Venom Ex Machina? Exploring the Potential of Cell-Free Protein Production for Venom Biodiscovery
Abstract
:1. Introduction
2. Results
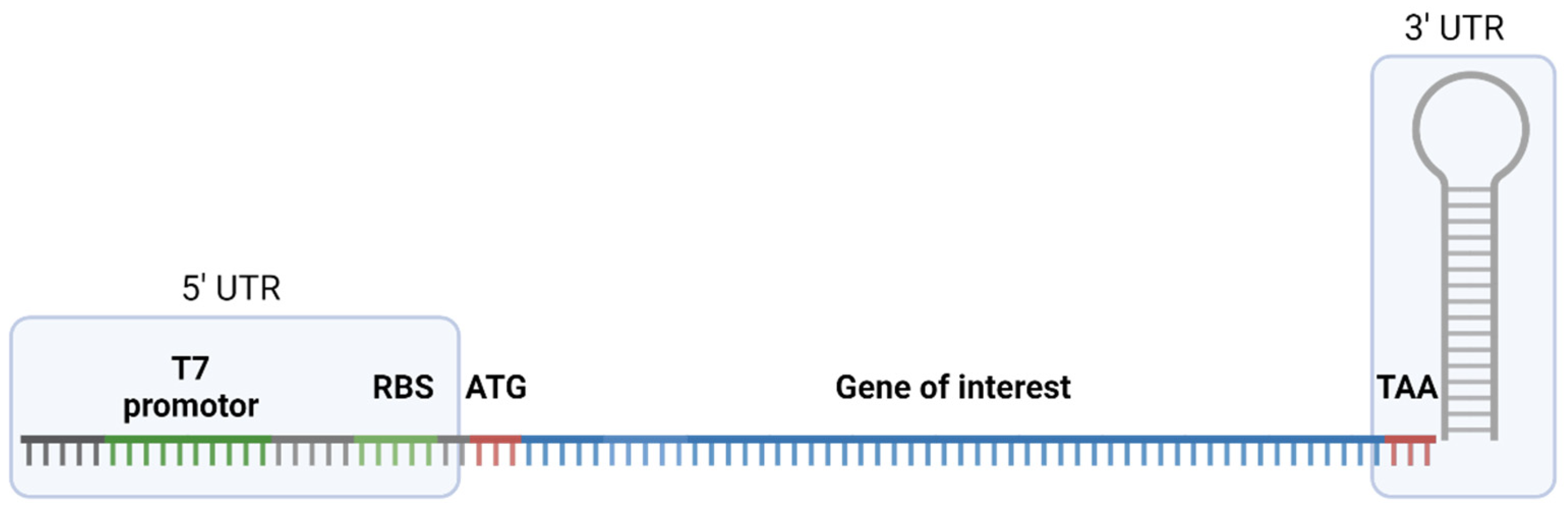
2.1. Gene Constructs for Cell-Free Production of Toxins
2.2. Limited Success Rates of Cell-Free Expression
3. Discussion
4. Materials and Methods
4.1. Selection of Candidates
4.2. Cell-Free Production and SDS-PAGE
4.3. Estimation of Expression Yields
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex Cocktails: The Evolutionary Novelty of Venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The Toxicogenomic Multiverse: Convergent Recruitment of Proteins into Animal Venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed]
- Gilding, E.K.; Jami, S.; Deuis, J.R.; Israel, M.R.; Harvey, P.J.; Poth, A.G.; Rehm, F.B.H.; Stow, J.L.; Robinson, S.D.; Yap, K.; et al. Neurotoxic Peptides from the Venom of the Giant Australian Stinging Tree. Sci. Adv. 2020, 6, eabb8828. [Google Scholar] [CrossRef] [PubMed]
- Schendel, V.; Rash, L.D.; Jenner, R.A.; Undheim, E.A.B. The Diversity of Venom: The Importance of Behavior and Venom System Morphology in Understanding Its Ecology and Evolution. Toxins 2019, 11, 666. [Google Scholar] [CrossRef] [PubMed]
- Herzig, V.; Cristofori-Armstrong, B.; Israel, M.R.; Nixon, S.A.; Vetter, I.; King, G.F. Animal Toxins—Nature’s Evolutionary-Refined Toolkit for Basic Research and Drug Discovery. Biochem. Pharmacol. 2020, 181, 114096. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, T.B.; Clark, R.J. Advances in Venom Peptide Drug Discovery: Where Are We at and Where Are We Heading? Expert. Opin. Drug Discov. 2021, 16, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Pennington, M.W.; Czerwinski, A.; Norton, R.S. Peptide Therapeutics from Venom: Current Status and Potential. Bioorg. Med. Chem. 2018, 26, 2738–2758. [Google Scholar] [CrossRef] [PubMed]
- Herzig, V.; King, G.F.; Undheim, E.A.B. Can We Resolve the Taxonomic Bias in Spider Venom Research? Toxicon 2019, 1, 100005. [Google Scholar] [CrossRef] [PubMed]
- Lüddecke, T.; Vilcinskas, A.; Lemke, S. Phylogeny-Guided Selection of Priority Groups for Venom Bioprospecting: Harvesting Toxin Sequences in Tarantulas as a Case Study. Toxins 2019, 11, 488. [Google Scholar] [CrossRef]
- von Reumont, B.M.; Campbell, L.I.; Jenner, R.A. Quo Vadis Venomics? A Roadmap to Neglected Venomous Invertebrates. Toxins 2014, 6, 3488–3551. [Google Scholar] [CrossRef]
- Erkoc, P.; von Reumont, B.M.; Lüddecke, T.; Henke, M.; Ulshöfer, T.; Vilcinskas, A.; Fürst, R.; Schiffmann, S. The Pharmacological Potential of Novel Melittin Variants from the Honeybee and Solitary Bees against Inflammation and Cancer. Toxins 2022, 14, 818. [Google Scholar] [CrossRef]
