lawson, T.M.; Lelotte, J.; Daoud, L.; Di Perri, D.; Whenham, N.; Duprez, T.; Tajeddine, N.; Tissir, F.; Raftopoulos, C. Long-term survival in patients with IDH-wildtype glioblastoma: Clinical and molecular characteristics. Acta Neurochir. 2023, 165, 1075–1085. [Google Scholar] [CrossRef]
- Ramos-Fresnedo, A.; Pullen, M.W.; Perez-Vega, C.; Domingo, R.A.; Akinduro, O.O.; Almeida, J.P.; Suarez-Meade, P.; Marenco-Hillembrand, L.; Jentoft, M.E.; Bendok, B.R.; et al. The survival outcomes of molecular glioblastoma IDH-wildtype: A multicenter study. J. Neuro-Oncol. 2022, 157, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Christians, A.; Adel-Horowski, A.; Banan, R.; Lehmann, U.; Bartels, S.; Behling, F.; Barrantes-Freer, A.; Stadelmann, C.; Rohde, V.; Stockhammer, F.; et al. The prognostic role of IDH mutations in homogeneously treated patients with anaplastic astrocytomas and glioblastomas. Acta Neuropathol. Commun. 2019, 7, 156. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, T.; Minami, Y.; Jinzaki, M.; Toda, M.; Yoshida, K.; Sasaki, H. Predictive markers for MGMT promoter methylation in glioblastomas. Neurosurg. Rev. 2019, 42, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, A.; Hachem, L.D.; Mansouri, S.; Nassiri, F.; Laperriere, N.J.; Xia, D.; Lindeman, N.I.; Wen, P.Y.; Chakravarti, A.; Mehta, M.P.; et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: Refining the approach based on emerging evidence and current challenges. Neuro-Oncology 2019, 21, 167–178. [Google Scholar] [CrossRef]
- Padin-Iruegas, E.; Chamorro-Petronacci, C.M.; Sines-Cajade, I.; Lorenzo-Pouso, A.I.; Blanco-Carrión, A.; Pérez-Jardón, A.; Gándara-Vila, P.; Pérez-Sayans, M. DNA Methylation by Bisulfite Next-Generation Sequencing for MLH1 and MGMT in Oral Squamous Cell Carcinomas and Potentially Malignant Disorders: An Integrative Analysis towards Field Cancerization. Medicina 2022, 58, 878. [Google Scholar] [CrossRef] [PubMed]
- Philteos, J.; Karmur, B.S.; Mansouri, A. MGMT Testing in Glioblastomas: Pitfalls and Opportunities. Am. J. Clin. Oncol. 2019, 42, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Arora, I.; Gurav, M.; Rumde, R.; Dhanavade, S.; Kadam, V.; Kurani, H.; Shetty, O.; Goda, J.; Shetty, P.; Moiyadi, A.; et al. MGMT gene promoter methylation and its correlation with clinicopathological parameters in glioblastomas. Neurol. India 2018, 66, 1106–1114. [Google Scholar] [CrossRef]
- Binabaj, M.M.; Bahrami, A.; ShahidSales, S.; Joodi, M.; Mashhad, M.J.; Hassanian, S.M.; Anvari, K.; Avan, A. The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J. Cell. Physiol. 2018, 233, 378–386. [Google Scholar] [CrossRef]
- Hosoya, T.; Takahashi, M.; Honda-Kitahara, M.; Miyakita, Y.; Ohno, M.; Yanagisawa, S.; Omura, T.; Kawauchi, D.; Tamura, Y.; Kikuchi, M.; et al. MGMT gene promoter methylation by pyrosequencing method correlates volumetric response and neurological status in IDH wild-type glioblastomas. J. Neuro-Oncol. 2022, 157, 561–571. [Google Scholar] [CrossRef]
- Villani, V.; Casini, B.; Pace, A.; Prosperini, L.; Carapella, C.M.; Vidiri, A.; Fabi, A.; Carosi, M. The Prognostic Value of Pyrosequencing-Detected MGMT Promoter Hypermethylation in Newly Diagnosed Patients with Glioblastoma. Dis. Markers 2015, 2015, 604719. [Google Scholar] [CrossRef]
