N-Terminal Amino Acid Affects the Translation Efficiency at Lower Temperatures in a Reconstituted Protein Synthesis System
Abstract
1. Introduction
2. Results
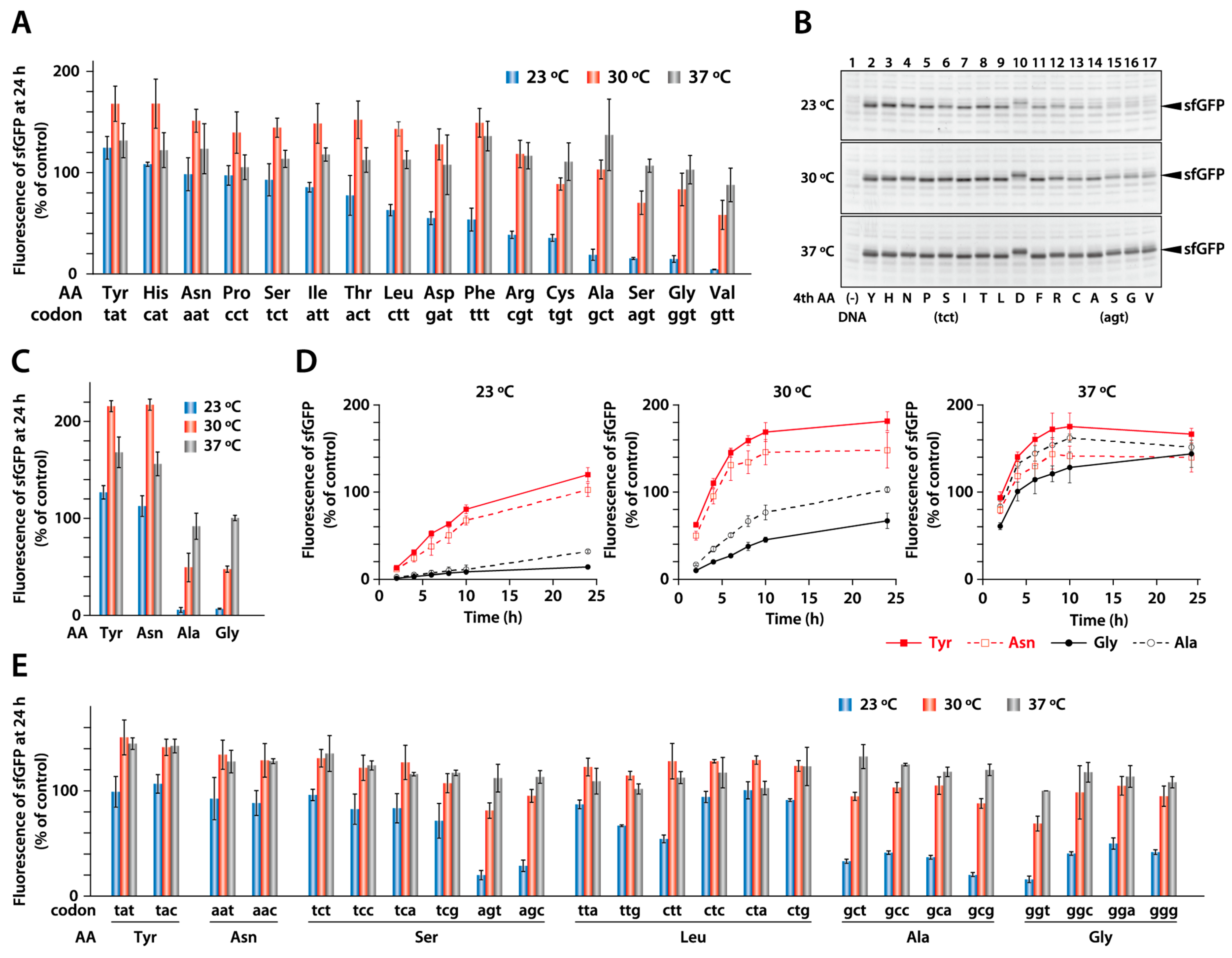
2.1. Synthesis of Model Proteins at Different Temperatures
2.2. Synthesis of sfGFP with a Different Fourth Amino Acid
2.3. Effect of a Synonymous Codon at the Fourth Position
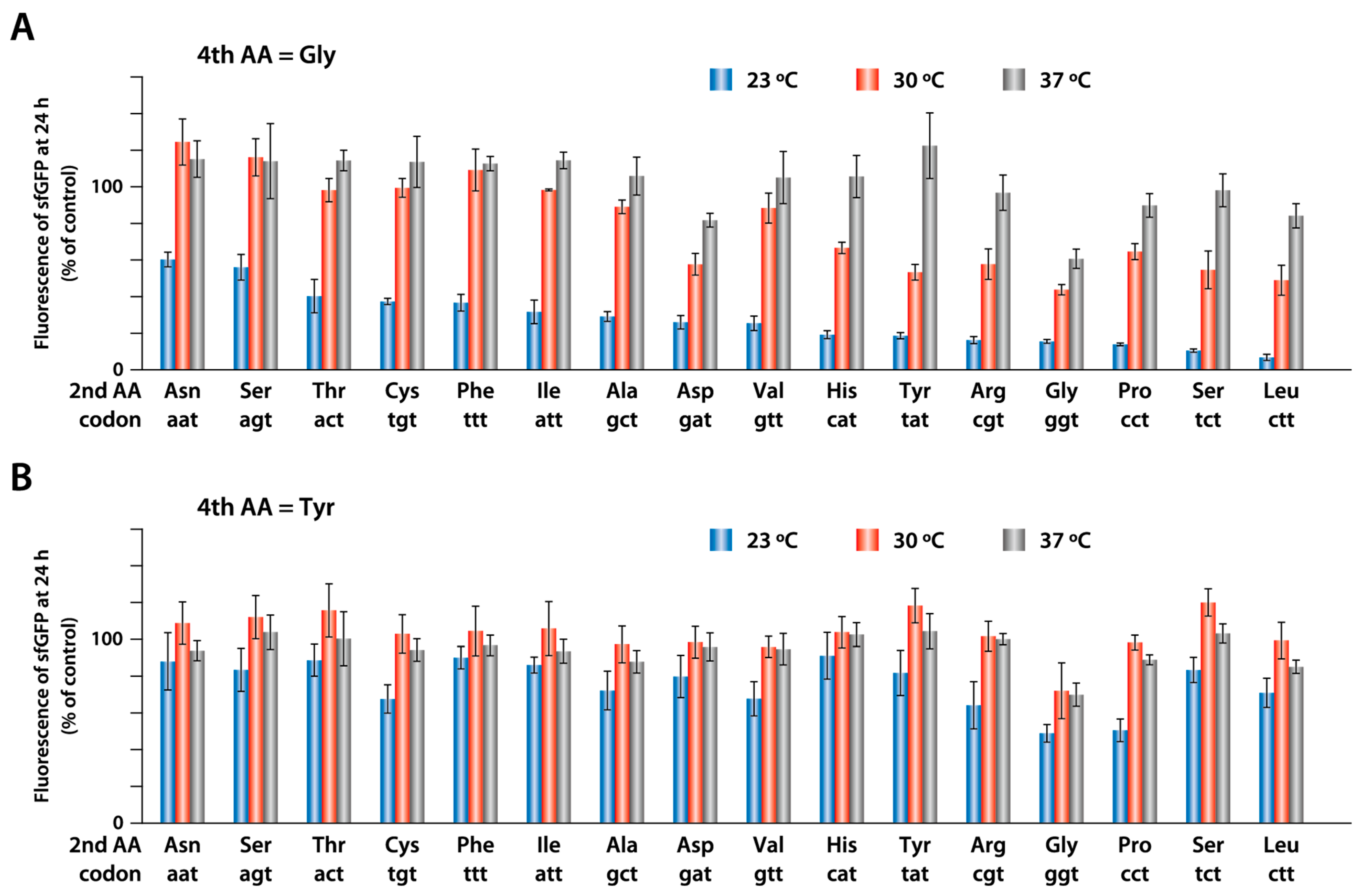
2.4. Synthesis of sfGFP with Different Amino Acids at the Second Position
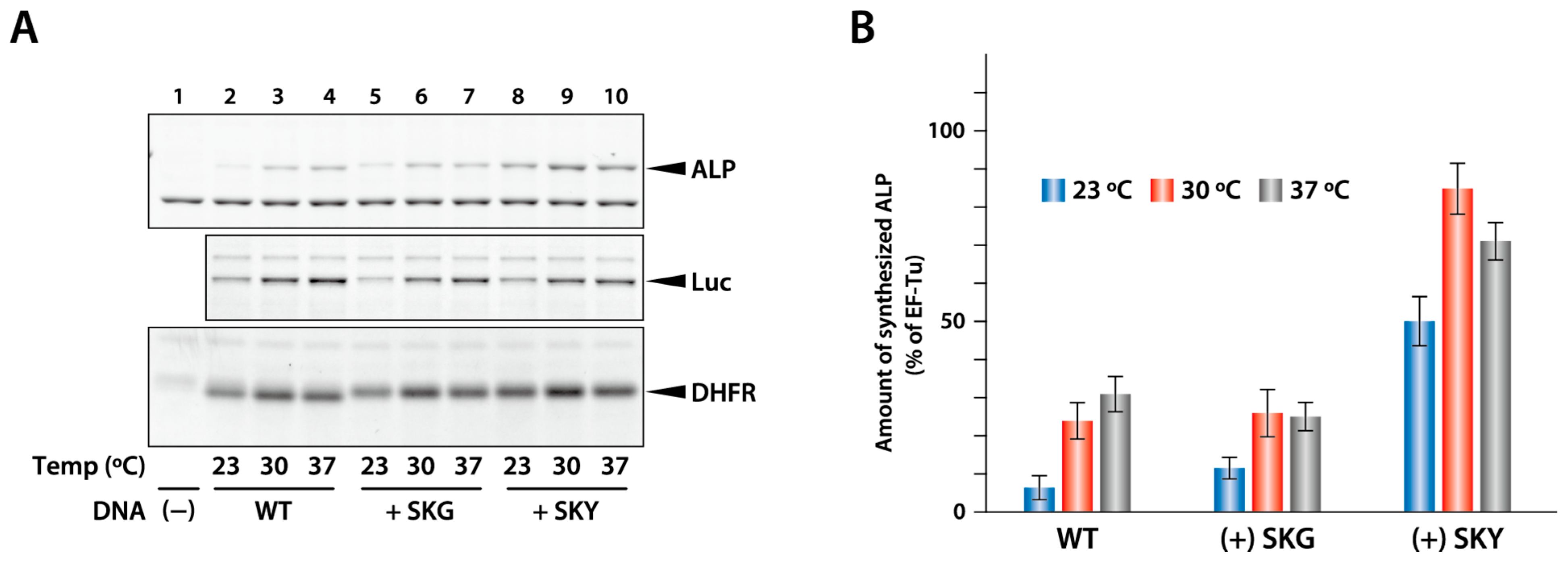
2.5. Effect of Addition of N-Terminal Sequence on the Productivity of Other Proteins
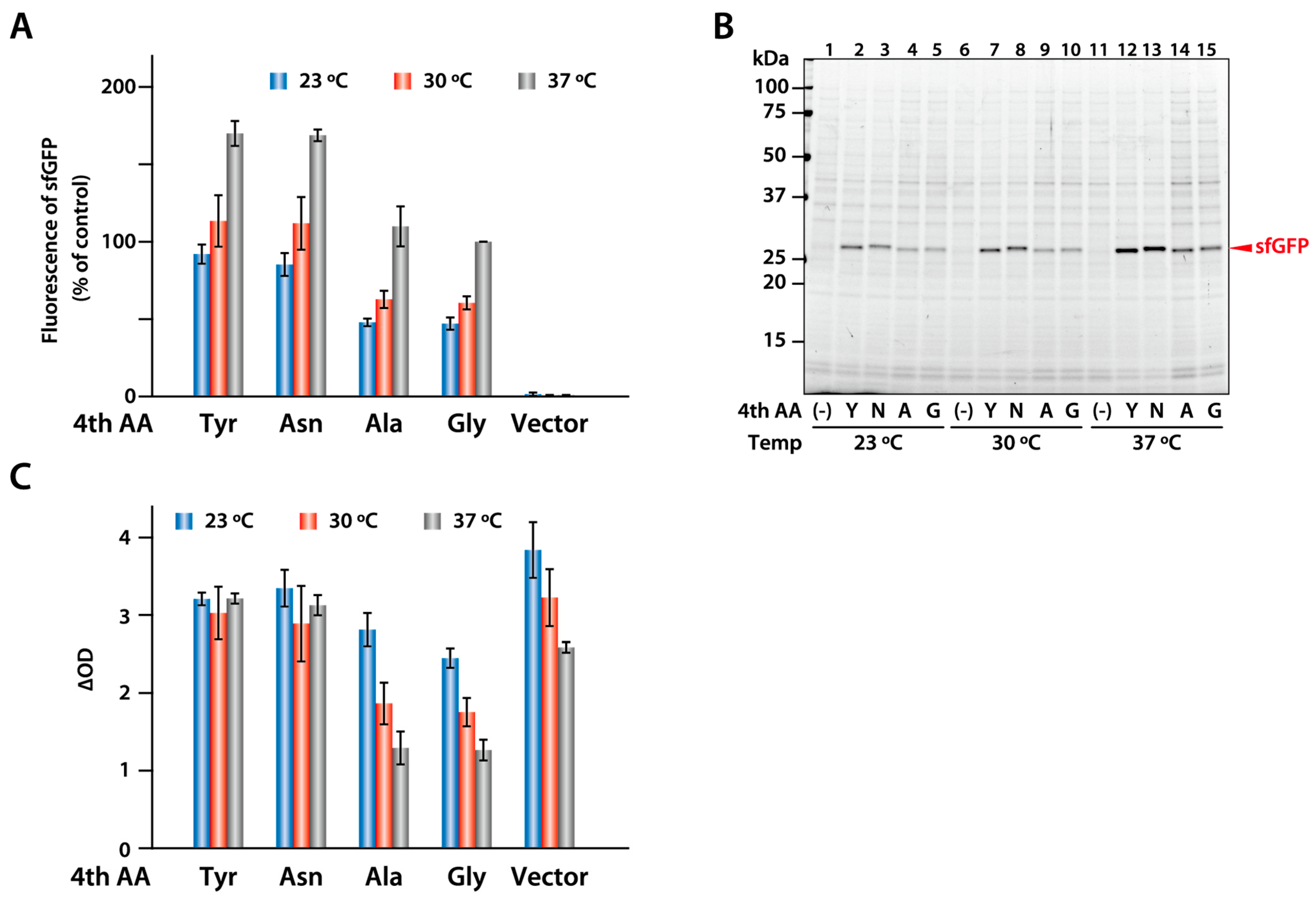
2.6. Expression of sfGFP Variants in E. coli
3. Discussion
4. Materials and Methods
4.1. Preparation of Template and Plasmid DNA for Cell-Free Protein Synthesis and Expression in E. coli, Respectively
4.2. Protein Synthesis Using the PURE System
4.3. Expression of sfGFP in E. coli
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-Free Translation Reconstituted with Purified Components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef]
- Chadani, Y.; Niwa, T.; Izumi, T.; Sugata, N.; Nagao, A.; Suzuki, T.; Chiba, S.; Ito, K.; Taguchi, H. Intrinsic Ribosome Destabilization Underlies Translation and Provides an Organism with a Strategy of Environmental Sensing. Mol. Cell 2017, 68, 528–539.e5. [Google Scholar] [CrossRef]
