Systems Genome: Coordinated Gene Activity Networks, Recurring Coordination Modules, and Genome Homeostasis in Developing Neurons
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
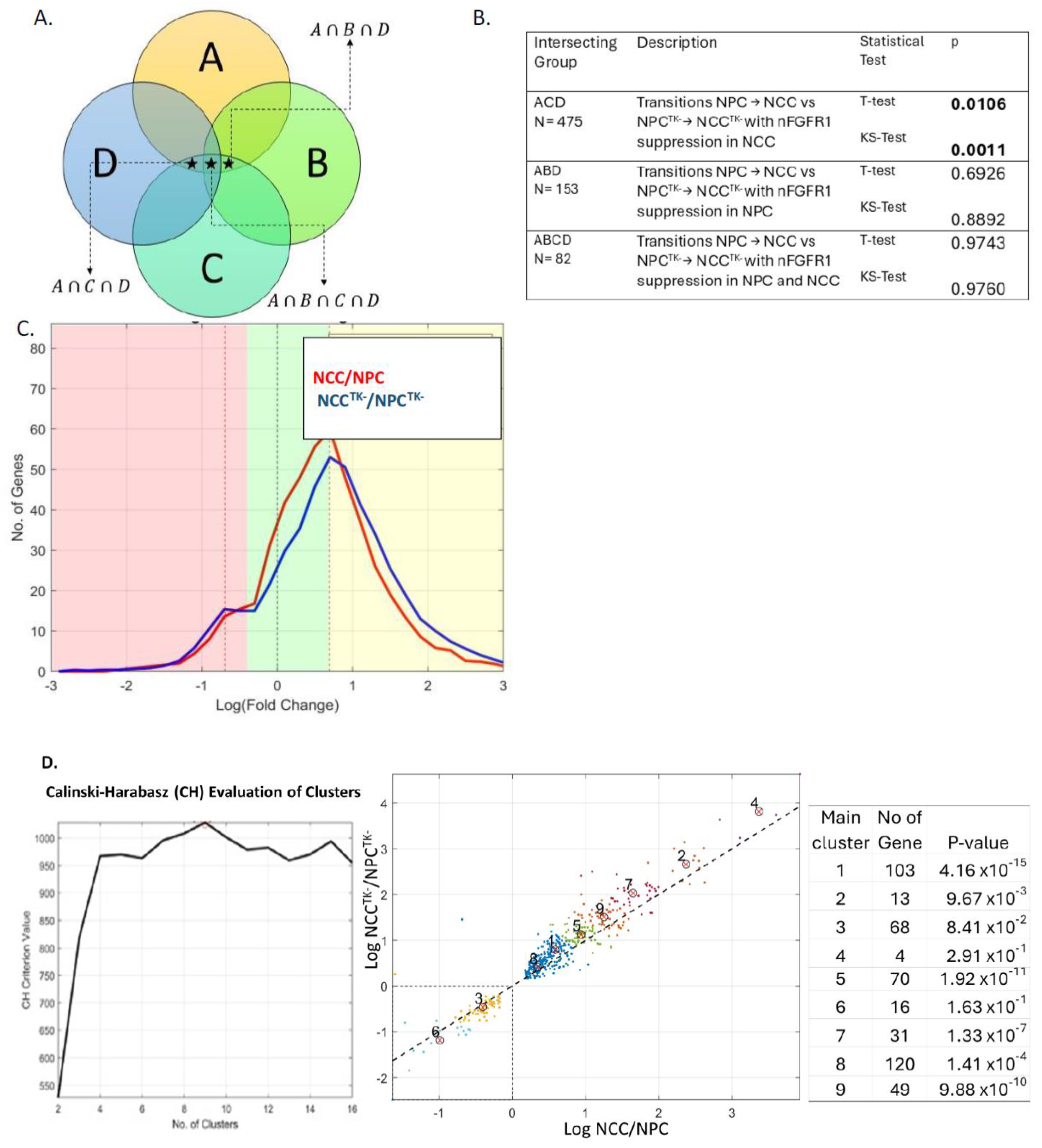
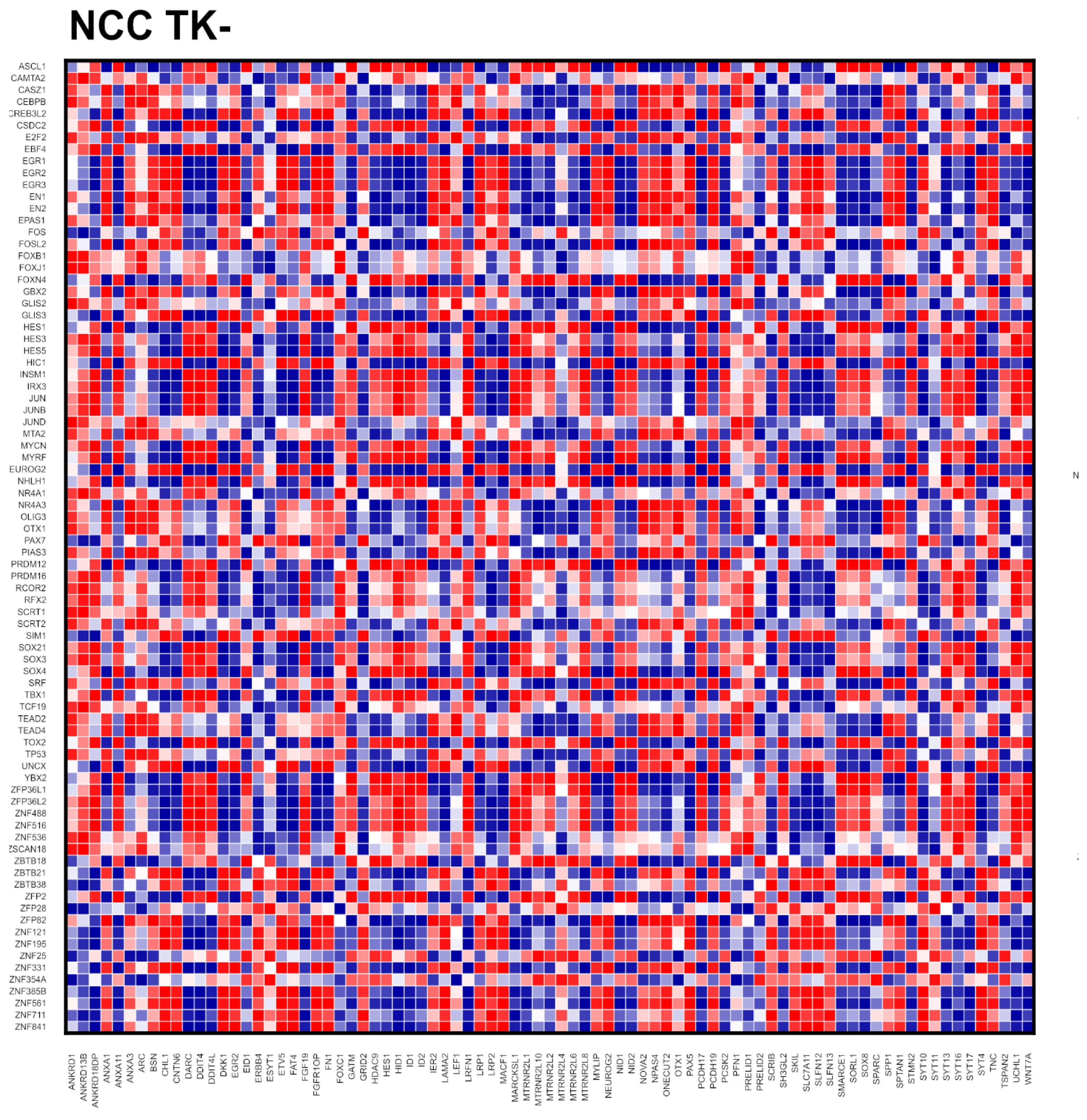
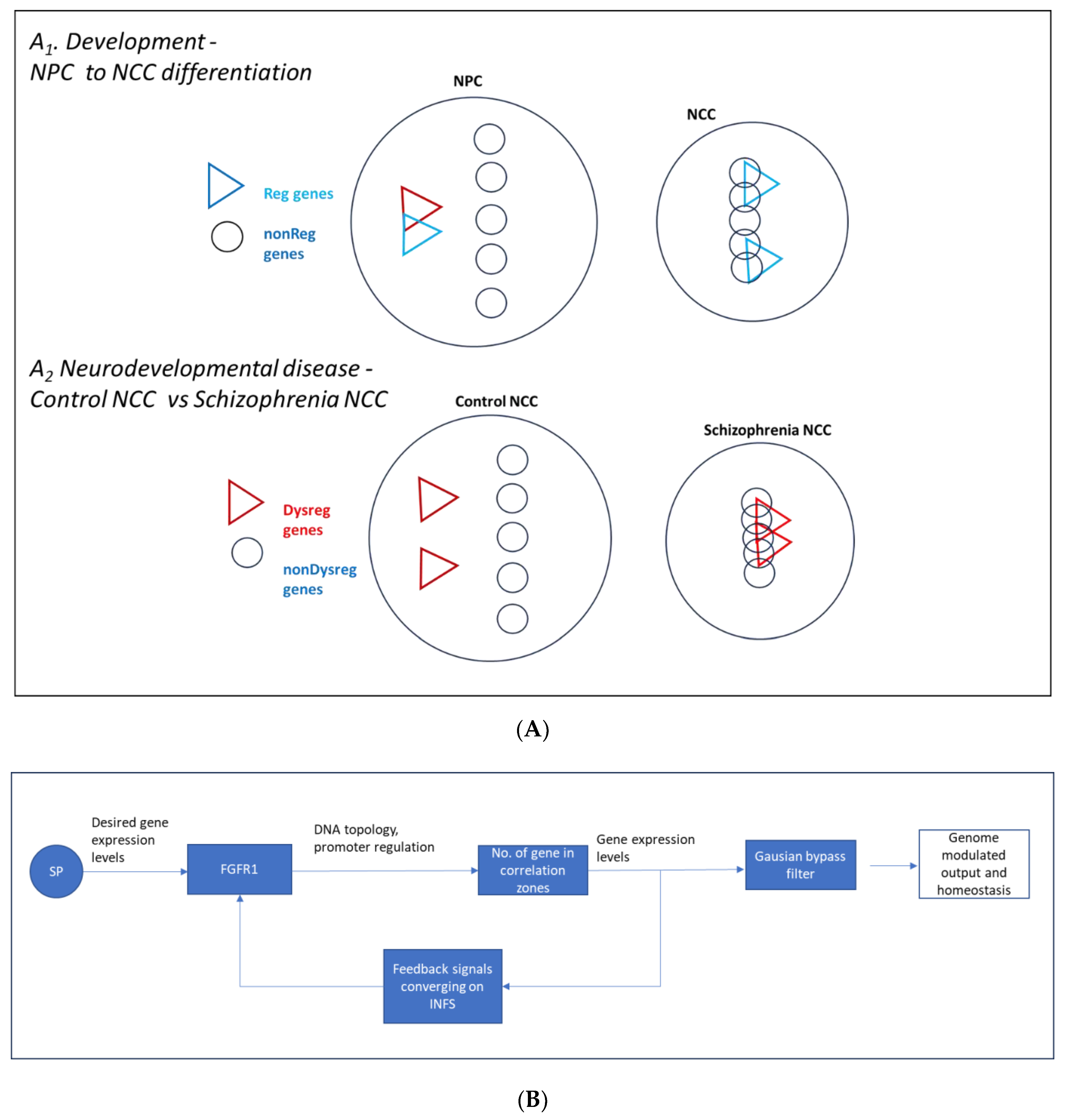