- van Dijken, B.R.J.; van der Hoorn, A. Radiology in the lead: Towards radiological profiling for precision medicine in glioblastoma patients? Editorial comment on Glioblastoma patients with a moderate vascular profile benefit the most from MGMT methylation. Eur. Radiol. 2021, 31, 1736–1737. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, V.E.; Dai, H.Y.; Stensjøen, A.L.; Berntsen, E.M.; Salvesen, Ø.; Solheim, O.; Torp, S.H. MGMT Promoter Methylation Status Is Not Related to Histological or Radiological Features in IDH Wild-type Glioblastomas. J. Neuropathol. Exp. Neurol. 2020, 79, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Li, X.; Li, H. Perfusion parameters derived from MRI for preoperative prediction of IDH mutation and MGMT promoter methylation status in glioblastomas. Magn. Reson. Imaging 2021, 83, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Limam, S.; Missaoui, N.; Abdessayed, N.; Mestiri, S.; Selmi, B.; Mokni, M.; Yacoubi, M.T. Prognostic significance of MGMT methylation and expression of MGMT, P53, EGFR, MDM2 and PTEN in glioblastoma multiforme. Ann. Biol. Clin. 2019, 77, 307–317. [Google Scholar] [CrossRef]
- Kessler, T.; Sahm, F.; Sadik, A.; Stichel, D.; Hertenstein, A.; Reifenberger, G.; Zacher, A.; Sabel, M.; Tabatabai, G.; Steinbach, J.; et al. Molecular differences in IDH wildtype glioblastoma according to MGMT promoter methylation. Neuro-Oncology 2018, 20, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Brat, D.J.; Aldape, K.; Bridge, J.A.; Canoll, P.; Colman, H.; Hameed, M.R.; Harris, B.T.; Hattab, E.M.; Huse, J.T.; Jenkins, R.B.; et al. Molecular Biomarker Testing for the Diagnosis of Diffuse Gliomas. Arch. Pathol. Lab. Med. 2022, 146, 547–574. [Google Scholar] [CrossRef]
- Caccese, M.; Simonelli, M.; Villani, V.; Rizzato, S.; Ius, T.; Pasqualetti, F.; Russo, M.; Rudà, R.; Amoroso, R.; Bellu, L.; et al. Definition of the Prognostic Role of MGMT Promoter Methylation Value by Pyrosequencing in Newly Diagnosed IDH Wild-Type Glioblastoma Patients Treated with Radiochemotherapy: A Large Multicenter Study. Cancer 2022, 14, 2425. [Google Scholar] [CrossRef]
- Jin, L.; Guo, S.; Zhang, X.; Mo, Y.; Ke, S.; Duan, C. Optimal treatment strategy for adult patients with newly diagnosed glioblastoma: A systematic review and network meta-analysis. Neurosurg. Rev. 2021, 44, 1943–1955. [Google Scholar] [CrossRef]
- Jovčevska, I. Genetic secrets of long-term glioblastoma survivors. Bosn. J. Basic Med. Sci. 2019, 19, 116–124. [Google Scholar] [CrossRef]
- Monica, R.D.; Cuomo, M.; Buonaiuto, M.; Costabile, D.; Franca, R.A.; De Caro, M.D.B.; Catapano, G.; Chiariotti, L.; Visconti, R. MGMT and Whole-Genome DNA Methylation Impacts on Diagnosis, Prognosis and Therapy of Glioblastoma Multiforme. Int. J. Mol. Sci. 2022, 23, 7148. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Shoaf, M.L.; Cioffi, G.; Waite, K.; Kruchko, C.; Wen, P.Y.; Brat, D.J.; Barnholtz-Sloan, J.S.; Iorgulescu, J.B. National-level overall survival patterns for molecularly-defined diffuse glioma types in the United States. Neuro-Oncology 2023, 25, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Quillien, V.; Lavenu, A.; Ducray, F.; Joly, M.J.; Chinot, O.; Fina, F.; Sanson, M.; Carpentier, C.; Karayan-Tapon, L.; Rivet, P.; et al. Validation of the high-performance of pyrosequencing for clinical MGMT testing on a cohort of glioblastoma patients from a prospective dedicated multicentric trial. Oncotarget 2016, 7, 61916–61929. [Google Scholar] [CrossRef]
