Abstract
In cervical biopsies, for diagnosis of Human Papilloma Virus (HPV) related conditions, the immunohistochemical staining for p16 has a diagnostic value only if diffusely and strongly positive, pattern named “block-like”. “Weak and/or focal (w/f) p16 expression” is commonly considered nonspecific. In our previous study, we demonstrated the presence of high-risk HPV (hrHPV) DNA by LiPa method in biopsies showing w/f p16 positivity. The aim of the present study was to investigate the presence of hrHPV-DNA by CISH in the areas showing w/f p16 expression. We assessed the presence of hrHPV16, 18, 31, 33, 51 by CISH in a group of 20 cervical biopsies showing w/f p16 expression, some with increased Ki67, and in 10 cases of block-like expression, employed as control. The immunohistochemical p16 expression was also assessed by digital pathology. hrHPV-CISH nuclear positivity was encountered in 12/20 cases of w/f p16 expression (60%). Different patterns of nuclear positivity were identified, classified as punctate, diffuse and mixed, with different epithelial distributions. Our results, albeit in a limited casuistry, show the presence of HPV in an integrated status highlighted by CISH in w/f p16 positive cases. This could suggest the necessity of a careful follow-up of the patients with “weak” and/or “focal” immunohistochemical patterns of p16, mainly in cases of increased Ki67 cell proliferation index, supplemented with molecular biology examinations.
1. Introduction
Human Papilloma Virus (HPV) is a double-strand DNA virus lacking a viral envelope and presenting an icosahedral structure with a specific tropism for the epithelium of skin and mucosa [1]. HPV plays a well-known role in cervical cancer. Persistent high-risk HPV (hrHPV) infection has been recognized to be responsible for high-grade squamous intraepithelial lesions (HSIL) of the uterine cervix, as well as a subgroup of cancers of the vulva, vagina, penis, and anus, and also for some neoplastic lesions of the head and neck [2,3]. Low-risk genotypes (lrHPV), such as HPV6 and 11, are associated with 90% of anogenital warts and recurrent respiratory papillomatosis in men and women [4]. Around 200 HPV types have been fully sequenced, characterized and numbered.
Based on the risk of progression to carcinomas, HPV genotypes have been distinguished in different groups by The International Agency for Research on Cancer (IARC). There are 12 hrHPV types recognized as human carcinogens (Group 1) [5]. The IARC Group 1 of hrHPVs includes genotypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59. Probable or possible hrHPV types (IARC Group 2A and 2B) comprise HPV26, 53, 66, 67, 68, 70, 73, and 82. The last group of lrHPV (IARC Group 3) include HPV6, 11 40, 42, 54, 55, 61, 62, 64, 69, 71, 72, 81, 83, 84, and 89 [5].
It has been reported that a successful infection of HPV requires access to the virus to actively replicate cells, specifically those in the mitosis phase of the cell cycle [6]. Therefore, HPV infection targets replicating cells at the squamocolumnar junction, or it is facilitated by microabrasions occurring on the squamous superficial layers, which expose actively dividing stem/basal epithelial cells [1]. Once started, the HPV infection cycle goes through phases closely linked to the differentiation of epithelial cells.
The early phase of the infection is localized in the basal layers of the squamous epithelium and is characterized by the expression of the early viral genes E1 and E2; the intermediate phase occurs in the intermediate layers, while the late phase, with the assembly and release of mature viral particles, occurs in the squamous superficial layers [7].
Transient HPV infection is cleared by the immune system of the immune-competent host and resolved without consequences. However, in other circumstances, the HPV infection may become persistent, establishing a substrate that favors the development of cancerous lesions [4]. Indeed, in cervical cancer, hrHPV is considered the driver of oncogenesis as it represents the main etiological agent (virtually 100%) of this neoplasia [8]. The integration of specific viral DNA fragments in the host genome leads to the overexpression of E6 and E7 oncoproteins, which can induce proliferation and malignant transformation by interacting with Tumor Protein 53 (TP53) and (Retinoblastoma protein) pRB, both regulators of the cell cycle [9].
The E6 oncoprotein induces the degradation of TP53, allowing cells to continue replicating. E7 causes disruption and loss of the RB/E2F repressor complex by releasing the E2F transcription factor, allowing it to be free to carry out its role in transcription.
This allows cells to enter the S phase of the cell cycle and leads to overexpression of the Cyclin-Dependent kinase inhibitor 2A (CDKN2A/p16) gene with over-expression of the p16 protein [10,11]. So, p16 immunohistochemical over-expression in cervical epithelium is considered a surrogate for transcriptionally active hrHPV infection [12].
Noteworthy, the immunohistochemical expression of p16 was considered to be of diagnostic value only when showing widespread and intense nuclear and cytoplasmic immunoreactivity in more than 50% of epithelial thickness. This type of expression has been called “block-like” p16 expression [13].
In contrast, p16 expression in more than 10 basal/parabasal cells but less than 50% of the epithelial thickness, called “weak and/or focal (w/f) p16 expression” has been considered a nonspecific finding, as it is sometimes detected in the reactive squamous epithelium [14]. Consequently, HPV molecular detection and follow-up are not indicated for cases with “w/f p16 expression”.
Ki67 is a marker of cell proliferation, usually over-expressed in neoplastic tissue. It is a very important prognostic factor and usually helps to establish the degree of differentiation of most tumors. Under physiological conditions, p16 and Ki67 are not co-expressed. Their co-expression correlated with high-grade cervical epithelial lesions and suggests deregulation of the cell cycle by hrHPV [15].
In our previous study, we assessed hrHPV presence by performing an HPV DNA LiPa test on paraffin-embedded cervical biopsies without obvious dysplasia and with w/f p16 expression. We defined them as “cervical atypical lesions” (CAL) with w/f p16 expression. Most showed a high proliferation rate, as detected by increased Ki67 immunostaining. HPV DNA LiPa test confirmed the presence of hrHPV genotypes (i.e., HPV 31, 51, 56, 59, 26, 53, 66, 73 and 82) in most cases [16].
Unfortunately, the PCR-based HPV DNA LiPa test is inadequate for determining whether HPV detected in tissue is transcriptionally active and whether it is playing the role of the oncogenetic driver or is just an innocent bystander [16]. Instead, chromogenic DNA in situ hybridization (DNA-CISH) for detecting HPV DNA in situ has the advantage of preserving the HPV DNA hybridization signals in the tissue morphological context. The different patterns of CISH signals obtained in the nuclei have been classified as “diffuse”, “punctate”, or “mixed” and have been previously correlated in the literature with the phases of the hrHPV infectious cycle [17,18,19].
The diffuse pattern has been correlated with the episomal status of HPV and with its role as a “passenger”. In contrast, the punctate pattern was correlated with the integrated status of HPV and with its role as a “driver” [18]. The punctate pattern is typically found in high-grade squamous intraepithelial lesions (H-SIL), where a low number of viral copies is usually present. The acquisition of viral integration in the basal layers is a key moment in the development of H-SIL, and the punctate pattern has been shown to be a marker of high-grade lesions [19]. In contrast, low-grade squamous intraepithelial lesions (L-SIL) are characterized by a diffuse pattern, consisting of an intense nuclear positivity of CISH signals in the superficial epithelial layers. This pattern corresponds in most cases to the episomal phase of the infection and a more significant number of viral copies, which probably will be eliminated with the desquamation of the superficial cells.
In several cases, a mixed pattern can be found, with a diffuse pattern in the superficial layers and a punctate pattern in the basal layers of the squamous epithelium.
The outcome of cases showing this pattern is less clear-cut and depends largely on the host’s immune response.
The aim of the present study is to investigate the presence of hrHPV-DNA in cervical biopsies with w/f p16 expression and to evaluate the oncogenetic status of the virus, using the DNA-CISH technique, to verify the significance of the w/f immunohistochemical pattern of p16. We propose a new diagnostic role for this immunohistochemical pattern, challenging its historical definition of a “nonspecific finding”.
2. Results
Two expert pathologists (DC and AGG) reviewed all the cases included in the study. They established the histological diagnosis based on morphological and immunohistochemical analysis of Ki67 expression, which allowed for the evaluation of proliferation index. The subjective evaluation of w/f p16 immunohistochemical expression was confirmed by digital pathology evaluation, which highlighted the presence of at least one p16-positive area at the intensity threshold of 0.5. HPV DNA test identified the specific HPV genotype, while the assessment of HPV signal pattern was analyzed by CISH in the morphological context of cervical epithelium. The cases were divided respectively into group 1 (20 cases) and group 2 (10 cases) based on the presence of w/f or block-like p16 expression. Clinical, histological, immunohistochemical and molecular data of the two groups are respectively reported in Table 1 and Table 2. Regarding morphology, w/f p16—group 1 included 4/20 cases (20%) of immature squamous metaplasia with minimal koilocytosis, without significant cellular atypia, 10/20 cases with L-SIL (50%) and 6/20 (30%) cases with H-SIL (Table 1). In group 2, all 10 cases were characterized by H-SIL and 2/10 also had focal areas of invasive squamous carcinoma (Table 2).

Table 1.
Clinical and histological data of patients with weak and/or focal p16 immunohistochemical positivity.

Table 2.
Clinical and histological data of patients with “block-like” p16 immunohistochemical positivity.