2.1. Gene Activity during NCC Differentiation: Genome Homeostasis and Role of nFGFR1
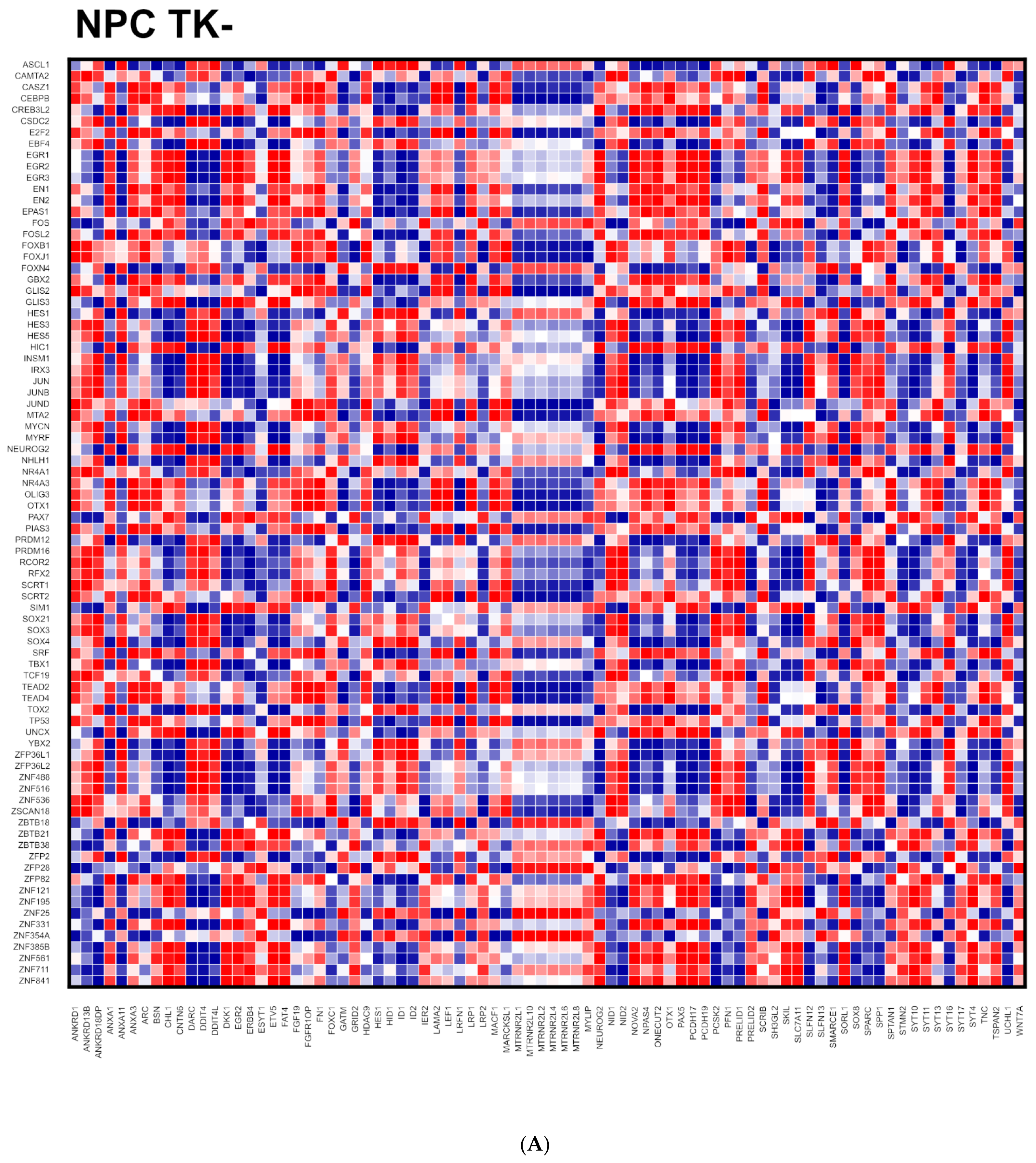
2.2. Coordination of Nervous System Development Genes during NPC → NCC Transition: Role of nFGFR1
2.3. nFGFR1 Coordinates the Entire Expressed Genome during NPC → NCC Transition
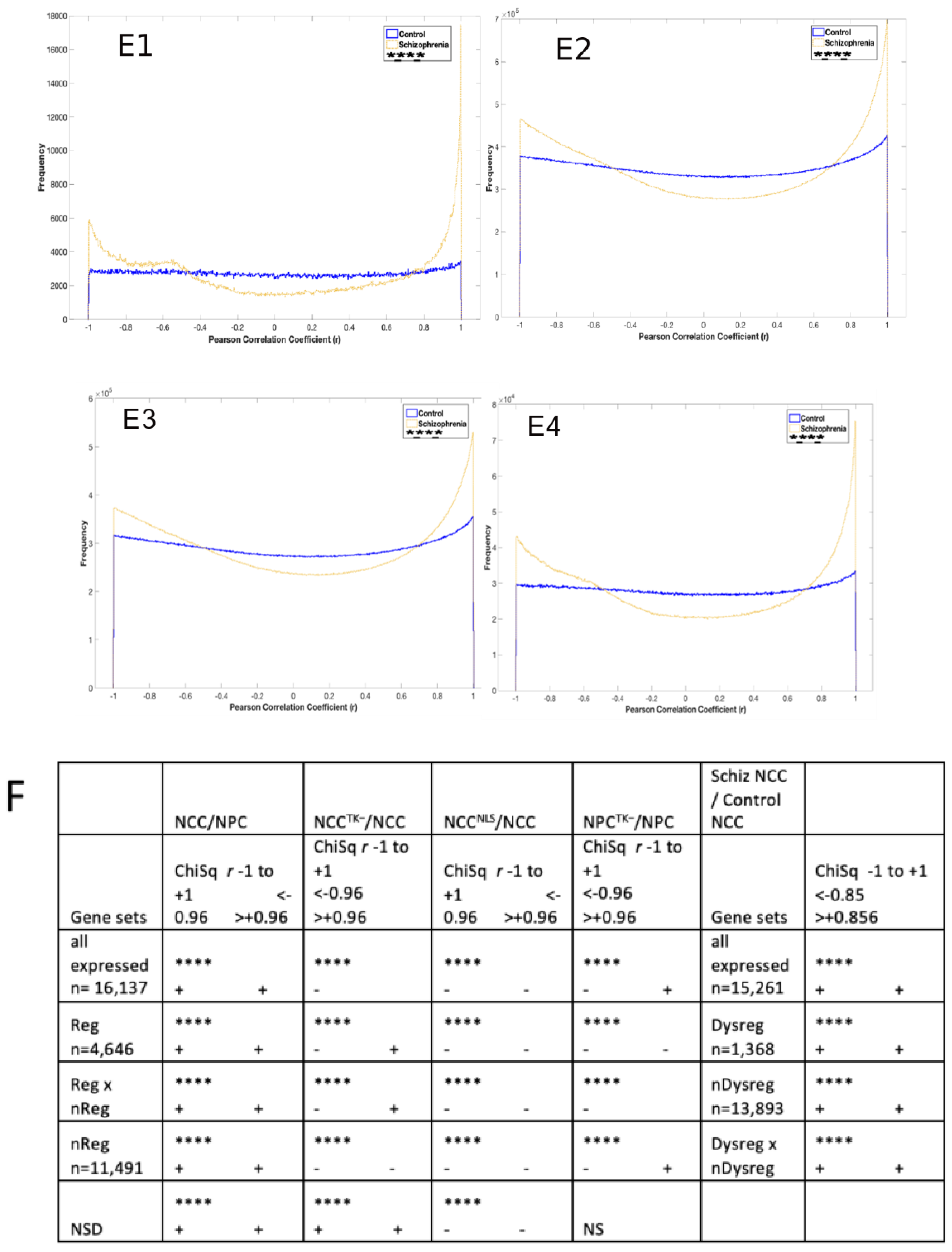
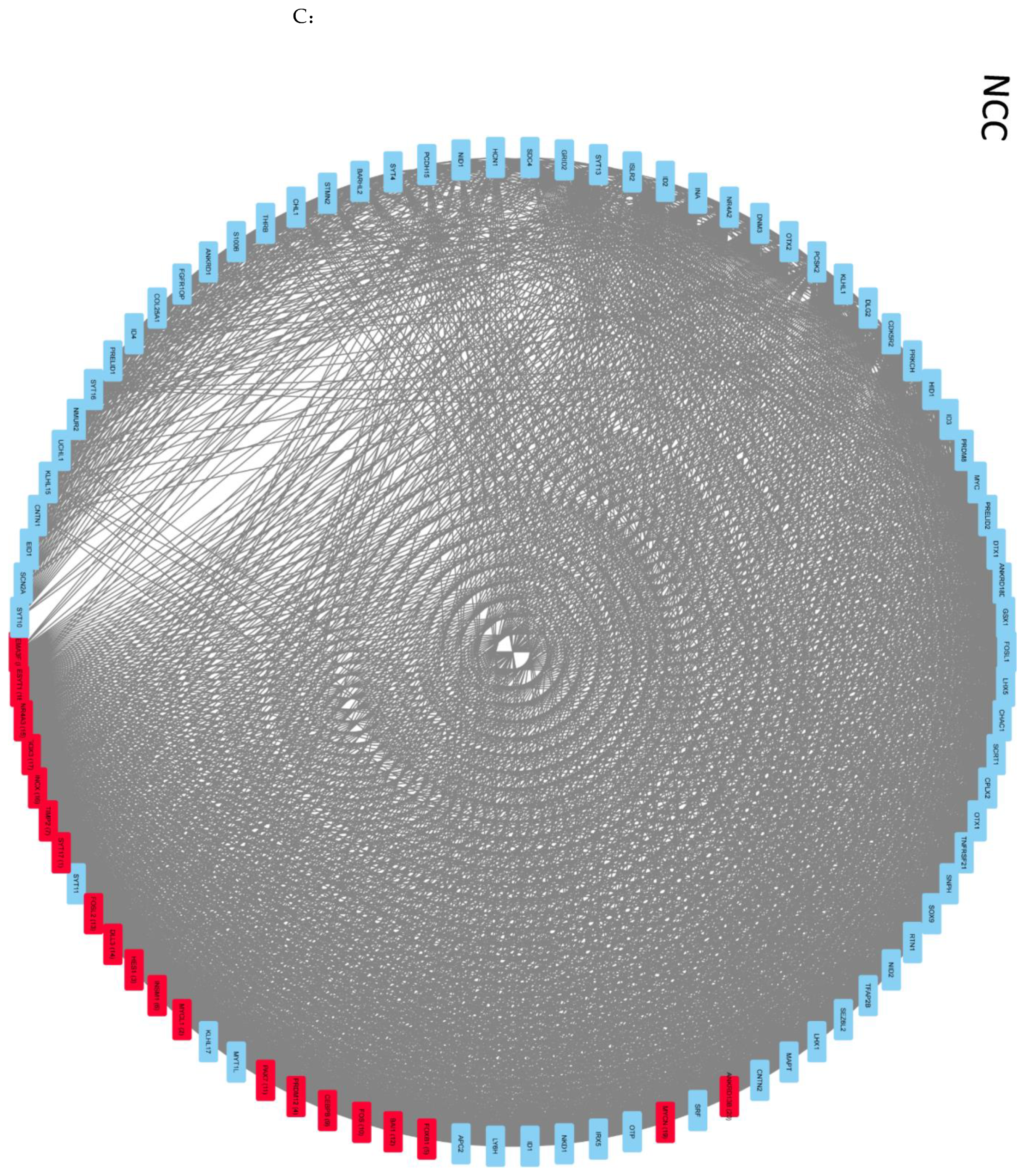