- Wang, S.; Song, C.; Zha, Y.; Li, L. The prognostic value of MGMT promoter status by pyrosequencing assay for glioblastoma patients’ survival: A meta-analysis. World J. Surg. Oncol. 2016, 14, 261. [Google Scholar] [CrossRef]
- Witthayanuwat, S.; Pesee, M.; Supaadirek, C.; Supakalin, N.; Thamronganantasakul, K.; Krusun, S. Survival Analysis of Glioblastoma Multiforme. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 2613. [Google Scholar] [PubMed]
- Dahlrot, R.H.; Larsen, P.; Boldt, H.B.; Kreutzfeldt, M.S.; Hansen, S.; Hjelmborg, J.B.; Kristensen, B.W. Posttreatment Effect of MGMT Methylation Level on Glioblastoma Survival. J. Neuropathol. Exp. Neurol. 2019, 78, 633–640. [Google Scholar] [CrossRef]
- Audureau, E.; Chivet, A.; Ursu, R.; Corns, R.; Metellus, P.; Zouaoui, S.; Guyotat, J.; Le Reste, P.J.; Faillot, T.; Litre, F.; et al. Prognostic factors for survival in adult patients with recurrent glioblastoma: A decision-tree-based model. J. Neuro-Oncol. 2018, 136, 565–576. [Google Scholar] [CrossRef]
- Gately, L.; Collins, A.; Murphy, M.; Dowling, A. Age alone is not a predictor for survival in glioblastoma. J. Neuro-Oncol. 2016, 129, 479–485. [Google Scholar] [CrossRef]
- Rautajoki, K.J.; Jaatinen, S.; Hartewig, A.; Tiihonen, A.M.; Annala, M.; Salonen, I.; Valkonen, M.; Simola, V.; Vuorinen, E.M.; Kivinen, A.; et al. Genomic characterization of IDH-mutant astrocytoma progression to grade 4 in the treatment setting. Acta Neuropathol. Commun. 2023, 11, 176. [Google Scholar] [CrossRef]
- Miller, J.J.; Gonzalez Castro, L.N.; McBrayer, S.; Weller, M.; Cloughesy, T.; Portnow, J.; Andronesi, O.; Barnholtz-Sloan, J.S.; Baumert, B.G.; Berger, M.S.; et al. Isocitrate dehydrogenase (IDH) mutant gliomas: A Society for Neuro-Oncology (SNO) consensus review on diagnosis, management, and future directions. Neuro-Oncology 2023, 25, 4–25. [Google Scholar] [CrossRef]
- Nakasu, S.; Deguchi, S.; Nakasu, Y. IDH wild-type lower-grade gliomas with glioblastoma molecular features: A systematic review and meta-analysis. Brain Tumor Pathol. 2023, 40, 143–157. [Google Scholar] [CrossRef]
- Barbagallo, G.M.V.; Paratore, S.; Caltabiano, R.; Palmucci, S.; Parra, H.S.; Privitera, G.; Motta, F.; Lanzafame, S.; Scaglione, G.; Longo, A.; et al. Long-term therapy with temozolomide is a feasible option for newly diagnosed glioblastoma: A single-institution experience with as many as 101 temozolomide cycles. Neurosurg. Focus 2014, 37, E4. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, V.G.; Doval, M.B.; Bellvert, C.G.; Goliney, V.G.; Asencio, O.S.; Martín, A.G.; Domínguez, J.I. Quantitative analysis of MGMT promoter methylation status changes by pyrosequencing in recurrent glioblastoma. Neuropathol. Off. J. Jpn. Soc. Neuropathol. 