- Chadani, Y.; Kanamori, T.; Niwa, T.; Ichihara, K.; Nakayama, K.I.; Matsumoto, A.; Taguchi, H. Mechanistic Dissection of Premature Translation Termination Induced by Acidic Residues-Enriched Nascent Peptide. Cell Rep. 2023, 42, 113569. [Google Scholar] [CrossRef]
- Shimizu, Y. ArfA Recruits RF2 into Stalled Ribosomes. J. Mol. Biol. 2012, 423, 624–631. [Google Scholar] [CrossRef]
- Niwa, T.; Ying, B.-W.; Saito, K.; Jin, W.; Takada, S.; Ueda, T.; Taguchi, H. Bimodal Protein Solubility Distribution Revealed by an Aggregation Analysis of the Entire Ensemble of Escherichia Coli Proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 4201–4206. [Google Scholar] [CrossRef]
- Niwa, T.; Kanamori, T.; Ueda, T.; Taguchi, H. Global Analysis of Chaperone Effects Using a Reconstituted Cell-Free Translation System. Proc. Natl. Acad. Sci. USA 2012, 109, 8937–8942. [Google Scholar] [CrossRef]
- Gersteuer, F.; Morici, M.; Gabrielli, S.; Fujiwara, K.; Safdari, H.A.; Paternoga, H.; Bock, L.V.; Chiba, S.; Wilson, D.N. The SecM Arrest Peptide Traps a Pre-Peptide Bond Formation State of the Ribosome. Nat. Commun. 2024, 15, 2431. [Google Scholar] [CrossRef]
- Steinkühler, J.; Peruzzi, J.A.; Krüger, A.; Villaseñor, C.G.; Jacobs, M.L.; Jewett, M.C.; Kamat, N.P. Improving Cell-Free Expression of Model Membrane Proteins by Tuning Ribosome Cotranslational Membrane Association and Nascent Chain Aggregation. ACS Synth. Biol. 2023, 13, 129–140. [Google Scholar] [CrossRef]