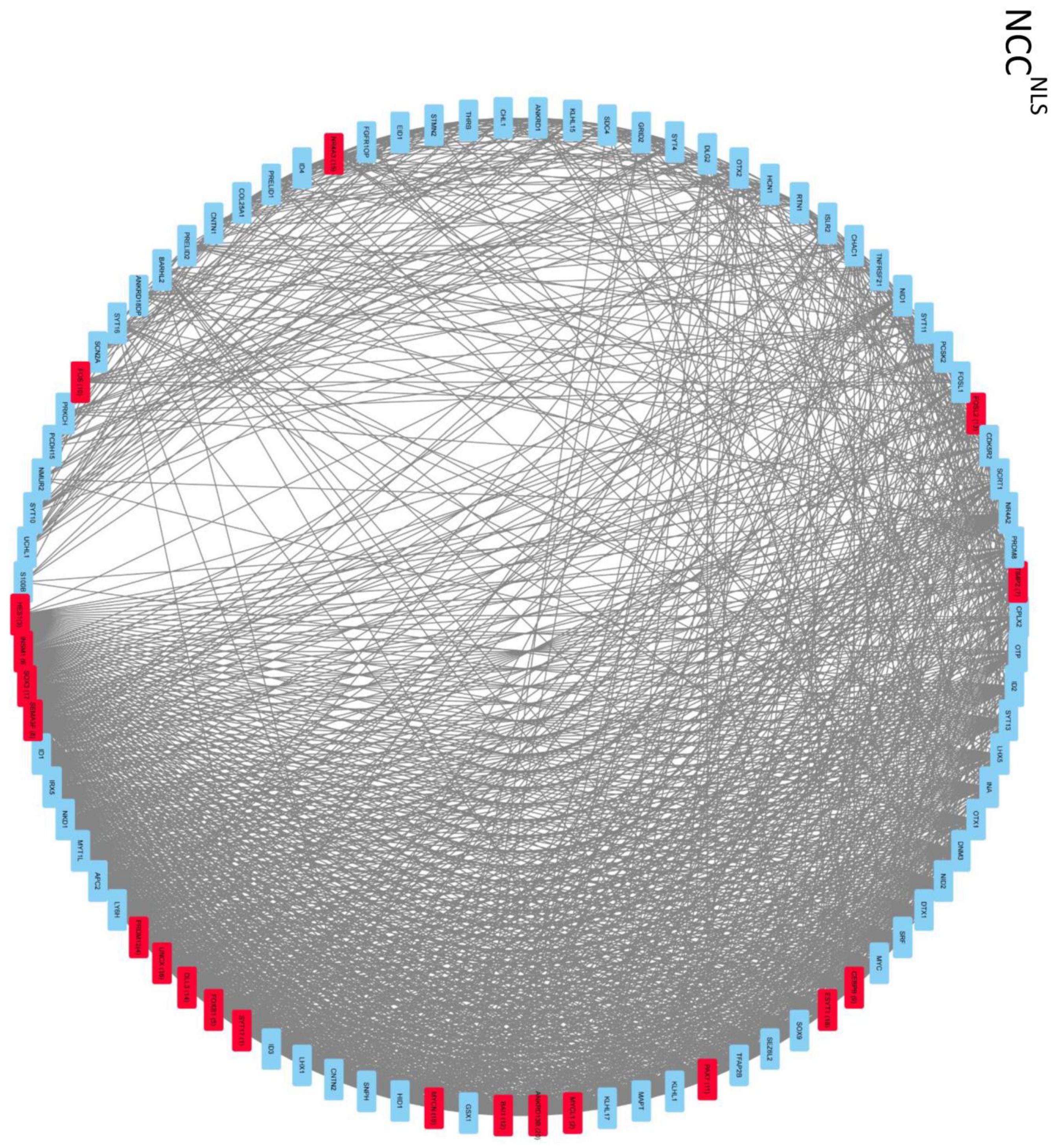
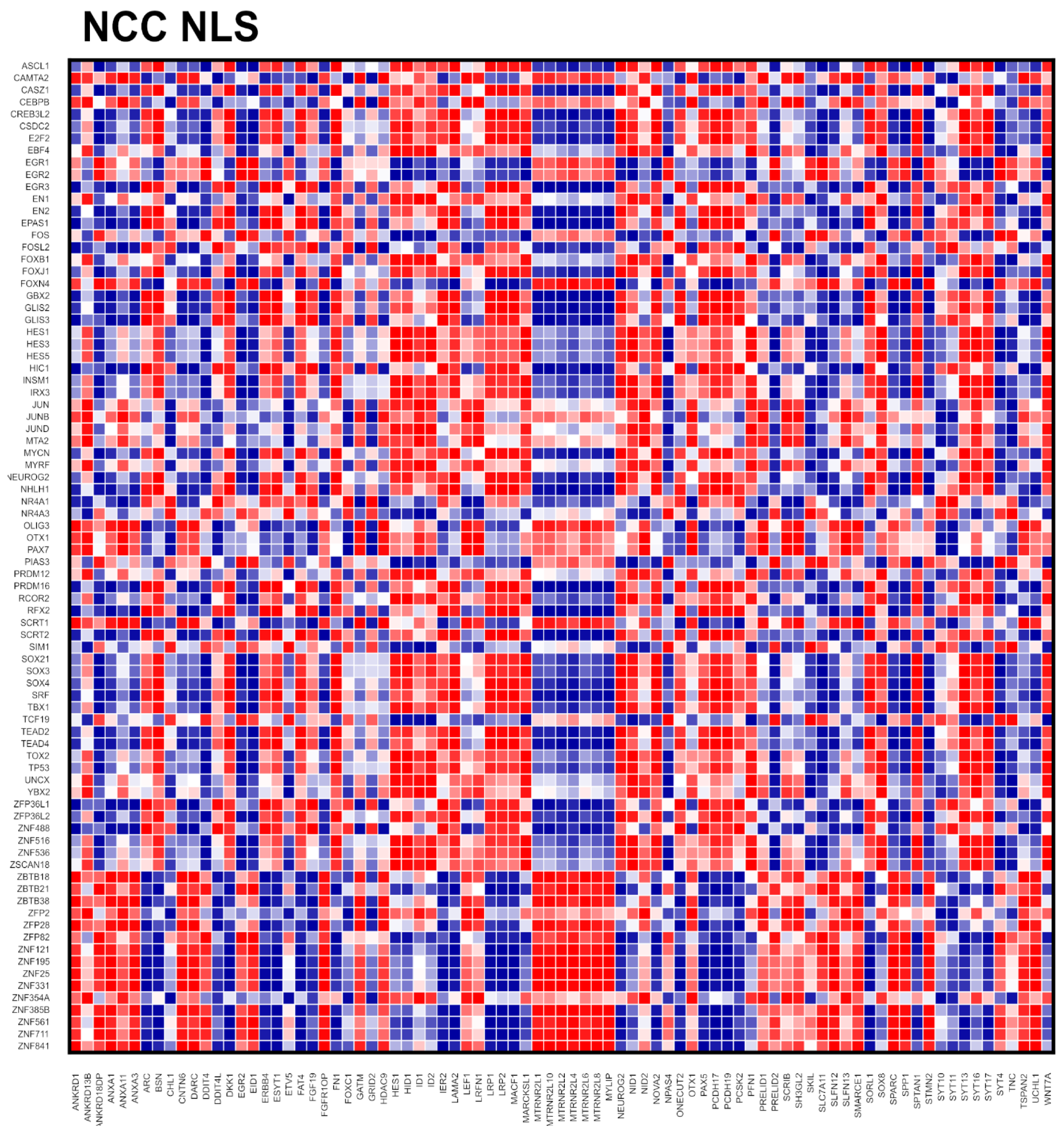
2.4. Global Genome Coordination in NCCs Is Altered in Schizophrenia
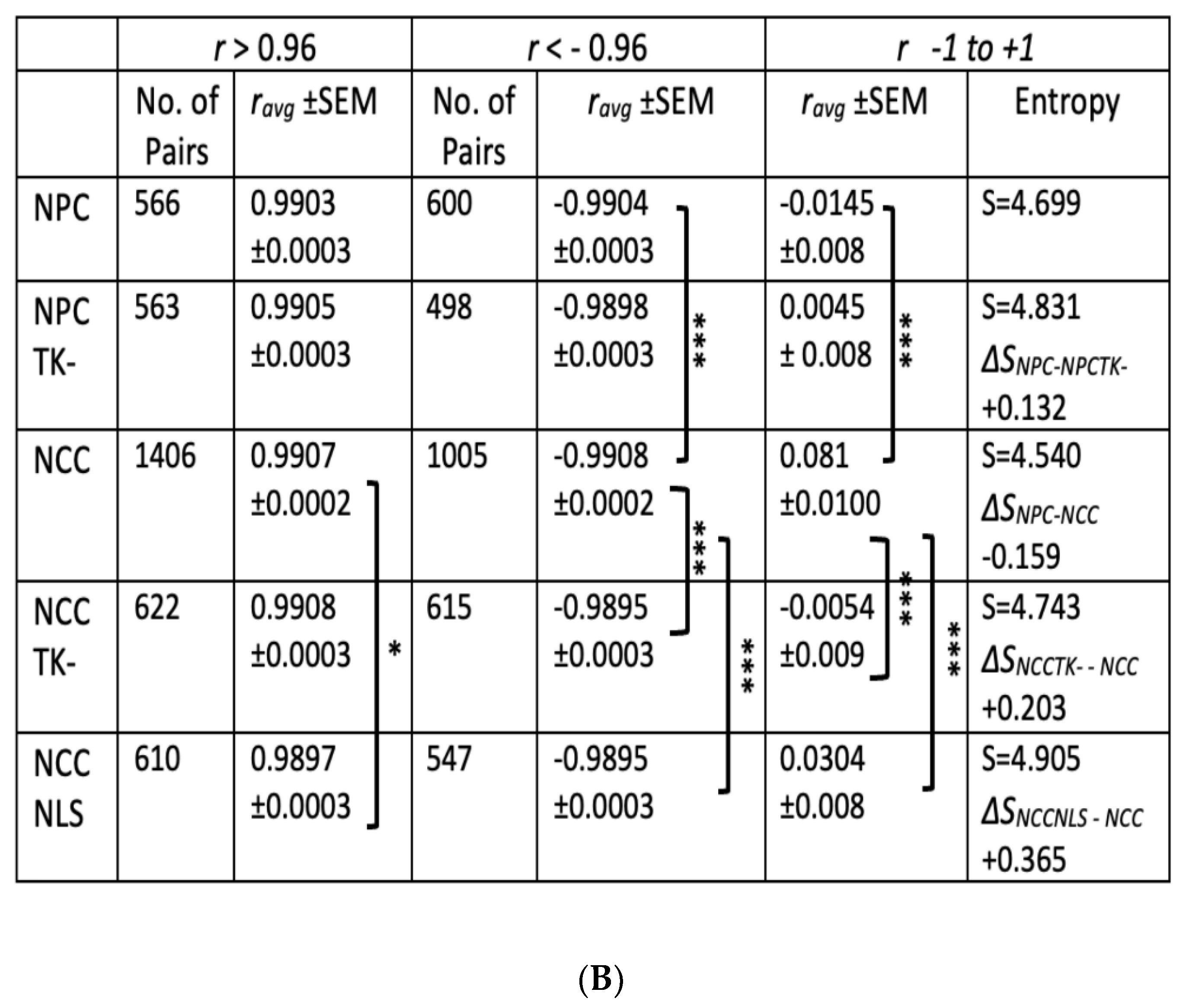
2.5. Entropy of Gene Correlation Is Influenced by NPC → NCC Transition, nFGFR1, and Schizophrenia
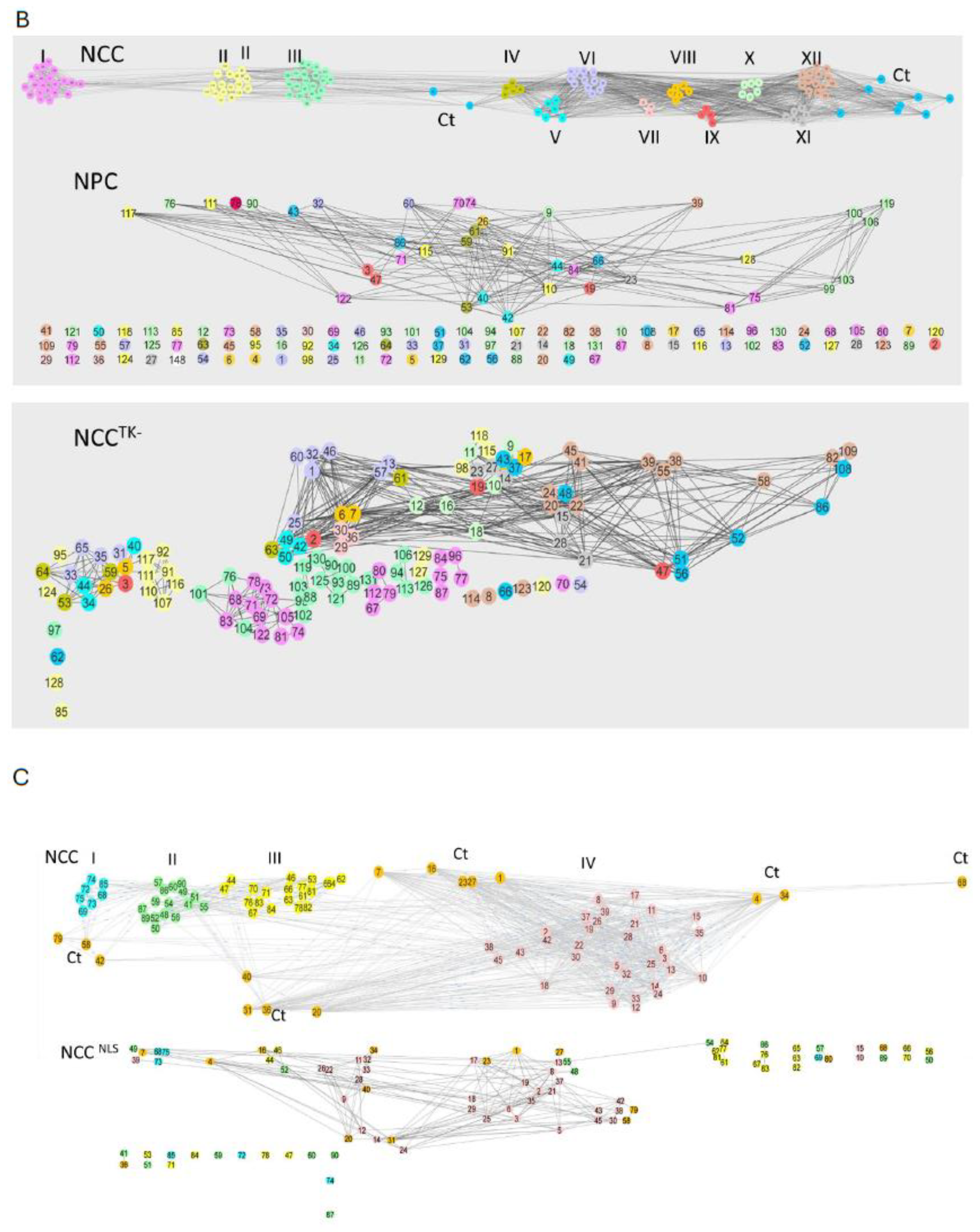