2023, 43, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Lalezari, S.; Chou, A.P.; Tran, A.; Solis, O.E.; Khanlou, N.; Chen, W.; Li, S.; Carrillo, J.A.; Chowdhury, R.; Selfridgeet, J.; et al. Combined analysis of O6-methylguanine-DNA methyltransferase protein expression and promoter methylation provides optimized prognostication of glioblastoma outcome. Neuro-Oncology 2013, 15, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Lattanzio, L.; Borgognone, M.; Mocellini, C.; Giordano, F.; Favata, E.; Fasano, G.; Vivenza, D.; Monteverde, M.; Tonissi, F.; Ghiglia, A.; et al. MGMT promoter methylation and glioblastoma: A comparison of analytical methods and of tumor specimens. Int. J. Biol. Markers 2015, 30, e208–e216. [Google Scholar] [CrossRef] [PubMed]
- Gurrieri, L.; De Carlo, E.; Gerratana, L.; De Maglio, G.; Macerelli, M.; Pisa, F.E.; Masiero, E.; Aprile, G.; Follador, A.; Puglisi, F.; et al. MGMT pyrosequencing-based cut-off methylation level and clinical outcome in patients with glioblastoma multiforme. Future Oncol. 2018, 14, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.Y.; Ho, H.L.; Lin, S.C.; Ho, T.D.H.; Ho, D.M.T. The MGMT promoter single-nucleotide polymorphism rs1625649 had prognostic impact on patients with MGMT methylated glioblastoma. PLoS ONE 2017, 12, e0186430. [Google Scholar] [CrossRef]
- Håvik, A.B.; Brandal, P.; Honne, H.; Dahlback, H.S.S.; Scheie, D.; Hektoen, M.; Meling, T.R.; Helseth, E.; Heim, S.; Lothe, R.A.; et al. MGMT promoter methylation in gliomas-assessment by pyrosequencing and quantitative methylation-specific PCR. J. Transl. Med. 2012, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Michaelsen, S.R.; Dyrbye, H.; Aslan, D.; Grunnet, K.; Christensen, J.I.; Poulsen, H.S.; Grønbæk, K.; Broholm, H. Assessment of Quantitative and Allelic MGMT Methylation Patterns as a Prognostic Marker in Glioblastoma. J. Neuropathol. Exp. Neurol. 2016, 75, 246–255. [Google Scholar] [CrossRef]
- Shen, D.; Liu, T.; Lin, Q.; Lu, X.; Wang, Q.; Lin, F.; Mao, W. MGMT promoter methylation correlates with an overall survival benefit in Chinese high-grade glioblastoma patients treated with radiotherapy and alkylating agent-based chemotherapy: A single-institution study. PLoS ONE 2014, 9, e107558. [Google Scholar] [CrossRef]
- Pareira, E.S.; Kitano, Y.; Ohara, K.; Kanazawa, T.; Nakagawa, Y.; Yoshida, K.; Sasaki, H. Immunohistochemistry for O6-methylguanin-DNA methyltransferase in glioblastomas defined by WHO2016: Correlation with promoter methylation status and patients’ progression-free survival with the cut-off value determined by ROC analysis. J. Clin. Neurosci. Off. J. Neurosurg. Soc. Australas. 2020, 73, 231–236. [Google Scholar] [CrossRef]
- Yuan, G.; Niu, L.; Zhang, Y.; Wang, X.; Ma, K.; Yin, H.; Dai, J.; Zhou, W.; Pan, Y. Defining optimal cutoff value of MGMT promoter methylation by ROC analysis for clinical setting in glioblastoma patients. J. Neuro-Oncol. 2017, 133, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Bienkowski, M.; Berghoff, A.S.; Marosi, C.; Wöhrer, A.; Heinzl, H.; Hainfellner, J.A.; Preusser, M. Clinical Neuropathology practice guide 5-2015: MGMT methylation pyrosequencing in glioblastoma: Unresolved issues and open questions. Clin. Neuropathol. 