- Daly, N.L.; Wilson, D. Structural Diversity of Arthropod Venom Toxins. Toxicon 2018, 152, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Vargas, J.M.; Possani, L.D.; Luna-Ramírez, K. Arthropod Toxins Acting on Neuronal Potassium Channels. Neuropharmacology 2017, 127, 139–160. [Google Scholar] [CrossRef] [PubMed]
- Uzair, B.; Bint-E-Irshad, S.; Khan, B.A.; Azad, B.; Mahmood, T.; Rehman, M.U.; Braga, V.A. Scorpion Venom Peptides as a Potential Source for Human Drug Candidates. Protein Pept. Lett. 2018, 25, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Lüddecke, T.; Paas, A.; Harris, R.J.; Talmann, L.; Kirchhoff, K.N.; Billion, A.; Hardes, K.; Steinbrink, A.; Gerlach, D.; Fry, B.G.; et al. Venom Biotechnology: Casting Light on Nature’s Deadliest Weapons Using Synthetic Biology. Front. Bioeng. Biotechnol. 2023, 11, 811905. [Google Scholar] [CrossRef] [PubMed]
- Rivera-de-Torre, E.; Rimbault, C.; Jenkins, T.P.; Sørensen, C.V.; Damsbo, A.; Saez, N.J.; Duhoo, Y.; Hackney, C.M.; Ellgaard, L.; Laustsen, A.H. Strategies for Heterologous Expression, Synthesis, and Purification of Animal Venom Toxins. Front. Bioeng. Biotechnol. 2022, 9, 1166601. [Google Scholar] [CrossRef] [PubMed]
- Carlson, E.D.; Gan, R.; Hodgman, C.E.; Jewett, M.C. Cell-Free Protein Synthesis: Applications Come of Age. Biotechnol. Adv. 2012, 30, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Tinafar, A.; Jaenes, K.; Pardee, K. Synthetic Biology Goes Cell-Free. BMC Biol. 2019, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Krinsky, N.; Kaduri, M.; Shainsky-Roitman, J.; Goldfeder, M.; Ivanir, E.; Benhar, I.; Shoham, Y.; Schroeder, A. A Simple and Rapid Method for Preparing a Cell-Free Bacterial Lysate for Protein Synthesis. PLoS ONE 2016, 11, e0165137. [Google Scholar] [CrossRef]
- Silverman, A.D.; Karim, A.S.; Jewett, M.C. Cell-Free Gene Expression: An Expanded Repertoire of Applications. Nat. Rev. Genet. 2020, 21, 151–170. [Google Scholar] [CrossRef]
- Lüddecke, T.; Paas, A.; Talmann, L.; Kirchhoff, K.N.; von Reumont, B.M.; Billion, A.; Timm, T.; Lochnit, G.; Vilcinskas, A. A Spider Toxin Exemplifies the Promises and Pitfalls of Cell-Free Protein Production for Venom Biodiscovery. Toxins 2021, 13, 575. [Google Scholar] [CrossRef] [PubMed]