2.6. Reconstruction of the Strongly Correlated Genome during NPC → NCC Transition and in Schizophrenia
2.7. NSD Coordinate GANs Are Reconstructed during NPC → NCC Transition: Role of nFGFR1
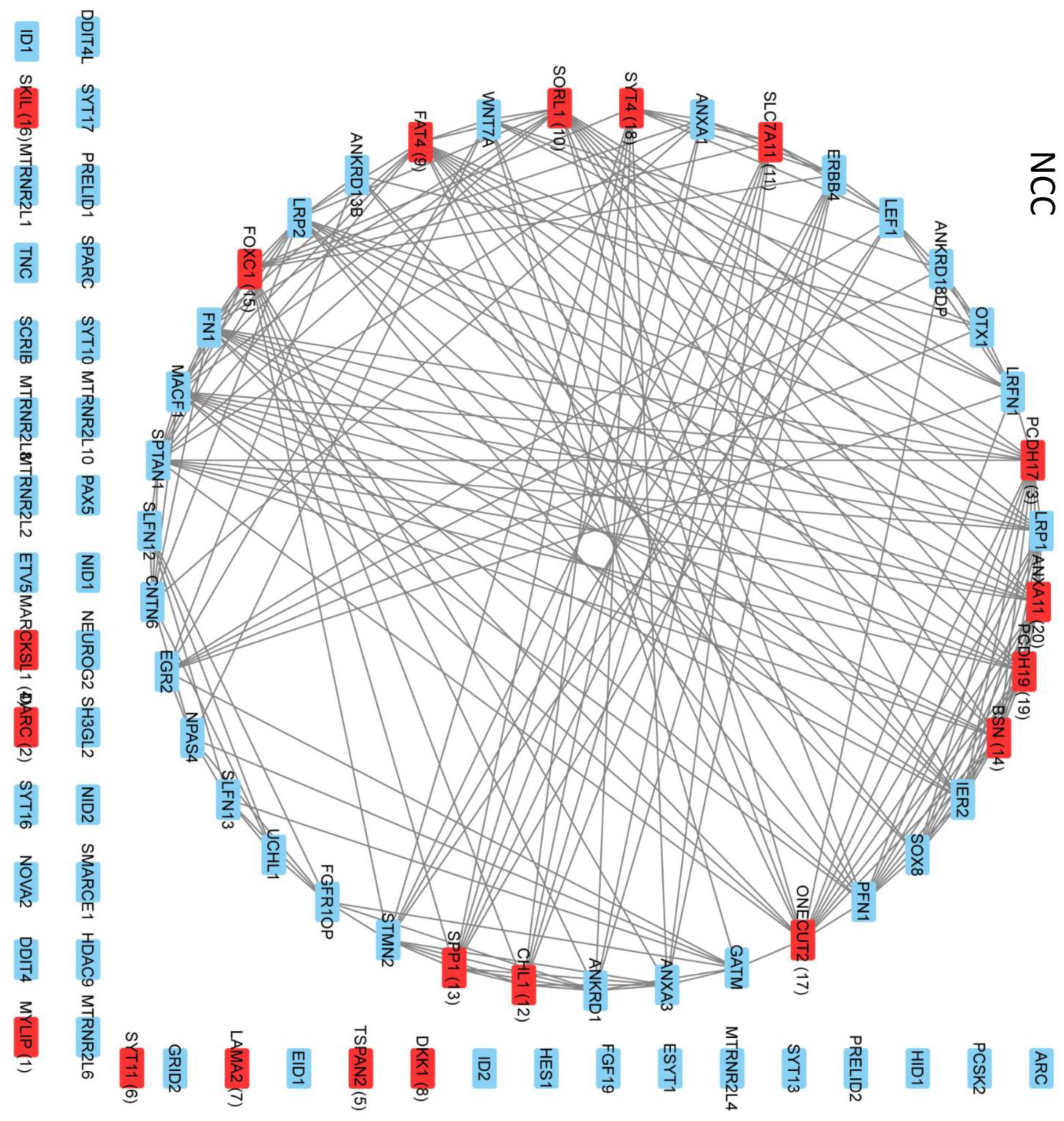
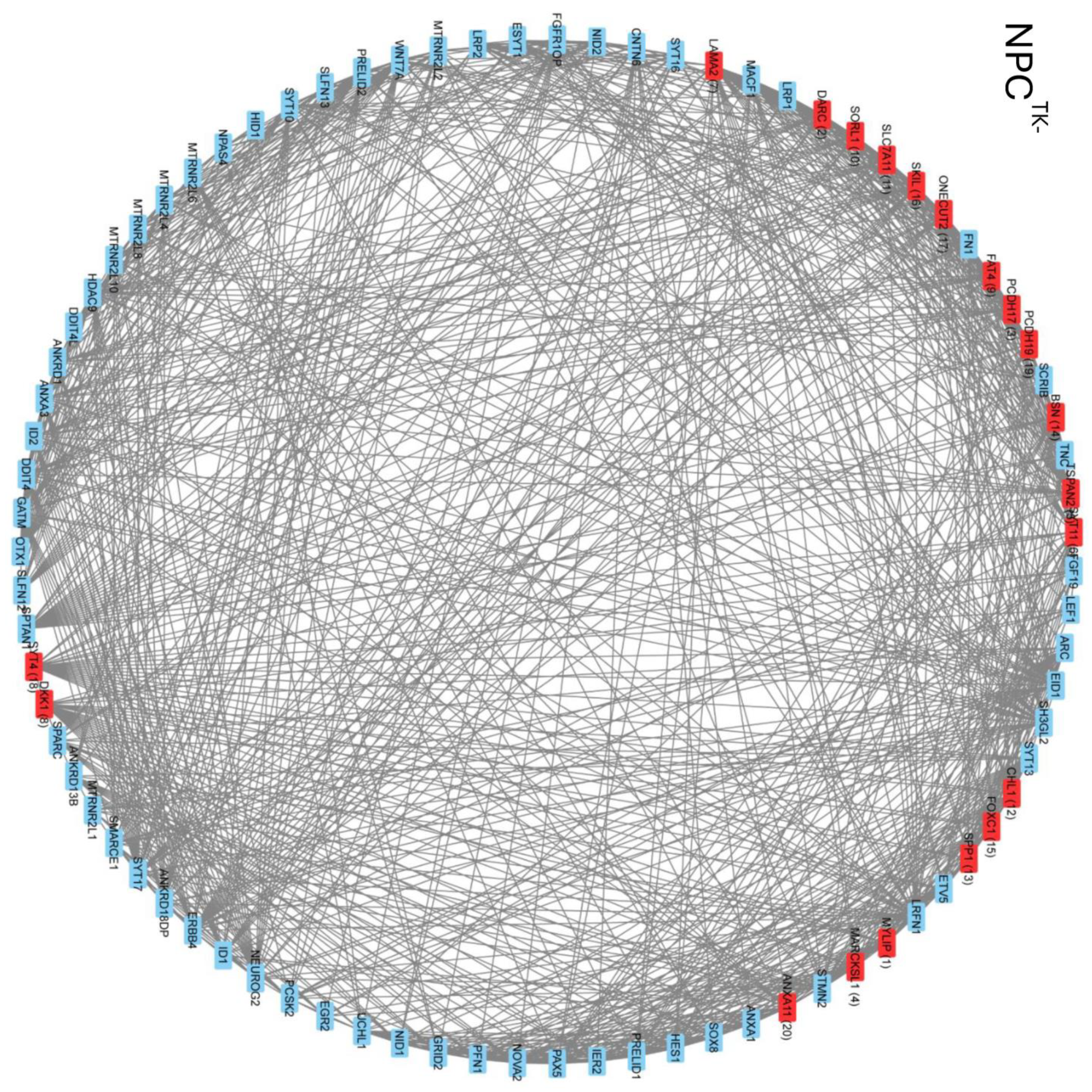
2.8. Intra- and Intercluster Synchronization of Functionally Related GAN Genes