- Matsumoto, R.; Niwa, T.; Shimane, Y.; Kuruma, Y.; Taguchi, H.; Kanamori, T. Regulated N-Terminal Modification of Proteins Synthesized Using a Reconstituted Cell-Free Protein Synthesis System. ACS Synth. Biol. 2023, 12, 1935–1942. [Google Scholar] [CrossRef]
- Aoyama, R.; Masuda, K.; Shimojo, M.; Kanamori, T.; Ueda, T.; Shimizu, Y. In Vitro Reconstitution of the Escherichia Coli 70S Ribosome with a Full Set of Recombinant Ribosomal Proteins. J. Biochem. 2021, 171, 227–237. [Google Scholar] [CrossRef]
- Hagino, K.; Ichihashi, N. In Vitro Transcription/Translation-Coupled DNA Replication through Partial Regeneration of 20 Aminoacyl-tRNA Synthetases. ACS Synth. Biol. 2023, 12, 1252–1263. [Google Scholar] [CrossRef]
- Eto, S.; Matsumura, R.; Shimane, Y.; Fujimi, M.; Berhanu, S.; Kasama, T.; Kuruma, Y. Phospholipid Synthesis inside Phospholipid Membrane Vesicles. Commun. Biol. 2022, 5, 1016. [Google Scholar] [CrossRef]
- Nakai, H.; Isshiki, K.; Hattori, M.; Maehira, H.; Yamaguchi, T.; Masuda, K.; Shimizu, Y.; Watanabe, T.; Hohsaka, T.; Shihoya, W.; et al. Cell-Free Synthesis of Human Endothelin Receptors and Its Application to Ribosome Display. Anal. Chem. 2022, 94, 3831–3839. [Google Scholar] [CrossRef]
- Ushiyama, R.; Nanjo, S.; Tsugane, M.; Sato, R.; Matsuura, T.; Suzuki, H. Identifying Conditions for Protein Synthesis Inside Giant Vesicles Using Microfluidics toward Standardized Artificial Cell Production. ACS Synth. Biol. 2023, 13, 68–76. [Google Scholar] [CrossRef]