2015, 34, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Poon, M.T.C.; Keni, S.; Vimalan, V.; Ip, C.; Smith, C.; Erridge, S.; Weir, C.J.; Brennan, P.M. Extent of MGMT promoter methylation modifies the effect of temozolomide on overall survival in patients with glioblastoma: A regional cohort study. Neuro-Oncol. Adv. 2021, 3, vdab171. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.C.; Kim, K.U.; Kim, Y.Z. Prognostic Role of Methylation Status of the MGMT Promoter Determined Quantitatively by Pyrosequencing in Glioblastoma Patients. J. Korean Neurosurg. Soc. 2016, 59, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Thon, N.; Thorsteinsdottir, J.; Eigenbrod, S.; Schüller, U.; Lutz, J.; Kreth, S.; Belka, C.; Tonn, J.C.; Niyazi, M.; Kreth, F.W. Outcome in unresectable glioblastoma: MGMT promoter methylation makes the difference. J. Neurol. 2017, 264, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Minniti, G.; Salvati, M.; Arcella, A.; Buttarelli, F.; D’Elia, A.; Lanzetta, G.; Esposito, V.; Scarpino, S.; Maurizi Enrici, R.; Giangaspero, F. Correlation between O6-methylguanine-DNA methyltransferase and survival in elderly patients with glioblastoma treated with radiotherapy plus concomitant and adjuvant temozolomide. J. Neuro-Oncol. 2011, 102, 311–316. [Google Scholar] [CrossRef]
- Ashkan, K.; Baig Mirza, A.; Soumpasis, C.; Syrris, C.; Kalaitzoglou, D.; Sharma, C.; James, Z.J.; Khoja, A.K.; Ahmed, R.; Vastani, A.; et al. MGMT Promoter Methylation: Prognostication beyond Treatment Response. J. Pers. Med. 2023, 13, 999. [Google Scholar] [CrossRef]
- Malmström, A.; Grønberg, B.H.; Marosi, C.; Stupp, R.; Frappaz, D.; Schultz, H.; Henriksson, R. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: The Nordic randomised, phase 3 trial. Lancet Oncol. 2012, 13, 916–926. [Google Scholar] [CrossRef]
- Nagane, M.; Kobayashi, K.; Ohnishi, A.; Shimizu, S.; Shiokawa, Y. Prognostic significance of O6-methylguanine-DNA methyltransferase protein expression in patients with recurrent glioblastoma treated with temozolomide. Jpn. J. Clin. Oncol. 2007, 37, 897–906. [Google Scholar] [CrossRef]
- Silber, J.R.; Bobola, M.S.; Blank, A.; Chamberlain, M.C. O(6)-methylguanine-DNA methyltransferase in glioma therapy: Promise and problems. Biochim. Biophys. Acta 2012, 1826, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.R.; Wang, M.; Aldape, K.D.; Stupp, R.; Hegi, M.E.; Jaeckle, K.A.; Armstrong, T.S.; Wefel, J.S.; Won, M.; Blumenthal, D.T.; et al. Dose-dense temozolomide for newly diagnosed glioblastoma: A randomized phase III clinical trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 4085–4091. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.; Pongor, L.; Su, Y.T.; Xi, L.; Raffeld, M.; Quezado, M.; Trepel, J.; Aldape, K.; Pommier, Y.; Wu, J. MGMT status as a clinical biomarker in glioblastoma. Trends Cancer 2020, 6, 380. [Google Scholar] [CrossRef] [PubMed]
- Gerstner, E.R.; Yip, S.; Wang, D.L.; Louis, D.N.; Iafrate, A.J.; Batchelor, T.T. Mgmt methylation is a prognostic biomarker in elderly patients with newly diagnosed glioblastoma. Neurology 2009, 73, 1509–1510. [Google Scholar] [CrossRef]
- Gibson, D.; Ravi, A.; Rodriguez, E.; Chang, S.; Oberheim Bush, N.; Taylor, J.; Clarke, J.; Solomon, D.; Scheffler, A.; Witte, J.; et al. Quantitative analysis of MGMT promoter methylation in glioblastoma suggests nonlinear prognostic effect. Neuro-Oncol. Adv. 2023, 5, vdad115. [Google Scholar] [CrossRef]