2.9. Cross-Coordination of Transcription Factor Genes and Neurodevelopmental Genes Is Changed during NPC → NCC Transition and Influenced by nFGFR1
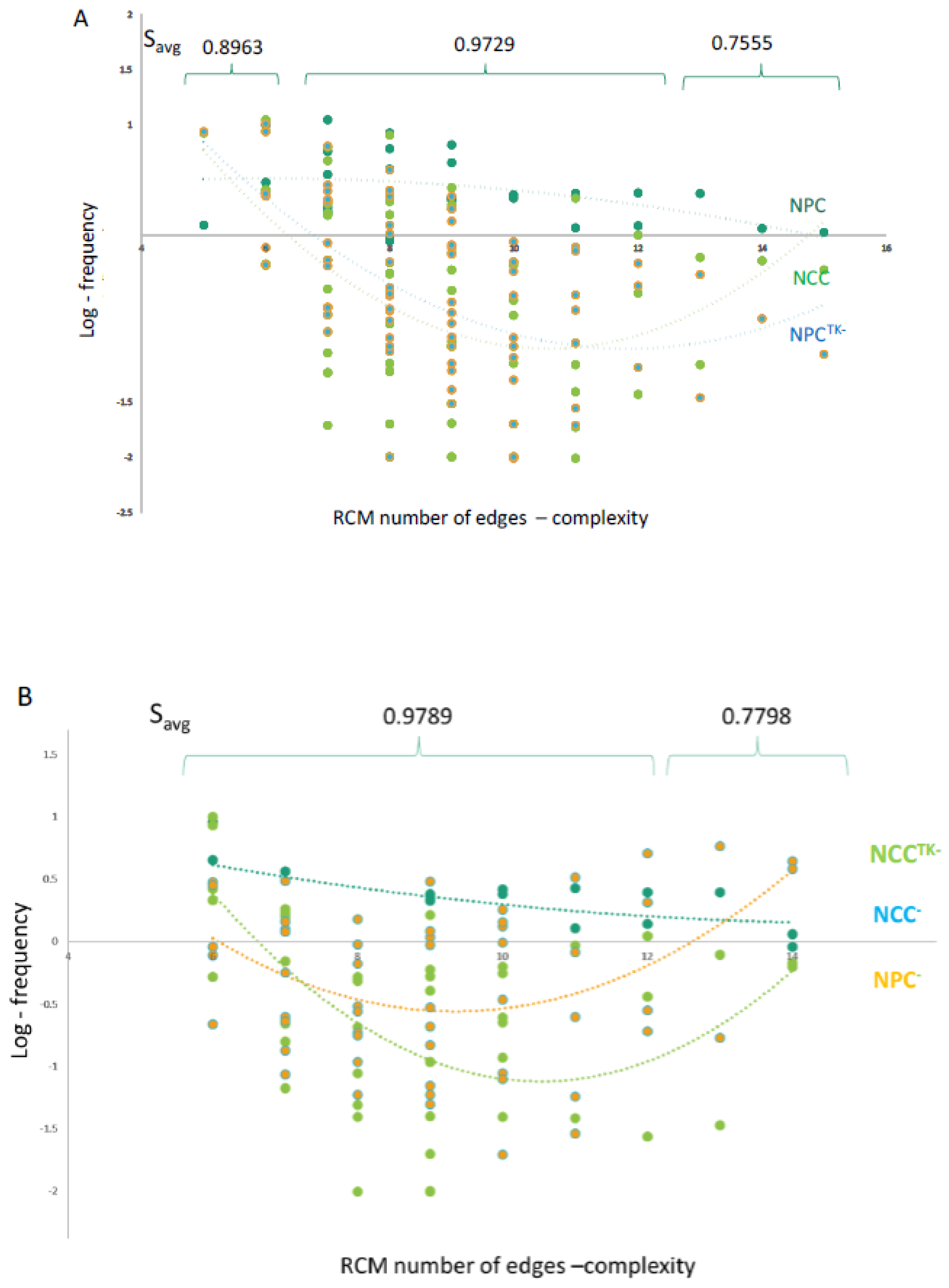
2.10. GANs Comprise Recurring Coordination Modules (RCMs): The Distribution of RCMs Changes during NPC → NCC Transition and Is Programmed by nFGFR1
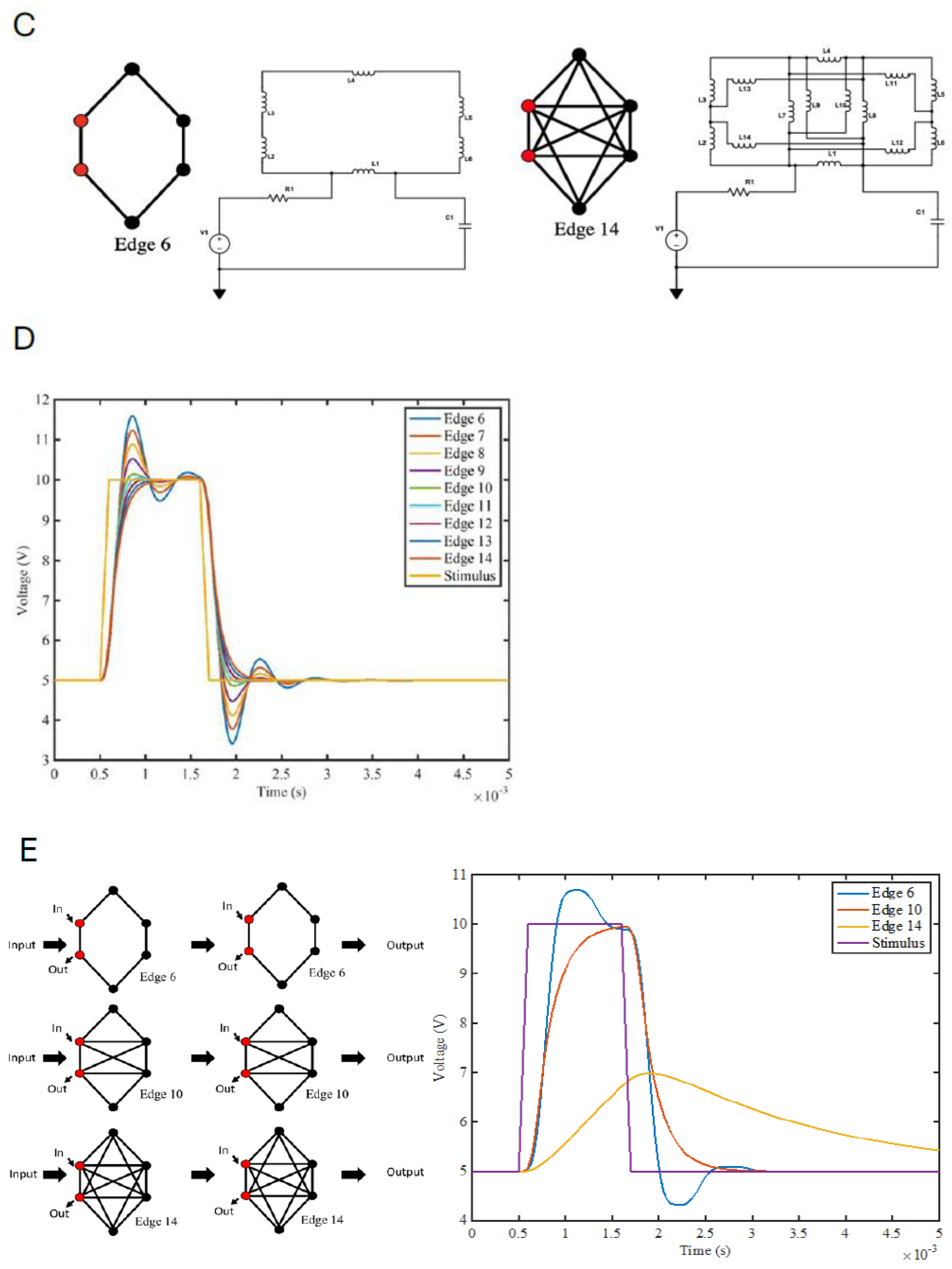
2.11. Modeling RCMs as Information-Processing Circuits: RCMs Influence Regulatory Noise and Signal Transmission
3. Discussion
3.1. Entropy-Based Genome Synchronization Model: Over-Synchronization in Schizophrenia
3.2. GANs
3.3. RCMs Are Kernels of GANs
3.4. nFGFR1 Coordination of Gene Activities May Reflect Control of Genome Topology