In group 1, most cases (16/20, 80%) had w/f p16 expression (score 1+) (Figure 1B). In the remaining 4/20 cases (20%), the prevalent areas of w/f p16 expression were associated with small areas of more intense and extensive p16 expression (score 3+), identifying the “mixed” pattern (score 1+/3+). This was evidenced in two cases of L-SIL and two cases of H-SIL (Figure 2B).
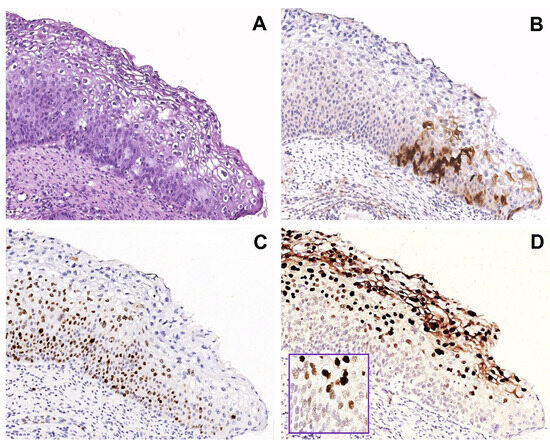
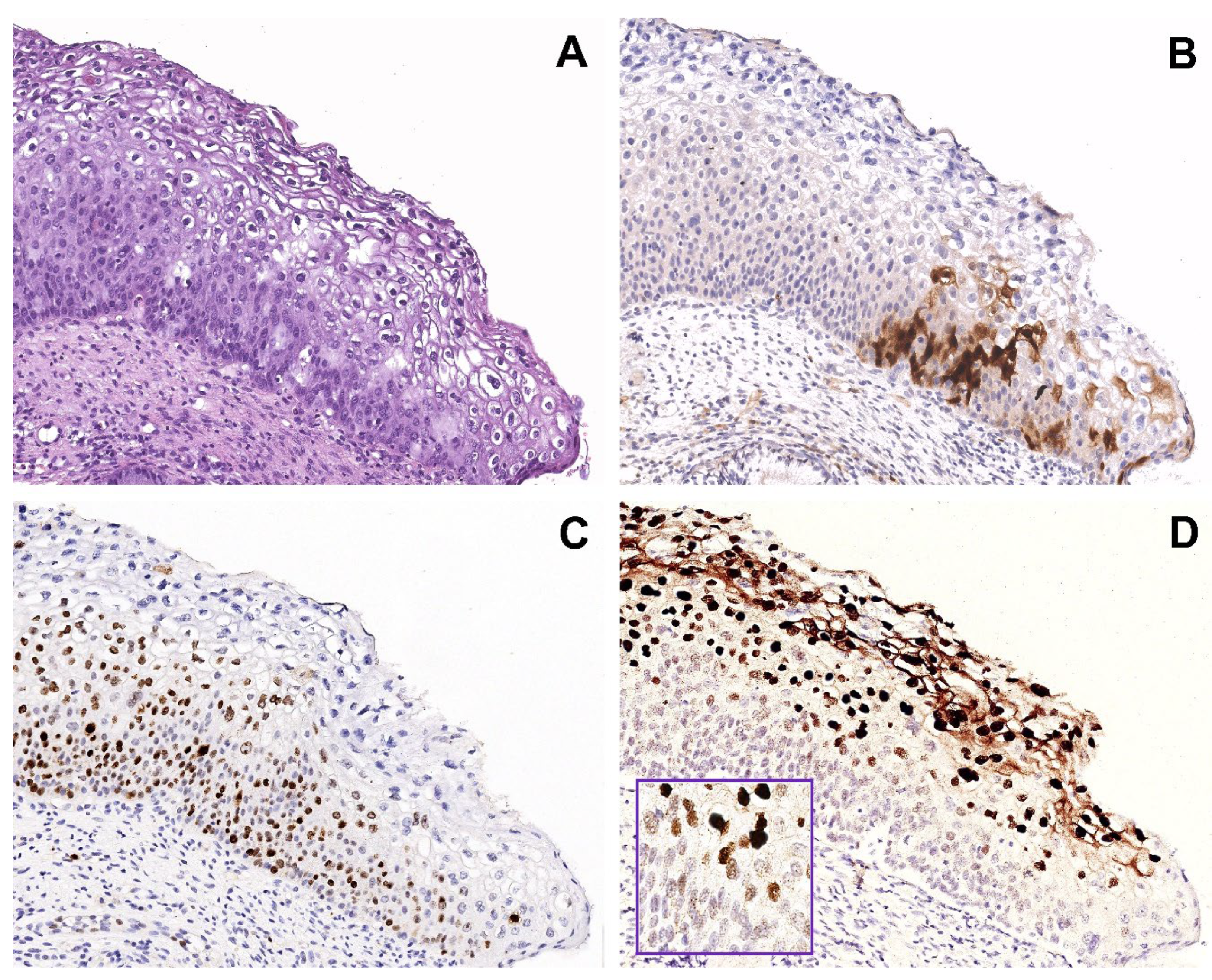
Figure 1.
Group 1 case: serial sections of w/f p16 positive/HPV51 positive case with H-SIL. (A) Biopsy shows H-SIL (CIN2). (B) Focal expression of p16 in a few basal/parabasal cells. (C) Ki67 immunohistochemical staining highlighting a high proliferation rate in the basal and intermediate squamous layers. (D) Mixed pattern of CISH signals: strong, diffuse pattern in the nuclei of the superficial squamous layer and punctate pattern in the nuclei of the basal layer, suggesting a different state of HPV integration. Original magnification (A–D) 100×, figure inserted in (D): 400×; (A) hematoxylin and eosin staining; (B,C) immunoperoxidase staining with 3,3’-diaminobenzidine (DAB) chromogen; (D) and insert, chromogenic in situ hybridization staining (CISH).
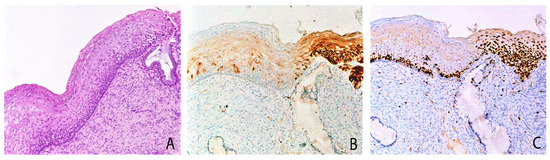
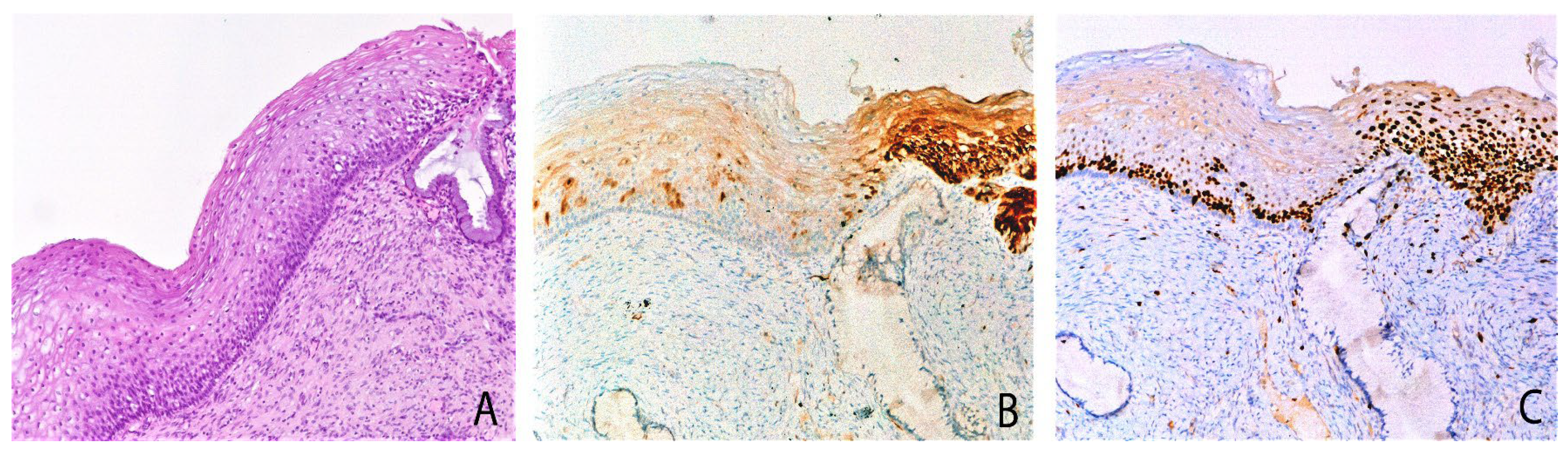
Figure 2.
Cervical biopsy with mild epithelial atypia, mixed pattern of P16 expression and Ki67 expression: serial sections. (A) The biopsy shows mild atypia. (B) On the left weak/focal p16 expression; on the right block-like p16 expression positivity, diffuse in all the squamous layers. (C) Ki67 immunohistochemical staining showed a mild increase of proliferating cells in w/f p16 area and a strong increase in block-like p16 positive area. Original magnifications: 100×; (A) hematoxylin and Eosin staining; (B,C) immunoperoxidase staining with 3,3’-diaminobenzidine (DAB) chromogen. Adapted from Cabibi et al., Diagnostics (Basel), 2021 [16].
Nuclear expression of Ki67 was limited to the lower squamous layers (score 1+) in 4/4 cases with immature metaplasia and 8/10 cases with L-SIL; the remaining 2/10 L-SIL cases showed Ki67 expression extended to the intermediate epithelial layers (score 2+). Finally, in 6/6 H-SIL cases the expression of Ki67 was widespread throughout the epithelial thickness (score 3+).