- Katsigiannis, S.; Grau, S.; Krischek, B.; Er, K.; Pintea, B.; Goldbrunner, R.; Stavrinou, P. MGMT-Positive vs MGMT-Negative Patients with Glioblastoma: Identification of Prognostic Factors and Resection Threshold. Neurosurgery 2021, 88, E323–E329. [Google Scholar] [CrossRef]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and elaboration. PLoS Med. 2012, 10, 51. [Google Scholar] [CrossRef]

| Qualitative Variables | Number of Patients | Average MGMT Methylation | |
|---|---|---|---|
| Gender | Male | 57.5% (45) | 20.13% |
| Female | 42.3% (33) | 22.19% | |
| Age | <70 years old | 61.5% (48) | 19.27% |
| ≥70 years old | 38.5% (30) | 22.10% | |
| Exitus | Alive | 12.8% (10) | 19.09% |
| Dead | 87.2% (68) | 31.23% | |
| Cut-Off | Methylated | Unmethylated |
|---|---|---|
| 5% | 67.7% (52) | 31.3% (26) |
| 9% | 51.3% (40) | 48.7% (38) |
| 11% | 47.4% (37) | 52.6% (41) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, F.F.V.e.; Di Domenico, M.; Caponio, V.C.A.; Pérez-Sayáns, M.; Camolesi, G.C.V.; Rojo-Álvarez, L.I.; Ballini, A.; García-García, A.; Padín-Iruegas, M.E.; Suaréz-Peñaranda, J.M. Pyrosequencing Analysis of O-6-Methylguanine-DNA Methyltransferase Methylation at Different Cut-Offs of Positivity Associated with Treatment Response and Disease-Specific Survival in Isocitrate Dehydrogenase-Wildtype Grade 4 Glioblastoma. Int. J. Mol. Sci. 2024, 25, 612. https://doi.org/10.3390/ijms25010612
Silva FFVe, Di Domenico M, Caponio VCA, Pérez-Sayáns M, Camolesi GCV, Rojo-Álvarez LI, Ballini A, García-García A, Padín-Iruegas ME, Suaréz-Peñaranda JM. Pyrosequencing Analysis of O-6-Methylguanine-DNA Methyltransferase Methylation at Different Cut-Offs of Positivity Associated with Treatment Response and Disease-Specific Survival in Isocitrate Dehydrogenase-Wildtype Grade 4 Glioblastoma. International Journal of Molecular Sciences. 2024; 25(1):612. https://doi.org/10.3390/ijms25010612
Chicago/Turabian StyleSilva, Fábio França Vieira e, Marina Di Domenico, Vito Carlo Alberto Caponio, Mario Pérez-Sayáns, Gisela Cristina Vianna Camolesi, Laura Isabel Rojo-Álvarez, Andrea Ballini, Abel García-García, María Elena Padín-Iruegas, and Jose Manuel Suaréz-Peñaranda. 2024. "Pyrosequencing Analysis of O-6-Methylguanine-DNA Methyltransferase Methylation at Different Cut-Offs of Positivity Associated with Treatment Response and Disease-Specific Survival in Isocitrate Dehydrogenase-Wildtype Grade 4 Glioblastoma" International Journal of Molecular Sciences 25, no. 1: 612. https://doi.org/10.3390/ijms25010612
APA StyleSilva, F. F. V. e., Di Domenico, M., Caponio, V. C. A., Pérez-Sayáns, M., Camolesi, G. C. V., Rojo-Álvarez, L. I., Ballini, A., García-García, A., Padín-Iruegas, M. E., & Suaréz-Peñaranda, J. M. (2024). Pyrosequencing Analysis of O-6-Methylguanine-DNA Methyltransferase Methylation at Different Cut-Offs of Positivity Associated with Treatment Response and Disease-Specific Survival in Isocitrate Dehydrogenase-Wildtype Grade 4 Glioblastoma. International Journal of Molecular Sciences, 25(1), 612. https://doi.org/10.3390/ijms25010612