3.5. Is nFGFR1 a Proportional-Integral-Derivative Controller?
4. Materials and Methods
4.1. Experimental Design
4.2. Cell Cultures and Treatments
4.3. Analysis of the Coordination of Gene Activities
4.4. Computational Methods and Statistical Analysis
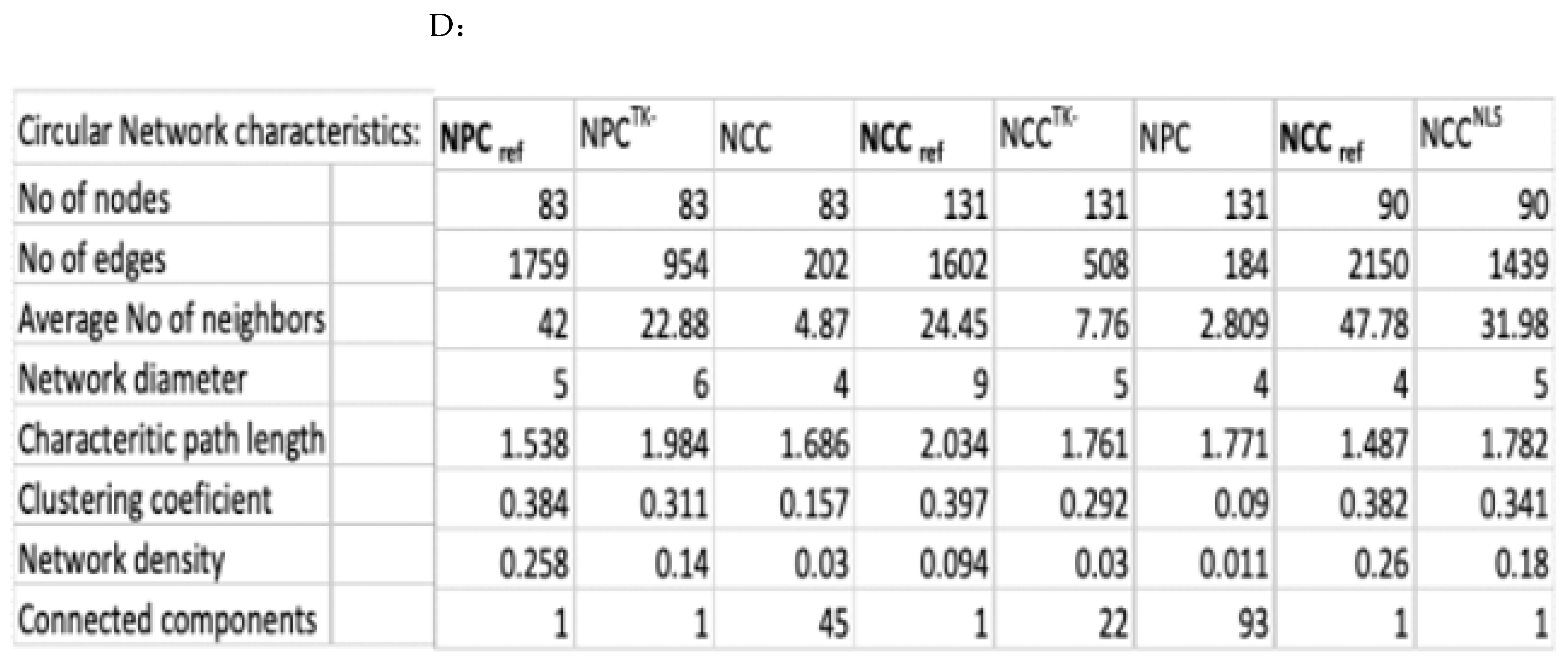
4.5. Complex Network Analyses
- Number of Nodes and Edges: These are the fundamental units of a network, representing entities and their connections, respectively.
- Average Number of Neighbors: This metric calculates the average connectivity of nodes, reflecting the typical structural environment of a node within the network.
- Network Diameter: Defined as the longest shortest path between any two nodes in the network, this metric indicates the “largeness” of the network’s scope.
- Shortest Path Length (Characteristic Path Length): This is a key metric in network theory, indicating the minimum path length between two nodes, and is crucial for understanding the efficiency of network connectivity. The average of these shortest paths across all pairs is the characteristic path length.
- Clustering Coefficient: This metric describes the likelihood that nodes adjacent to any given node are also connected to each other, forming a cluster.
- -
- is the actual number of edges among the neighbors of ;
- -
- is the total possible number of edges that could exist among the neighbors.
- Network Density: This normalized metric reflects the ratio of actual connections to possible connections in the network, providing insight into how densely the network is populated with edges.