Of note, among the 6 H-SIL cases, only 2/6 cases showed the mixed p16 pattern, while in the remaining 4/6 H-SIL cases the w/f p16 pattern persisted, although they clearly showed morphological characteristics of H- SIL and high proliferation rate, as evidenced by Ki67 score 3+.
In group 2, where all cases had p16 block-like expression by definition, Ki67 expression was always increased in all layers, indicating a high proliferation rate, in line with their H-SIL phenotype.
Three different HPV genotypes were identified in the w/f 1 group: 10/20 cases (50%) harbored HPV16, 6/20 cases (30%) harbored HPV31, 4/20 cases (20%) harbored HPV51.
Surprisingly, all 4/4 HPV51 positive cases coincided with the 4 H-SIL cases with w/f p16 expression, and Ki67 increased (Figure 1A–C).
By contrast, in group 1 no H-SIL cases with w/f p16 expression resulted positive to HPV16; only L-SIL cases harbored HPV16 genotype.
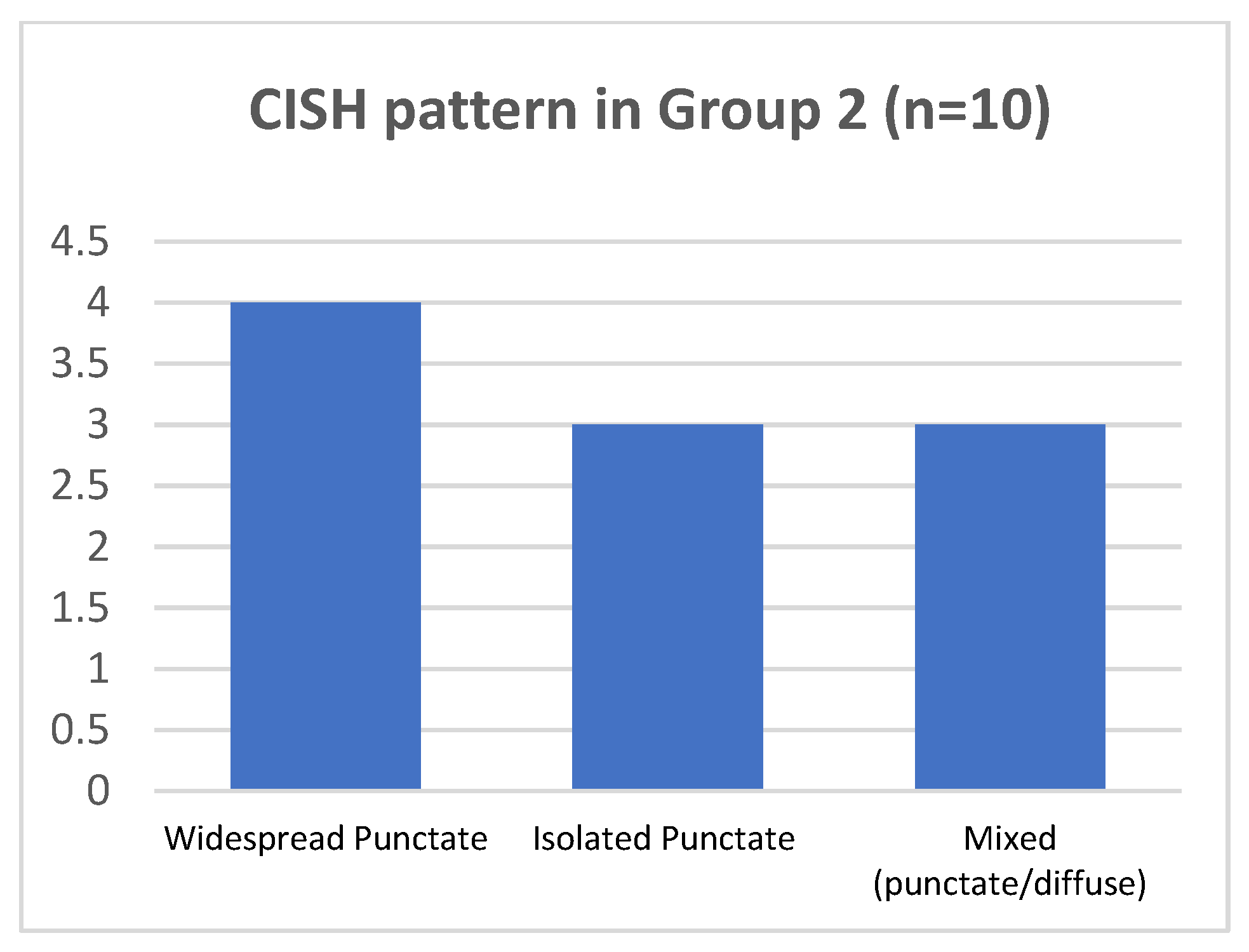
In the group 2, all cases consisted of H-SIL lesions with p16 block like expression and increased Ki67 nuclear positivity (3+) (Figure 3). HPV16 and HPV31 were the most represented genotypes: HPV16 was detected in 8 out of 10 cases and HPV31 in 3 out of 10 cases, co-expressed with other genotypes, namely HPV16, 33, 51.
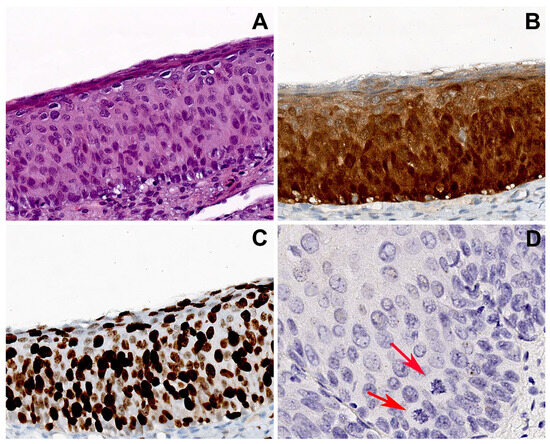
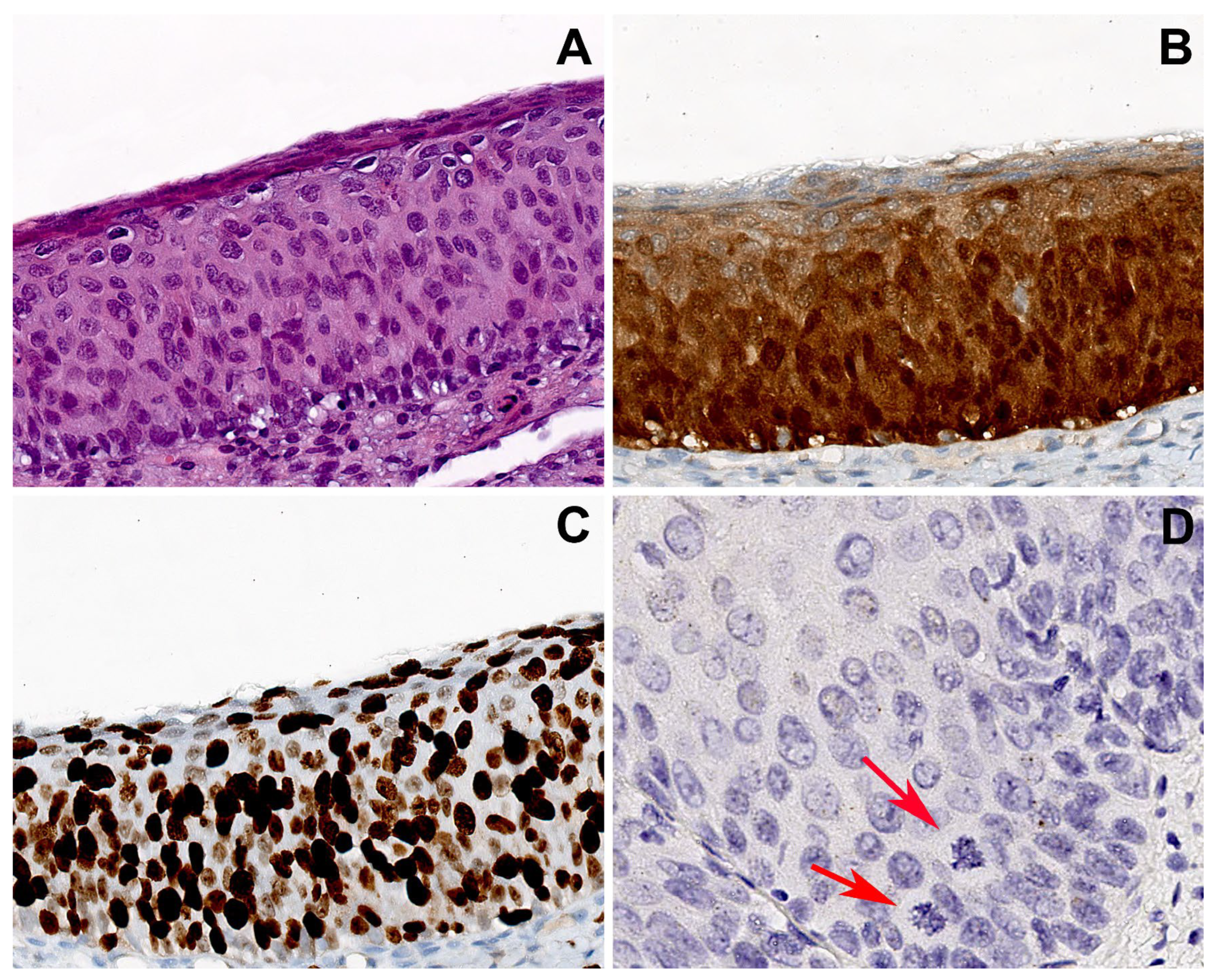
Figure 3.