- Vlasak, R.; Kreil, G. Nucleotide Sequence of Cloned cDNAs Coding for Preprosecapin, a Major Product of Queen-Bee Venom Glands. Eur. J. Biochem. 1984, 145, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, W.; Kou, X.; Luo, Y.; Zhang, Y.; Ma, B.; Wang, M.; Huang, K. Establishment and Optimization of a Wheat Germ Cell-Free Protein Synthesis System and Its Application in Venom Kallikrein. Protein Expr. Purif. 2012, 84, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cui, Z.; Huang, Y.-H.; de Veer, S.J.; Aralov, A.V.; Guo, Z.; Moradi, S.V.; Hinton, A.O.; Deuis, J.R.; Guo, S.; et al. Towards a Generic Prototyping Approach for Therapeutically-Relevant Peptides and Proteins in a Cell-Free Translation System. Nat. Commun. 2022, 13, 260. [Google Scholar] [CrossRef] [PubMed]
- Lüddecke, T.; von Reumont, B.M.; Förster, F.; Billion, A.; Timm, T.; Lochnit, G.; Vilcinskas, A.; Lemke, S. An Economic Dilemma between Molecular Weapon Systems May Explain an Arachno-Atypical Venom in Wasp Spiders (Argiope Bruennichi). Biomolecules 2020, 10, 978. [Google Scholar] [CrossRef] [PubMed]
- Gremski, L.H.; da Justa, H.C.; da Silva, T.P.; Polli, N.L.C.; Antunes, B.C.; Minozzo, J.C.; Wille, A.C.M.; Senff-Ribeiro, A.; Arni, R.K.; Veiga, S.S. Forty Years of the Description of Brown Spider Venom Phospholipases-D. Toxins 2020, 12, 164. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Moreira, D.; Gremski, L.H.; de Moraes, F.R.; Vuitika, L.; Wille, A.C.M.; Hernández González, J.E.; Chaim, O.M.; Senff-Ribeiro, A.; Arni, R.K.; Veiga, S.S. Brown Spider Venom Phospholipase-D Activity upon Different Lipid Substrates. Toxins 2023, 15, 109. [Google Scholar] [CrossRef]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Research 2023, 51, D523–D531. [Google Scholar] [CrossRef]

| Toxin Group | Toxin Name | UniProt ID or Source | Source Organism | Lineage | Size [kDa] |
|---|---|---|---|---|---|
| Cysteine-rich neurotoxins | U-Asilidin(1)-Mar1a | P0DQI8 | Machimus arthriticus (Zeller, 1840) | Robber fly | 3.1 |
| U-Asilidin(1)-Eru1a | P0DQJ1 | Eutolmus rufibarbis (Meigen, 1820) | Robber fly | 3.1 | |
| Delta-miturgitoxin-Cp1a | C0HKG7 | Cheiracanthium punctorium (Villers, 1789) | Spider | 15.1 | |
| Omega Hexatoxin Hi1a | P0C2L5 | Hadronyche infensa (Hickman, 1964) | Spider | 3.9 | |
| Omega-theraphotoxin-Cc1a | D5J6X1 | Pelinobius muticus (Karsch, 1885) | Spider | 4.3 | |
| U11-pisautoxin-Dm1a | S5MK94 | Dolomedes mizhoanus (Kishida, 1936) | Spider | 6.8 | |
| U18-barytoxin-Tl1a | W4VRU3 | Trittame loki (Raven, 1990) | Spider | 7.5 | |
| U1-barytoxin-Tl1a | W4VRV2 | Trittame loki (Raven, 1990) | Spider | 11.1 | |
| U1-oxotoxin-Ot1a | W0LQ84 | Oxyopes takobius (Andreeva & Tyschchenko, 1969) | Spider | 13.7 | |
| U1-pisautoxin-Dm1a | S5N3Q8 | Dolomedes mizhoanus (Kishida, 1936) | Spider | 11.4 | |
| U1-TRTX-Lp1b | P61506 | Lasiodora parahybana (Mello-Leitao, 1917) | Spider | 5.7 | |
| Protease inhibitor | Hirudin | P28509 | Hirudo medicinalis (Linnaeus, 1758) | Leech | 7.0 |