- Kohyama, S.; Frohn, B.P.; Babl, L.; Schwille, P. Machine Learning-Aided Design and Screening of an Emergent Protein Function in Synthetic Cells. Nat. Commun. 2024, 15, 2010. [Google Scholar] [CrossRef]
- Nagumo, Y.; Fujiwara, K.; Horisawa, K.; Yanagawa, H.; Doi, N. PURE mRNA Display for in Vitro Selection of Single-Chain Antibodies. J. Biochem. 2015, 159, 519–526. [Google Scholar] [CrossRef]
- Kazuta, Y.; Matsuura, T.; Ichihashi, N.; Yomo, T. Synthesis of Milligram Quantities of Proteins Using a Reconstituted in Vitro Protein Synthesis System. J. Biosci. Bioeng. 2014, 118, 554–557. [Google Scholar] [CrossRef]
- Goodman, D.B.; Church, G.M.; Kosuri, S. Causes and Effects of N-Terminal Codon Bias in Bacterial Genes. Science 2013, 342, 475–479. [Google Scholar] [CrossRef]
- Sato, T.; Terabe, M.; Watanabe, H.; Gojobori, T.; Hori-Takemoto, C.; Ki, M. Codon and Base Biases after the Initiation Codon of the Open Reading Frames in the Escherichia Coli Genome and Their Influence on the Translation Efficiency. J. Biochem. 2001, 129, 851–860. [Google Scholar] [CrossRef]
- Verma, M.; Choi, J.; Cottrell, K.A.; Lavagnino, Z.; Thomas, E.N.; Pavlovic-Djuranovic, S.; Szczesny, P.; Piston, D.W.; Zaher, H.S.; Puglisi, J.D.; et al. A Short Translational Ramp Determines the Efficiency of Protein Synthesis. Nat. Commun. 2019, 10, 5774. [Google Scholar] [CrossRef]