Group 2 case: serial sections of cervical biopsy with H-SIL, block-like p16 expression, and HPV16. (A) Biopsy shows H-SIL. (B) Block-like p16 positivity, strong and diffuse in all the squamous layers. (C) Ki67 immunohistochemical staining highlighting a high proliferation index with widespread positivity extended to the upper epithelial layers. (D) CISH showed an isolate punctate pattern of positivity in rare nuclei of the lower third of the epithelium, suggesting the integration of HPV. Atypical mitoses are visible (arrows). Original magnifications: (A–C) 200×; (D) 400×; (A) hematoxylin and eosin staining; (B,C) immunoperoxidase staining with 3,3’-diaminobenzidine (DAB) chromogen; (D) chromogenic in situ hybridization staining (CISH).
Finally, HPV-DNA CISH allowed us to relate the p16 patterns observed in the two groups with the presence and distribution of hrHPV in the morphological context of the epithelium. The different patterns of CISH signals observed were classified as punctate, diffuse and mixed, the last one with strong, diffuse nuclear positivity in the superficial epithelial layer and a punctate pattern in the basal layer.
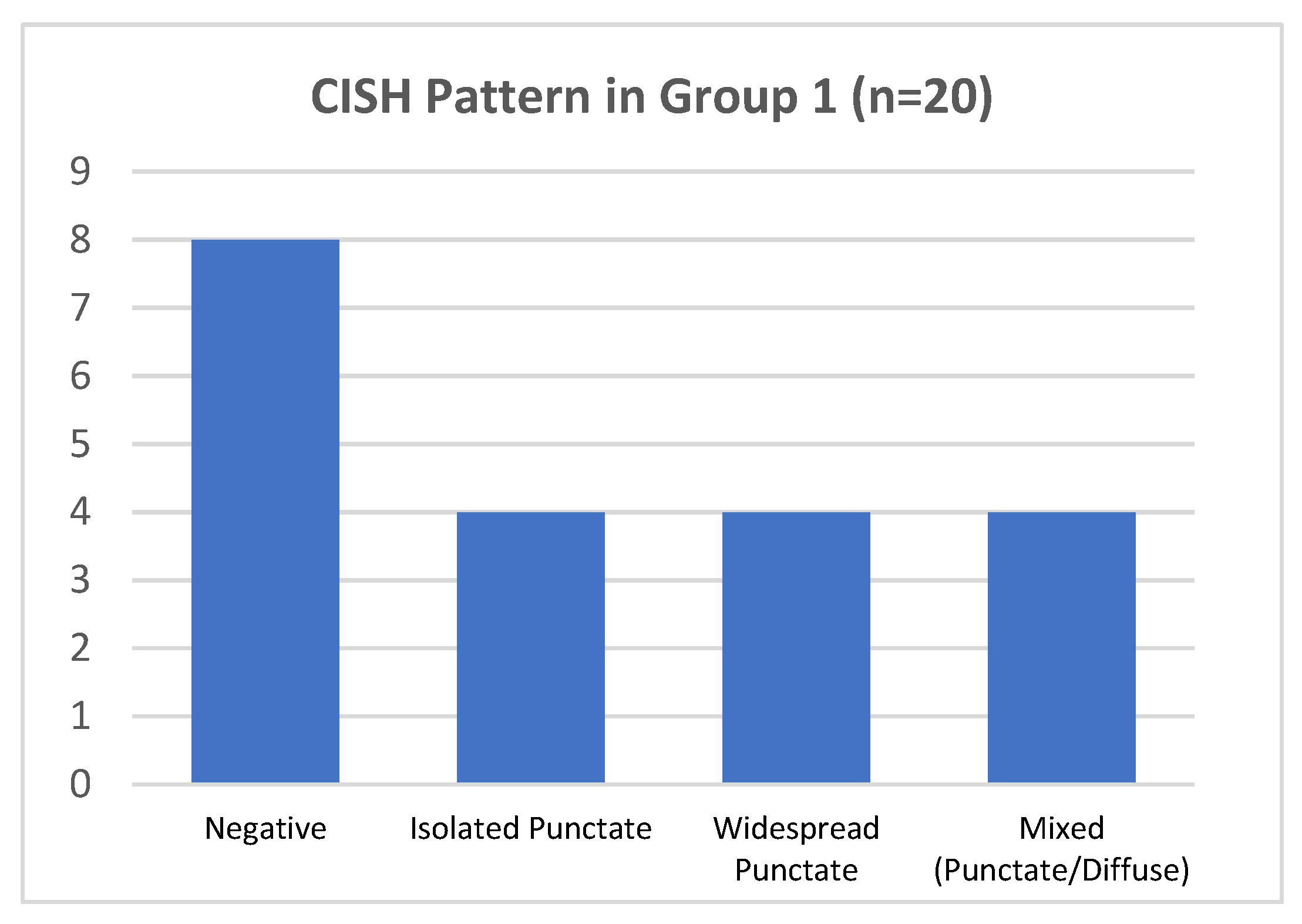
In group 1, hrHPV-CISH nuclear signals was encountered in 12/20 cases (60%) (Figure 4). The various patterns were associated to HPV genotypes, as follows:
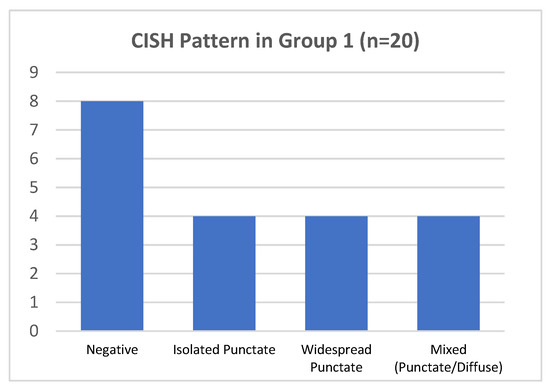
Figure 4.
CISH pattern in group 1.
- -
- In 4/20 cases harboring HPV 16 by DNA-test, the isolate punctate pattern was present in rare nuclei;
- -
- In 4/20 cases, harboring HPV 31, the widespread punctate CISH pattern was encountered;
- -
- In 4/20 cases (two with HPV31 and two with HPV51) a mixed CISH pattern was evidenced (Figure 1D).
Unexpectedly, the remaining 8/20 cases (40%) were negative for CISH HPV DNA, despite the DNA test indicating the presence of hrHPV genotypes (HPV16 in 6/8 and HPV51 in 2/8 cases), 4/8 cases presented increased Ki67 expression and one of them also showed clear morphological features of H-SIL (CIN2) (Table 1).
Interestingly, these 4 HPV31/HPV51 positive cases of group 1 were reported above with w/f p16 expression and high Ki67 expression in the whole thickness of the epithelium.
Figure 1 shows a representative case harboring the HPV51 genotype, with p16 w/f expression, increased ki67 expression, and mixed pattern of CISH signaling.
Of note, in serial sections there is a lack of strict correspondence between the w/f p16 positive areas and the areas with high proliferation rate.
As regard p16-block-like group 2, in 7/10 cases punctate pattern was present. In 4/7 cases (two of them with HPV16 and the two with HPV 31, together with other genotypes), the punctate pattern was widespread in all the epithelial layers; in 3/7 cases, harboring HPV16, the isolated punctate pattern was present. The mixed pattern was observed in 3 of 10 cases, harboring HPV16. No cases presented the CISH diffuse pattern (Figure 5 and Table 2).
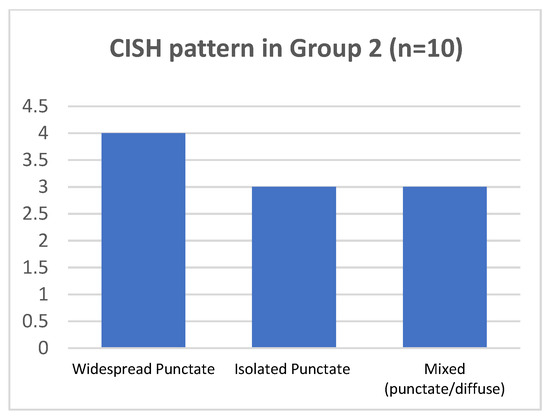
Figure 5.
CISH pattern in group 2.
3. Discussion
p16 expression, when strong and widespread (block-like expression), is considered a marker of transcriptionally active HPV infection [12]. W/f positivity for p16 is a frequent finding that, based on most literature, is considered nonspecific [20,21]. For these cases, HPV-DNA test and/or follow-up are not foreseen [13,22].
We previously assessed whether HPV DNA was indeed absent in such w/f p16-positive biopsies. We highlighted the presence of different hrHPV genotypes. The most frequent were HPV31 and HPV51. They belong to the IARC group 1 genotypes and have often been associated with an increase in the cell proliferation index (Ki67) [16].