| Antimicrobial peptide | Lycotoxin | B6DD06 | Lycosa singoriensis (Laxmann, 1770) | Spider | 4.8 |
| (Putative) Enzyme | Phospholipase D | A0A1L4BJ98 | Hemiscorpius lepturus (Peters, 1861) | Scorpion | 33.0 |
| Phospholipase D | Q8I914 | Loxosceles laeta (Nicolet, 1849) | Spider | 32.1 | |
| Phospholipase D | Q1KY80 | Loxosceles laeta (Nicolet, 1849) | Spider | 32.0 | |
| Phospholipase D | Q1KY79 | Loxosceles laeta (Nicolet, 1849) | Spider | 32.6 | |
| Phospholipase D | C0JB23 | Loxosceles laeta (Nicolet, 1849) | Spider | 31.5 | |
| Phospholipase D | Q8I912 | Loxosceles laeta (Nicolet, 1849) | Spider | 31.7 | |
| CAP | [25] | Argiope bruennichi (Scopoli, 1772) | Spider | 45.3 | |
| Three-finger toxin (3Ftx) | Long neurotoxin 1 | P34074 | Naja annulata (Peters, 1876) | Snake | 7.8 |
| Delta-elapitoxin-Cb1a | P0DL82 | Calliophis bivirgatus (Boie, 1827) | Snake | 6.7 | |
| Scutelatoxin | Q4VRI0 | Oxyuranus s. scutellatus (Peters, 1876) | Snake | 6.6 | |
| Cytotoxin 1 | P01455 | Naja annulifera (Peters, 1854) | Snake | 6.7 | |
| Cytotoxin sagitoxin | P83345 | Naja sagittifera (Wall, 1913) | Snake | 6.8 | |
| Others | Pimplin | Q8WPC8 | Pimpla hypochondriaca (Retzius, 1783) | Wasp | 13.1 |
| Ryncolin-1 | D8VNS7 | Cerberus rynchops (Schneider, 1799) | Snake | 36.4 | |
| Ryncolin-2 | D8VNS8 | Cerberus rynchops (Schneider, 1799) | Snake | 36.6 | |
| Ryncolin-3 | D8VNS9 | Cerberus rynchops (Schneider, 1799) | Snake | 36.4 | |
| Ryncolin-4 | D8VNT0 | Cerberus rynchops (Schneider, 1799) | Snake | 36.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paas, A.; Dresler, J.; Talmann, L.; Vilcinskas, A.; Lüddecke, T. Venom Ex Machina? Exploring the Potential of Cell-Free Protein Production for Venom Biodiscovery. Int. J. Mol. Sci. 2024, 25, 8286. https://doi.org/10.3390/ijms25158286
Paas A, Dresler J, Talmann L, Vilcinskas A, Lüddecke T. Venom Ex Machina? Exploring the Potential of Cell-Free Protein Production for Venom Biodiscovery. International Journal of Molecular Sciences. 2024; 25(15):8286. https://doi.org/10.3390/ijms25158286
Chicago/Turabian StylePaas, Anne, Josephine Dresler, Lea Talmann, Andreas Vilcinskas, and Tim Lüddecke. 2024. "Venom Ex Machina? Exploring the Potential of Cell-Free Protein Production for Venom Biodiscovery" International Journal of Molecular Sciences 25, no. 15: 8286. https://doi.org/10.3390/ijms25158286
APA StylePaas, A., Dresler, J., Talmann, L., Vilcinskas, A., & Lüddecke, T. (2024). Venom Ex Machina? Exploring the Potential of Cell-Free Protein Production for Venom Biodiscovery. International Journal of Molecular Sciences, 25(15), 8286. https://doi.org/10.3390/ijms25158286