- Moreira, M.H.; Barros, G.C.; Requião, R.D.; Rossetto, S.; Domitrovic, T.; Palhano, F.L. From Reporters to Endogenous Genes: The Impact of the First Five Codons on Translation Efficiency in Escherichia Coli. RNA Biol. 2019, 16, 1806–1816. [Google Scholar] [CrossRef] [PubMed]
- Osterman, I.A.; Chervontseva, Z.S.; Evfratov, S.A.; Sorokina, A.V.; Rodin, V.A.; Rubtsova, M.P.; Komarova, E.S.; Zatsepin, T.S.; Kabilov, M.R.; Bogdanov, A.A.; et al. Translation at First Sight: The Influence of Leading Codons. Nucleic Acids Res. 2020, 48, 6931–6942. [Google Scholar] [CrossRef] [PubMed]
- Bivona, L.; Zou, Z.; Stutzman, N.; Sun, P.D. Influence of the Second Amino Acid on Recombinant Protein Expression. Protein Expr. Purif. 2010, 74, 248–256. [Google Scholar] [CrossRef]
- Ojima-Kato, T.; Nagai, S.; Nakano, H. N-Terminal SKIK Peptide Tag Markedly Improves Expression of Difficult-to-Express Proteins in Escherichia Coli and Saccharomyces Cerevisiae. J. Biosci. Bioeng. 2017, 123, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Takai, K. CodHonEditor: Spreadsheets for Codon Optimization and Editing of Protein Coding Sequences. Nucleosides Nucleotides Nucleic Acids 2016, 35, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Matsumoto, R.; Kanamori, T. Constructive Approach for Synthesis of a Functional IgG Using a Reconstituted Cell-Free Protein Synthesis System. Sci. Rep. 2019, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Wang, T.; Lehmann, J. Peptidyl Transferase Center Decompaction and Structural Constraints during Early Protein Elongation on the Ribosome. Sci. Rep. 2021, 11, 24061. [Google Scholar] [CrossRef]