The HPV DNA test, while allowing precise genotyping of the virus, is not adequate to establish the carcinogenic potential of the presence of HPV. Indeed, our previous study, based on the detection of hrHPV by HPV DNA testing, was unable to specify what role the virus played at that time.
The role of the virus has been hypothesized, at least for some cases, by an inference based on the presence of a high proliferation rate, evidenced by the high expression of Ki67 [16].
CISH for in situ HPV-DNA detection has the advantage of preserving the morphological context of HPV DNA signals and predicting whether the virus is a “driver” or a “passenger” [18].
The purpose of the present study was to demonstrate, through HPV-DNA CISH, if the virus is observable in w/f p16 positive biopsies and to assess its pattern of nuclear positivity.
Our work confirmed the presence of hrHPV DNA by using CISH method in 12/20 (60%) w/f p16 biopsies, all of them showing the punctate or mixed pattern, suggesting the integrated status of the virus. The increased expression of Ki67 and the morphological characteristics of H-SIL in many of these w/f p16 positive cases supports the hypothesis that in these cases the virus has begun its role as oncogenic driver [23]. So, in many cases, the mechanism of cellular proliferation could have been triggered before the occurrence of block-like p16 positivity, and sometimes independently from p16 expression. In the serial sections of Figure 1 the absence of a strict correspondence between the w/f p16 positive area and the areas with high proliferation rate supports this hypothesis.
In other cases, the coexistence of both p16 w/f positive areas and block p16 positive areas in the same histological section suggests that the w/f pattern could be a precursor of block-like positivity (Figure 2). Thus, both the p16 w/f pattern and Ki67 over-expression could represent early events in carcinogenesis.
In H-SIL cases, harboring hrHPV DNA and with increased expression of ki67 but without the block-like pattern of p16 expression, we can hypothesize further mechanisms of carcinogenesis, different from the more well-known mechanisms of HPV16 and HPV18, leading to p53 inactivation and Rb blocking, the last one determining overexpression of p16 [24,25]. Consistent with this hypothesis, none of the group 1 cases with H-SIL/Ki67 3+ and w/f p16 expression harbored HPV 16, while all were positive for HPV51, one of the less frequent hrHPV genotypes. So, in group 1, HPV 16 was evidenced only in L-SIL cases.
On the contrary, in group 2, including H-SIL cases with block-like p16 expression and increased Ki67 (3+), HPV 16 was the most frequent genotype (7/10 cases), in keeping with the hypothesis previously proposed that the oncogenic effects of HPV16 are strictly dependent on the inactivation of Rb, with consequent quick increase of p16 transcription, resulting in early block-like p16 over-expression [16].
The assessment of the integration status of a virus by CISH is cost-effective and feasible for use in clinical practice. However, the interpretation of HPV-DNA CISH is sometimes difficult. Thus, the reliability of the results could depend on individual skills and visual assessments that might cause discrepancies between observers [26]. Thus, the occurrence of HPV-DNA test positive/CISH-negative cases could be explained by the low sensitivity of CISH method.
As reported by Evans et al., Although CISH is less sensitive than PCR, the non-detection of HPV DNA by CISH cannot be taken as proof of an HPV negative or passenger HPV associated lesion, as it is also possible that the driver HPV DNA load might be below the threshold of CISH detection [18]. Subsequently CISH is not reliable as screening method. In this regard, the use of HPV RNA CISH is suggested [17,27,28].
The implementation of Artificial Intelligence (AI) platforms capable of interpreting p16 patterns, could increase the sensitivity of CISH. The cytological screening of cervical pap smears has been the first pioneering application of software-based solutions with PAPnet [29], and recently novel progress in the field has been accomplished [30,31]. Immunohistochemical analysis of p16 expression and CISH signals through AI in cervical biopsies could improve the comprehension of w/f p16 expression and its predictive potential.
Furthermore, in biopsies showing H-SIL but with HPV CISH and HPV-DNA test negative results, a “hit and run” carcinogenic mechanism could be hypothesized [32]. Indeed, the damage caused by HPV infection could persists, leading to the development of epithelial dysplasia, even after the virus has been cleared by the host immune system [32].
According to the literature, the most represented CISH pattern in our study is the punctate pattern, which is the one related to the integrated status of the virus, the initiation mechanisms of carcinogenesis and the development of H-SIL/carcinoma. On the contrary, the episomal status, typical of the transient phase of HPV infection, results in diffuse CISH signal, and correlates with the productive phenotype of HPV and koilocytosis, observed in L-SIL [19,33,34]. The two patterns of CISH reactivity can be observed alone or admixed in the same lesion.
Interestingly, it has been recently showed that through dual-color fluorescence in situ hybridization, coupling probes for HPV DNA with probes for chromosomal sites specific of recurrent HPV integration, enables to better distinguish integrated from episomal HPV [35]. Techniques of this type should be enforced to improve the possibility of properly identifying integrated HPV in cervical biopsies, since the event of integration is considered one of the main triggers leading to aberrant expression of viral oncoprotein [36].
In our study, the p16 “block” control group showed mostly punctate signals in the basal layer, indicating integrated HPV status, typical of high-grade lesions.
Of special note and in disagreement with scientific literature that only considers block-like p16 expression as index of transcriptionally active HPV infection, we found the punctate pattern also in 12/20 biopsies with w/f p16.
So, this pattern must not be underestimated, mainly if HPV DNA test highlight the presence of HPV51 genotype. Omori et al. reported that CIN2 lesions with isolated punctate CISH pattern have a high risk of progression to CIN3, and none of them regress. The risk of progression is lower in cases of mixed signal, nevertheless, 21% of the cases progress [37].
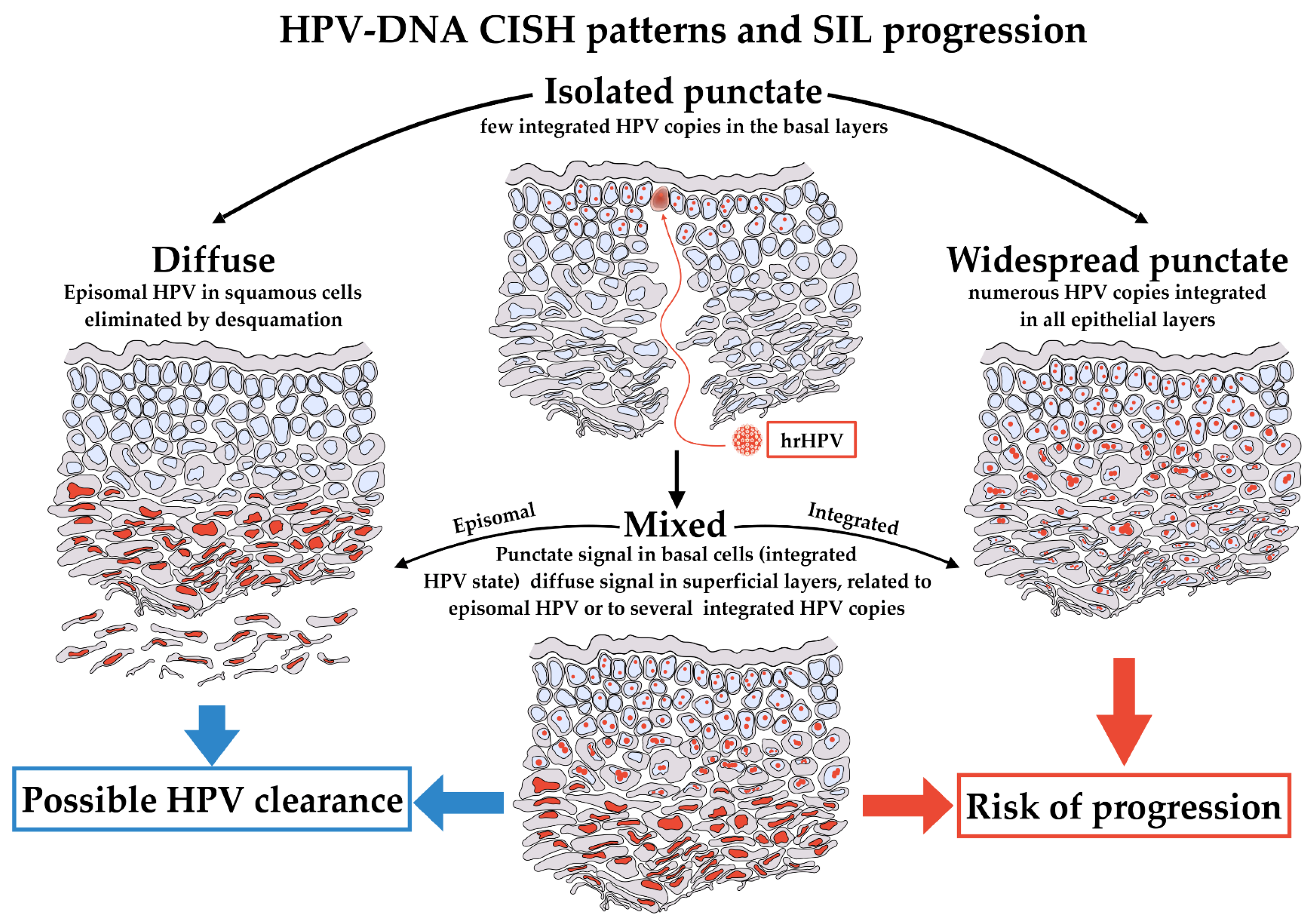
The finding of isolated punctate CISH signal is thought to be associated to a risk of progression or is indicative of a non-regressing lesion. Conversely, it remains unclear whether the mixed CISH pattern represents a process of transition from an episomal status to an integrated status or the opposite. Thus, the clinical significance of the mixed CISH pattern has not yet clarified [34]. Figure 6 shows the patterns of CISH signals and their proposed meanings.
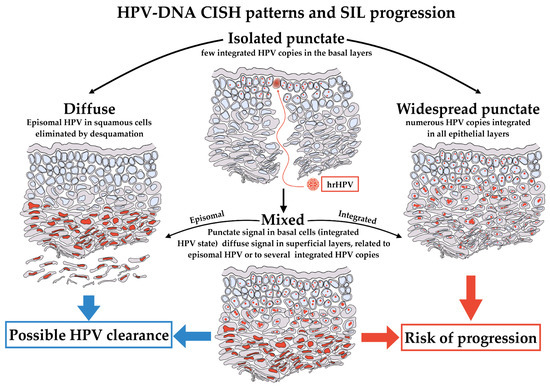
Figure 6.
HPV-DNA CISH patterns and their proposed meaning. hrHPV infection starts in basal cells of epithelium. The first phases of infection should result in an isolated punctate pattern of CISH, limited exclusively to this type of cells (top). The stages of infection could evolve in three different patterns of CISH, the diffuse, the widespread punctate and the mixed. A diffuse pattern indicates the presence of the virus in the squamous cells, and it is a pattern associated to a potential successful intervention of immune system, definitively eliminating HPV (left). Conversely, a widespread punctate pattern in all epithelial layers could be observed if HPV is stably integrated in the almost totality of cells. This is the pattern more likely to evolve toward progression (right). Lastly, the mixed pattern presents intermediate features, with isolated punctate pattern in basal and para-basal cells, becoming diffuse in the superficial layers (bottom). The mixed pattern could evolve toward either diffuse or widespread punctate and retains potential features of clearance or progression.
The present study, albeit performed in a limited casuistry, confirms that in biopsies with w/f p16 immunoreactivity, HPV may be present in integrated status, highlighted by punctate or mixed CISH pattern.
We are aware that w/f p16 cannot be proposed as a reference diagnostic marker for HPV presence and risk of progression, due to the limited number of our casuistry and to the reduced specificity of this finding. However, we think that in w/f p16 positive cases showing punctate or mixed CISH signal, especially in cases with high proliferation index, a follow-up is necessary due to the potential risk of progression, regardless of the presence of block-like p16 expression. As previously pointed out, it could be theorized that HPV integration of less frequent genotypes may be not strictly related to p16 immunoreactivity [16]. Importantly, w/f p16 reported a sensitivity of sensitivity of 80% (16/20 cases) and a specificity of 95% (19/20 cases) in hrHPV identification, considering HPV DNA test as reference method [16]. Moreover, we hypothesize that w/f p16 expression could potentially be beneficial to stratify LSILs into groups for high and low progression risk, verifying this hypothesis with an appropriate follow up. Our investigations will be further extended performing HPV RNA CISH for E6 and E7 mRNAs in w/f p16 positive cervical biopsies, to confirm the potential role of hrHPVs in this histological subgroup. To our knowledge, this is the first study correlating the w/f positive p16 immunohistochemical expression, highlighted using digital pathology, and hrHVP status, assessed by CISH.
We argue that most of w/f p16 positive low-grade lesions spontaneously regress in few months, probably due to the effective activation of the immune system but an increased clinical attention could reduce the number of underestimated cases with risk of progression, sensitively improving patient’s health.
4. Materials and Methods
4.1. Samples Collection
The study was retrospectively performed from a broader casuistry of 200 formalin-fixed, paraffin-embedded cervical biopsies received from June 2022 to December 2023 at the Unit of Anatomic Pathology of the Department of Health Promotion, Mother and Childcare, Internal Medicine and Medical Specialties, P. Giaccone University Hospital of Palermo. From this broader casuistry, we retrospectively selected 30 cases based on the criterion of hrHPV positivity from the HPV DNA test (see Section 4.2) performed by the Microbiology and Virology Unit of the same Department. The age of the patients ranged from 26 to 76 years, with an average age of 45.4 years. Two expert pathologists, AGG and DC, performed the histological evaluation of all biopsies, reporting them as Low-grade or High-grade Squamous Intraepithelial Lesions (L-SILs or H-SILs, respectively) or Metaplasia and Koilocytosis. Based on the study’s main aim, investigating the real meaning of p16 positivity in cervical intraepithelial lesions, we classified the biopsies exclusively according to morphological characteristics, thus not considering the p16 pattern as a classification parameter, differently from current LAST criteria [38]. We first performed p16 and ki67 immunohistochemistry on 20 consecutively selected cases with “w/f p16 immunohistochemical positivity”, constituting the case series. A group of 10 cases with “block-like” p16 positivity constituted the control group. An HPV-DNA test and the immunohistochemical assay were performed as reported below.
4.2. HPV Detection and Genotyping
For the HPV-DNA test, 5 µm-paraffin embedded sections of each biopsy were placed in a microcentrifuge tube with 200 µL of Feoli-Fonseca buffer (0.5% Tween 20, Tris-HCl 50 mM pH 8.5 and 1 mM ethylene diaminetetraacetic-acid), containing 300 µg/mL of proteinase K. The mixture was incubated at 56 °C overnight.
Proteinase K was then denatured at 95 °C for 10 min. After centrifugation at 13,000 rpm for 5 min, the supernatant was purified with QIAmp DNA mini kit (QIAgen, Hilden, Germany), following the protocol for DNA purification from blood or body fluids [39].
To control the extraction and to obtain a qualitative evaluation of the nucleic acid, we performed a spectrophotometric measurement evaluating the absorbance at the wavelength of 260 and 280 nm. The ratio between 1.8 and 2.0 guarantees the quality of the extracted DNA. The extracted DNA was stored at −20 °C or processed immediately.
HPV-DNA detection and genotyping was carried out using INNOLiPA® HPV Genotyping Extra II (Fujirebio, Tokyo, Japan), which identifies 23 genotypes classified as hrHPV or probable hrHPV (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 67, 68, 26, 53, 66, 70, 73, and 82) and 9 types classified as lrHPV (HPV6, 11, 40, 42, 43, 44, 54, 61, 62, 81, 83, and 89). A 65 bp fragment of the L1 gene [23,40], generated by an SPF10 PCR, was subjected to reverse-dot blot hybridization assay to identify the genotype, according to the manufacturer’s instructions.
4.3. Immunohistochemical Assay
Immunohistochemical stains were previously carried out with a BOND III automated slide staining system (Leica Biosystems, Newcastle, UK) according to the manufacturer’s instructions, using p16 monoclonal primary antibody (clone 6H12, Leica Biosystems, Newcastle, UK) and Ki67 monoclonal primary antibody (clone MIB-1, Agilent Technologies, Carpinteria, CA, USA). The 3,3-diaminobenzidine kit was used as chromogen. The slides were observed on a Leica DM2000 microscope (Leica Microsystems, Wetzlar, Germany).
P16 immunostaining was defined “weak and/or focal” when weak/focal nuclear and cytoplasmic positivity was present in more than 10 basal/parabasal cells and less than 50% of the epithelium thickness. A “block-like” p16 pattern was considered when strong/diffuse nuclear and cytoplasmic immunoreactivity represented greater than or equal to 50% reactivity in all the epithelium layers.
All slides were independently reviewed by two pathologists (DC and AGG).