- Nagao, A.; Nakanishi, Y.; Yamaguchi, Y.; Mishina, Y.; Karoji, M.; Toya, T.; Fujita, T.; Iwasaki, S.; Miyauchi, K.; Sakaguchi, Y.; et al. Quality Control of Protein Synthesis in the Early Elongation Stage. Nat. Commun. 2023, 14, 2704. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuse-Murakami, T.; Matsumoto, R.; Kanamori, T. N-Terminal Amino Acid Affects the Translation Efficiency at Lower Temperatures in a Reconstituted Protein Synthesis System. Int. J. Mol. Sci. 2024, 25, 5264. https://doi.org/10.3390/ijms25105264
Fuse-Murakami T, Matsumoto R, Kanamori T. N-Terminal Amino Acid Affects the Translation Efficiency at Lower Temperatures in a Reconstituted Protein Synthesis System. International Journal of Molecular Sciences. 2024; 25(10):5264. https://doi.org/10.3390/ijms25105264
Chicago/Turabian StyleFuse-Murakami, Tomoe, Rena Matsumoto, and Takashi Kanamori. 2024. "N-Terminal Amino Acid Affects the Translation Efficiency at Lower Temperatures in a Reconstituted Protein Synthesis System" International Journal of Molecular Sciences 25, no. 10: 5264. https://doi.org/10.3390/ijms25105264
APA StyleFuse-Murakami, T., Matsumoto, R., & Kanamori, T. (2024). N-Terminal Amino Acid Affects the Translation Efficiency at Lower Temperatures in a Reconstituted Protein Synthesis System. International Journal of Molecular Sciences, 25(10), 5264. https://doi.org/10.3390/ijms25105264