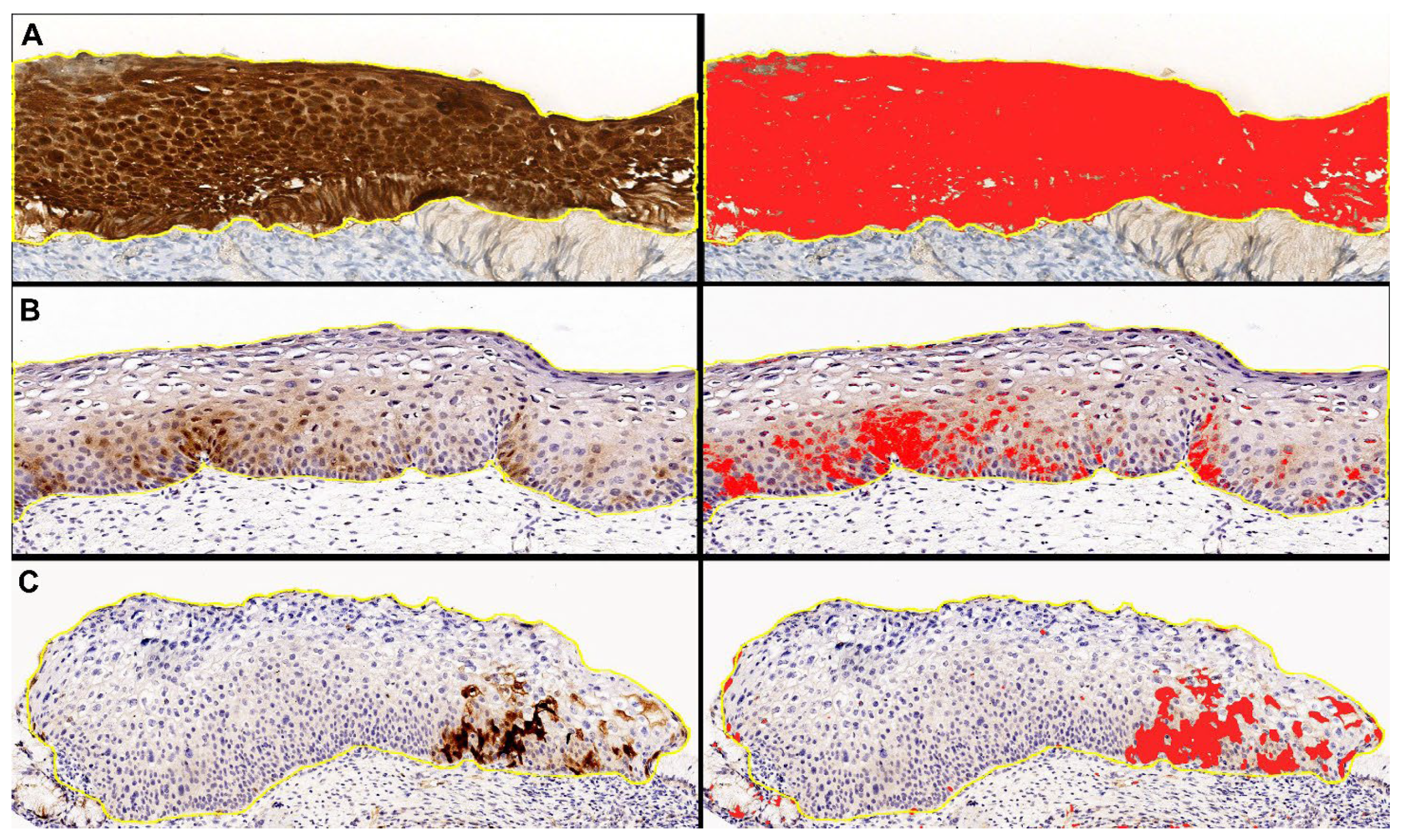
To standardize the selection of w/f p16 positive cases and eliminate any interobserver variability, we performed digital image analysis with QuPath software package v. 0.4.3 [41]. The immunohistochemical intensity threshold for p16 was set at the value of 0.5, since at this value the digital analysis correctly highlighted the “block-like” staining of the control cases, according to the previous definition in the literature (Figure 7).
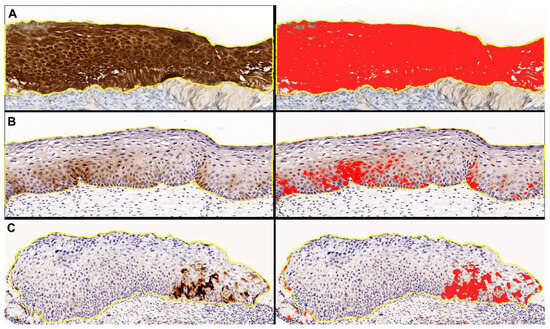
Figure 7.
Representative images of “digital pathology evaluation” performed with QuPath software on p16 digital slides, with original DAB-stained images (left) and the relative digital detection of DAB signal in red (right). (A) Block-like positive case. (B,C) Weak and/or focal positive cases.
Cases in which the digital analysis highlighted at the above-reported threshold, at least one spotty area of any size, were considered w/f p16 positive cases and were included in the study.
Noteworthy, the subjective inclusion criteria of the pathologists’ evaluation and the digital analysis criteria were concordant in all cases (k = 0.9).
Ki67 results were classified 1+ when nuclear expression was limited to the lower epithelial layers, 2+ when extended to the intermediate epithelial layers and 3+ when widespread expressed throughout the epithelial thickness.
4.4. Chromogenic DNA In Situ Hybridization (DNA-CISH) Assay
HPV DNA-CISH assay was performed on 20 cases previously resulted positive for at least one of the hrHPV genotypes (16, 18, 31, 33, and 51) by HPV-DNA test and with “w/f p16 positivity”. Five-micrometer-thick histological sections of cervical biopsy were prepared and subjected to HPV CISH, with probes for hrHPV16, 18, 31, 33, and 51 (Leica Biosystems). CISH was performed on Leica BOND III automatic slide stainer, according to manufacturer’s protocols. The above-mentioned probes were used because they allow detecting the most frequent hrHPV genotypes with a standardized procedure. Negative controls were included in each run.
Ten cases harboring at least one of the above reported hrHPV genotypes by HPV-DNA test and with “block-like” p16-positive immunostaining constituted the control group.
A positive HPV ISH test result was defined as positive if any of the cells showed brown, dot-like nuclear positivity (punctate pattern) or strong homogeneous, nuclear positivity (diffuse pattern) or both of them (mixed pattern).
Author Contributions
Conceptualization and methodology, D.C. and A.G.G.; software, A.G.G., V.B. and A.C.; validation, D.C. and G.C.; formal analysis, D.C., A.G.G. and A.Q.; data curation, E.F., R.P. and A.Q.; writing—original draft preparation, R.L.C. and A.G.G.; writing—review and editing, D.C., A.G.G. and A.Q.; supervision, D.C., A.G.G. and G.C.; funding acquisition, D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research has received funding from the European Union—NextGenerationEU through the Italian Ministry of University and Research under PNRR-M4C2-I1.3 Project PE_00000019 “HEAL ITALIA” to Prof. Daniela Cabibi CUP B73C220012500.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on reasonable request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Graham, S.V. The human papillomavirus replication cycle, and its links to cancer progression: A comprehensive review. Clin. Sci. 2017, 131, 2201–2221. [Google Scholar] [CrossRef] [PubMed]
- Scurry, J.; Wilkinson, E.J. Review of terminology of precursors of vulvar squamous cell carcinoma. J. Low. Genit. Tract Dis. 2006, 10, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Partridge, J.M.; Hughes, J.P.; Feng, Q.; Winer, R.L.; Weaver, B.A.; Xi, L.F.; Stern, M.E.; Lee, S.; O’reilly, S.F.; Hawes, S.E.; et al. Genital human papillomavirus infection in men: Incidence and risk factors in a cohort of university students. J. Infect. Dis. 2007, 196, 1128–1136. [Google Scholar] [CrossRef]
- Hartwig, S.; Syrjänen, S.; Dominiak-Felden, G.; Brotons, M.; Castellsagué, X. Estimation of the epidemiological burden of human papillomavirus-related cancers and non-malignant diseases in men in Europe: A review. BMC Cancer 2012, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Bzhalava, D.; Eklund, C.; Dillner, J. International standardization and classification of human papillomavirus types. Virology 2015, 476, 341–344. [Google Scholar] [CrossRef]
- Pyeon, D.; Pearce, S.M.; Lank, S.M.; Ahlquist, P.; Lambert, P.F. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009, 5, e1000318. [Google Scholar] [CrossRef]
- Kadaja, M.; Silla, T.; Ustav, E.; Ustav, M. Papillomavirus DNA replication—From initiation to genomic instability. Virology 2009, 384, 360–368. [Google Scholar] [CrossRef]
- Mpunga, T.; Chantal Umulisa, M.; Tenet, V.; Rugwizangoga, B.; Milner, D.A., Jr.; Munyanshongore, C.; Heideman, D.A.; Bleeker, M.C.; Tommasino, M.; Franceschi, S.; et al. Human papillomavirus genotypes in cervical and other HPV-related anogenital cancer in Rwanda, according to HIV status. Int. J. Cancer 2020, 146, 1514–1522. [Google Scholar] [CrossRef]
- Hebner, C.M.; Laimins, L.A. Human papillomaviruses: Basic mechanisms of pathogenesis and oncogenicity. Rev. Med. Virol. 2006, 16, 83–97. [Google Scholar] [CrossRef]
- Mantovani, F.; Banks, L. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 2001, 20, 7874–7887. [Google Scholar] [CrossRef]
- Mabruk, M. The mystery of human papillomaviruses in carcinogenesis. Expert Rev. Mol. Diagn. 2008, 8, 1–4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- da Mata, S.; Ferreira, J.; Nicolás, I.; Esteves, S.; Esteves, G.; Lérias, S.; Silva, F.; Saco, A.; Cochicho, D.; Cunha, M.; et al. P16 and HPV Genotype Significance in HPV-Associated Cervical Cancer-A Large Cohort of Two Tertiary Referral Centers. Int. J. Mol. Sci. 2021, 22, 2294. [Google Scholar] [CrossRef] [PubMed]
- Darragh, T.M.; Colgan, T.J.; Thomas Cox, J.; Heller, D.S.; Henry, M.R.; Luff, R.D.; McCalmont, T.; Nayar, R.; Palefsky, J.M.; Stoler, M.H.; et al. The Lower Anogenital Squamous Terminology Standardization project for HPV-associated lesions: Background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. Int. J. Gynecol. Pathol. 2013, 32, 76–115. [Google Scholar] [CrossRef] [PubMed]
- Skapa, P.; Robova, H.; Rob, L.; Zamecnik, J. p16(INK4a) immunoprofiles of squamous lesions of the uterine cervix-implications for the reclassification of atypical immature squamous metaplasia. Pathol. Oncol. Res. 2013, 19, 707–714. [Google Scholar] [CrossRef]
- Yu, L.; Fei, L.; Liu, X.; Pi, X.; Wang, L.; Chen, S. Application of p16/Ki-67 dual-staining cytology in cervical cancers. J. Cancer 2019, 10, 2654–2660. [Google Scholar] [CrossRef]
- Cabibi, D.; Napolitano, C.; Giannone, A.G.; Micciulla, M.C.; Porcasi, R.; Lo Coco, R.; Bosco, L.; Vinciguerra, M.; Capra, G. Predictive Role of the p16 Immunostaining Pattern in Atypical Cervical Biopsies with Less Common High Risk HPV Genotypes. Diagnostics 2021, 11, 1947. [Google Scholar] [CrossRef]
- Kelesidis, T.; Aish, L.; Steller, M.A.; Aish, I.S.; Shen, J.; Foukas, P.; Panayiotides, J.; Petrikkos, G.; Karakitsos, P.; Tsiodras, S. Human papillomavirus (HPV) detection using in situ hybridization in histologic samples: Correlations with cytologic changes and polymerase chain reaction HPV detection. Am. J. Clin. Pathol. 2011, 136, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.F.; Matthews, A.; Kandil, D.; Adamson, C.S.; Trotman, W.E.; Cooper, K. Discrimination of ‘driver’ and ‘passenger’ HPV in tonsillar carcinomas by the polymerase chain reaction, chromogenic in situ hybridization, and p16(INK4a) immunohistochemistry. Head Neck Pathol. 2011, 5, 344–348. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kalof, A.N.; Evans, M.F.; Simmons-Arnold, L.; Beatty, B.G.; Cooper, K. p16INK4A immunoexpression and HPV in situ hybridization signal patterns: Potential markers of high-grade cervical intraepithelial neoplasia. Am. J. Surg. Pathol. 2005, 29, 674–679. [Google Scholar] [CrossRef]
- Klaes, R.; Friedrich, T.; Spitkovsky, D.; Ridder, R.; Rudy, W.; Petry, U.; Dallenbach-Hellweg, G.; Schmidt, D.; von Knebel Doeberitz, M. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int. J. Cancer 2001, 92, 276–284. [Google Scholar] [CrossRef]
- Tsoumpou, I.; Arbyn, M.; Kyrgiou, M.; Wentzensen, N.; Koliopoulos, G.; Martin-Hirsch, P.; Malamou-Mitsi, V.; Paraskevaidis, E. p16(INK4a) immunostaining in cytological and histological specimens from the uterine cervix: A systematic review and meta-analysis. Cancer Treat. Rev. 2009, 35, 210–220. [Google Scholar] [CrossRef]
- Liu, Y.; Alqatari, M.; Sultan, K.; Ye, F.; Gao, D.; Sigel, K.; Zhang, D.; Kalir, T. Using p16 immunohistochemistry to classify morphologic cervical intraepithelial neoplasia 2: Correlation of ambiguous staining patterns with HPV subtypes and clinical outcome. Hum. Pathol. 2017, 66, 144–151. [Google Scholar] [CrossRef]
- Cabibi, D.; Giovannelli, L.; Martorana, A.; Migliore, M.C.; Tripodo, C.; Campione, M.; Ammatuna, P.; Aragona, F. Predictive role of histological features and Ki67 pattern on high-risk HPV presence in atypical cervical lesions. Histopathology 2007, 51, 713–716. [Google Scholar] [CrossRef]
- Boyer, S.N.; Wazer, D.E.; Band, V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996, 56, 4620–4624. [Google Scholar]
- Giarrè, M.; Caldeira, S.; Malanchi, I.; Ciccolini, F.; Leão, M.J.; Tommasino, M. Induction of pRb degradation by the human papillomavirus type 16 E7 protein is essential to efficiently overcome p16INK4a-imposed G1 cell cycle Arrest. J. Virol. 2001, 75, 4705–4712. [Google Scholar] [CrossRef]
- Fatima, N.; Cohen, C.; Lawson, D.; Siddiqui, M.T. Automated and manual human papilloma virus in situ hybridization and p16 immunohistochemistry: Comparison in metastatic oropharyngeal carcinoma. Acta Cytol. 2013, 57, 633–640. [Google Scholar] [CrossRef]
- Chi, J.; Preeshagul, I.R.; Sheikh-Fayyaz, S.; Teckie, S.; Kohn, N.; Ziemba, Y.; Laser, A.; Frank, D.; Ghaly, M.; Kamdar, D.; et al. Evaluating of HPV-DNA ISH as an adjunct to p16 testing in oropharyngeal cancer. Future Sci. OA 2020, 6, Fso606. [Google Scholar] [CrossRef]
- Zito Marino, F.; Ronchi, A.; Stilo, M.; Cozzolino, I.; La Mantia, E.; Colacurci, N.; Colella, G.; Franco, R. Multiplex HPV RNA in situ hybridization/p16 immunohistochemistry: A novel approach to detect papillomavirus in HPV-related cancers. A novel multiplex ISH/IHC assay to detect HPV. Infect. Agent. Cancer 2020, 15, 46. [Google Scholar] [CrossRef]
- Mango, L.J. Computer-assisted cervical cancer screening using neural networks. Cancer Lett. 1994, 77, 155–162. [Google Scholar] [CrossRef]
- Tan, S.L.; Selvachandran, G.; Ding, W.; Paramesran, R.; Kotecha, K. Cervical Cancer Classification From Pap Smear Images Using Deep Convolutional Neural Network Models. Interdiscip Sci 2024, 16, 16–38. [Google Scholar] [CrossRef]
- Sompawong, N.; Mopan, J.; Pooprasert, P.; Himakhun, W.; Suwannarurk, K.; Ngamvirojcharoen, J.; Vachiramon, T.; Tantibundhit, C. Automated Pap Smear Cervical Cancer Screening Using Deep Learning. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; Volume 2019, pp. 7044–7048. [Google Scholar] [CrossRef]
- Ferreira, D.A.; Idris, A.; McMillan, N.A.J. Analysis of a hit-and-run tumor model by HPV in oropharyngeal cancers. J. Med. Virol. 2023, 95, e28260. [Google Scholar] [CrossRef]
- Cooper, K.; Evans, M.; Mount, S. Biology and evolution of cervical squamous intraepithelial lesions: A hypothesis with diagnostic prognostic implications. Adv. Anat. Pathol. 2003, 10, 200–203. [Google Scholar] [CrossRef]
- Zappacosta, R.; Colasante, A.; Viola, P.; D’Antuono, T.; Lattanzio, G.; Capanna, S.; Gatta, D.M.P.; Rosini, S. Chromogenic in situ hybridization and p16/Ki67 dual staining on formalin-fixed paraffin-embedded cervical specimens: Correlation with HPV-DNA test, E6/E7 mRNA test, and potential clinical applications. Biomed. Res. Int. 2013, 2013, 453606. [Google Scholar] [CrossRef]
- Xiong, J.; Cheng, J.; Shen, H.; Ren, C.; Wang, L.; Gao, C.; Zhu, T.; Li, X.; Ding, W.; Zhu, D.; et al. Detection of HPV and Human Chromosome Sites by Dual-Color Fluorescence In Situ Hybridization Reveals Recurrent HPV Integration Sites and Heterogeneity in Cervical Cancer. Front. Oncol. 2021, 11, 734758. [Google Scholar] [CrossRef]
- Pett, M.; Coleman, N. Integration of high-risk human papillomavirus: A key event in cervical carcinogenesis? J. Pathol. 2007, 212, 356–367. [Google Scholar] [CrossRef]
- Omori, M.; Hashi, A.; Nakazawa, K.; Yuminamochi, T.; Yamane, T.; Hirata, S.; Katoh, R.; Hoshi, K. Estimation of prognoses for cervical intraepithelial neoplasia 2 by p16INK4a immunoexpression and high-risk HPV in situ hybridization signal types. Am. J. Clin. Pathol. 2007, 128, 208–217. [Google Scholar] [CrossRef]
- Nuño, T.; García, F. The Lower Anogenital Squamous Terminology Project and its implications for clinical care. Obstet. Gynecol. Clin. N. Am. 2013, 40, 225–233. [Google Scholar] [CrossRef]
- Giovannelli, L.; Lama, A.; Capra, G.; Giordano, V.; Aricò, P.; Ammatuna, P. Detection of human papillomavirus DNA in cervical samples: Analysis of the new PGMY-PCR compared to the hybrid capture II and MY-PCR assays and a two-step nested PCR assay. J. Clin. Microbiol. 2004, 42, 3861–3864. [Google Scholar] [CrossRef][Green Version]
- Safaeian, M.; Herrero, R.; Hildesheim, A.; Quint, W.; Freer, E.; Van Doorn, L.J.; Porras, C.; Silva, S.; González, P.; Bratti, M.C.; et al. Comparison of the SPF10-LiPA system to the Hybrid Capture 2 Assay for detection of carcinogenic human papillomavirus genotypes among 5,683 young women in Guanacaste, Costa Rica. J. Clin. Microbiol. 2007, 45, 1447–1454. [Google Scholar] [CrossRef][Green Version]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).