- Connected Components: A measure of the network’s overall connectivity, indicating how many sub-networks exist that are not interconnected.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GAN | gene activity network |
| RCM | recurring coordination module |
| R-C-L | resistor-capacitor-inductor electrical circuit |
| nFGFR1 | nuclear form of FGFR1 |
| FGFR1(SP-/NLS) | recombinant constitutively active nuclear FGFR1 |
| FGFR1(SP-/NLS)(TK-) | recombinant dominant negative nuclear FGFR1 with tyrosine kinase domain deleted |
| INFS | Integrative NuclearFGFR1 Signaling |
| NPC | neural progenitor cell |
| NPCTK− | NPC transfected with FGFR1(SP-/NLS)(TK-) |
| NCC | neuronal committed cell (early neurons, differentiated from NPCs) |
| NCCTK− | NPC transfected with FGFR1(SP-/NLS)(TK-) and stimulated to differentiate to NCC |
| NCCNLS | NPC transfected with FGFR1(SP-/NLS) and stimulated to differentiate to NCC |
| NSD | nervous system development (GO category) |
| Reg | regulated genes whose average activity changes during NPC → NCC transition |
| nonReg | non-regulated genes whose activity does not change during NPC → NCC transition |
| Dysreg | genes whose activity is dysregulated in NCCs derived from schizophrenia patients |
| nonDysreg | genes whose activity is not dysregulated in NCCs derived from schizophrenia patients |
References
- Kesic, S. Systems biology, emergence and antireductionism. Saudi J. Biol. Sci. 2016, 23, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Drack, M.; Wolkenhauer, O. System approaches of Weiss and Bertalanffy and their relevance for systems biology today. Semin. Cancer Biol. 2011, 21, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing chromosome conformation. Science 2002, 295, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Zakrzewski, W.; Dobrzynski, M.; Szymonowicz, M.; Rybak, Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar] [CrossRef]
- Stachowiak, M.K.; Stachowiak, E.K. Evidence-Based Theory for Integrated Genome Regulation of Ontogeny--An Unprecedented Role of Nuclear FGFR1 Signaling. J. Cell. Physiol. 2016, 231, 1199–1218. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, E.K.; Benson, C.A.; Narla, S.T.; Dimitri, A.; Chuye, L.E.B.; Dhiman, S.; Harikrishnan, K.; Elahi, S.; Freedman, D.; Brennand, K.J.; et al. Cerebral organoids reveal early cortical maldevelopment in schizophrenia-computational anatomy and genomics, role of FGFR1. Transl. Psychiatry Nat. 2017, 7, 6. [Google Scholar] [CrossRef]
- Fang, X.; Stachowiak, E.K.; Dunham-Ems, S.M.; Klejbor, I.; Stachowiak, M.K. Control of CREB-binding protein signaling by nuclear fibroblast growth factor receptor-1: A novel mechanism of gene regulation. J. Biol. Chem. 2005, 280, 28451–28462. [Google Scholar] [CrossRef]
- Jornet, J.M.; Bae, Y.; Handelmann, C.R.; Decker, B.; Balcerak, A.; Sangwan, A.; Miao, P.; Desai, A.; Feng, L.; Stachowiak, E.K. Optogenomic Interfaces: Bridging Biological Networks With the Electronic Digital World. Proc. IEEE 2019, 107, 1387–1401. [Google Scholar] [CrossRef]
- Narla, S.T.; Lee, Y.W.; Benson, C.A.; Sarder, P.; Brennand, K.J.; Stachowiak, E.K.; Stachowiak, M.K. Common developmental genome deprogramming in schizophrenia-Role of Integrative Nuclear FGFR1 Signaling (INFS). Schizophr. Res. 2017, 185, 17–32. [Google Scholar] [CrossRef]
- Peng, H.; Moffett, J.; Myers, J.; Fang, X.; Stachowiak, E.K.; Maher, P.; Kratz, E.; Hines, J.; Fluharty, S.J.; Mizukoshi, E.; et al. Novel nuclear signaling pathway mediates activation of fibroblast growth factor-2 gene by type 1 and type 2 angiotensin II receptors. Mol. Biol. Cell 2001, 12, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Reilly, J.F.; Maher, P.A. Importin beta-mediated nuclear import of fibroblast growth factor receptor: Role in cell proliferation. J. Cell Biol. 2001, 152, 1307–1312. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, M.K.; Birkaya, B.; Aletta, J.M.; Narla, S.T.; Benson, C.A.; Decker, B.; Stachowiak, E.K. Nuclear FGF receptor-1 and CREB binding protein: An integrative signaling module. J. Cell. Physiol. 2015, 230, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, M.K.; Maher, P.A.; Joy, A.; Mordechai, E.; Stachowiak, E.K. Nuclear accumulation of fibroblast growth factor receptors is regulated by multiple signals in adrenal medullary cells. Mol. Biol. Cell 1996, 7, 1299–1317. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, E.K.; Maher, P.A.; Tucholski, J.; Mordechai, E.; Joy, A.; Moffett, J.; Stachowiak, M.K. Nuclear accumulation of fibroblast growth factor receptors in human glial cells—association with cell proliferation. Oncogene 1997, 14, 2201–2211. [Google Scholar] [CrossRef]
- Egbivwie, N.; Cockle, J.V.; Humphries, M.; Ismail, A.; Esteves, F.; Taylor, C.; Brüning-Richardson, A. FGFR1 Expression and Role in Migration in Low and High Grade Pediatric Gliomas. Front. Oncol. 2019, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Coleman, S.J.; Chioni, A.M.; Ghallab, M.; Anderson, R.K.; Lemoine, N.R.; Kocher, H.M.; Grose, R.P. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol. Med. 2014, 6, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Formisano, L.; Stauffer, K.M.; Young, C.D.; Bhola, N.E.; Guerrero-Zotano, A.L.; Jansen, V.M.; Estrada, M.M.; Hutchinson, K.E.; Giltnane, J.M.; Schwarz, L.J.; et al. Association of FGFR1 with ERα Maintains Ligand-Independent ER Transcription and Mediates Resistance to Estrogen Deprivation in ER+ Breast Cancer. Clin. Cancer Res. 2017, 23, 6138–6150. [Google Scholar] [CrossRef] [PubMed]
- Decker, B.; Liput, M.; Abdellatif, H.; Yergeau, D.; Bae, Y.; Jornet, J.M.; Stachowiak, E.K.; Stachowiak, M.K. Global Genome Conformational Programming during Neuronal Development Is Associated with CTCF and Nuclear FGFR1-The Genome Archipelago Model. Int. J. Mol. Sci. 2020, 22, 347. [Google Scholar] [CrossRef]
- Maher, P.A. Nuclear Translocation of fibroblast growth factor (FGF) receptors in response to FGF-2. J. Cell Biol. 1996, 134, 529–536. [Google Scholar] [CrossRef]
- Gonzalez, A.M.; Berry, M.; Maher, P.A.; Logan, A.; Baird, A. A comprehensive analysis of the distribution of FGF-2 and FGFR1 in the rat brain. Brain Res. 1995, 701, 201–226. [Google Scholar] [CrossRef] [PubMed]
- Klimaschewski, L.; Meisinger, C.; Grothe, C. Localization and regulation of basic fibroblast growth factor (FGF-2) and FGF receptor-1 in rat superior cervical ganglion after axotomy. J. Neurobiol. 1999, 38, 499–506. [Google Scholar] [CrossRef]
- Chaudhuri, T.R.; Lin, Q.; Stachowiak, E.K.; Rosario, S.R.; Spernyak, J.A.; Ma, W.W.; Stachowiak, M.K.; Greene, M.K.; Quinn, G.P.; McDade, S.S.; et al. Dual-hit strategy for therapeutic targeting of pancreatic cancer in patient-derived xenograft tumors. Clin. Cancer Res. 2024, 30, 1367–1381. [Google Scholar] [CrossRef] [PubMed]
- Tuzon, C.T.; Rigueur, D.; Merrill, A.E. Nuclear Fibroblast Growth Factor Receptor Signaling in Skeletal Development and Disease. Curr. Osteoporos. Rep. 2019, 17, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Szymczyk, J.; Sluzalska, K.D.; Materla, I.; Opalinski, L.; Otlewski, J.; Zakrzewska, M. FGF/FGFR-Dependent Molecular Mechanisms Underlying Anti-Cancer Drug Resistance. Cancers 2021, 13, 5796. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.; Kim, D.H.; Kim, S.J.; Cho, N.C.; Lee, Y.H.; Jang, J.H.; Surh, Y.J. Nuclear Localization of Fibroblast Growth Factor Receptor 1 in Breast Cancer Cells Interacting with Cancer Associated Fibroblasts. J. Cancer Prev. 2022, 27, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Berlow, N.E.; Lathara, M.; Bharathy, N.; Martin, L.R.; Purohit, R.; Cleary, M.M.; Liu, Q.; Michalek, J.E.; Srinivasa, G.; et al. Sensitization of osteosarcoma to irradiation by targeting nuclear FGFR1. Biochem. Biophys. Res. Commun. 2022, 621, 101–108. [Google Scholar] [CrossRef]
- Servetto, A.; Kollipara, R.; Formisano, L.; Lin, C.-C.; Lee, K.-M.; Sudhan, D.R.; Gonzalez-Ericsson, P.I.; Chatterjee, S.; Guerrero-Zotano, A.; Mendiratta, S.; et al. Nuclear FGFR1 Regulates Gene Transcription and Promotes Antiestrogen Resistance in ER+ Breast Cancer. Clin. Cancer Res. 2021, 27, 4379–4396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, J.; Hanspers, K.; Xu, D.; Chen, L.; Pico, A.R. NOA: A cytoscape plugin for network ontology analysis. Bioinformatics 2013, 29, 2066–2067. [Google Scholar] [CrossRef]
- Alon, U. Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 2007, 8, 450–461. [Google Scholar] [CrossRef]
- Kashtan, N.; Itzkovitz, S.; Milo, R.; Alon, U. Efficient sampling algorithm for estimating subgraph concentrations and detecting network motifs. Bioinformatics 2004, 20, 1746–1758. [Google Scholar] [CrossRef] [PubMed]
- Wernicke, S.; Rasche, F. FANMOD: A tool for fast network motif detection. Bioinformatics 2006, 22, 1152–1153. [Google Scholar] [CrossRef] [PubMed]
- Chalancon, G.; Ravarani, C.N.; Balaji, S.; Martinez-Arias, A.; Aravind, L.; Jothi, R.; Babu, M.M. Interplay between gene expression noise and regulatory network architecture. Trends Genet. 2012, 28, 221–232. [Google Scholar] [CrossRef]
- Mousa, A.; Bakhiet, M. Role of cytokine signaling during nervous system development. Int. J. Mol. Sci. 2013, 14, 13931–13957. [Google Scholar] [CrossRef]
- Dawidowski, B.; Górniak, A.; Podwalski, P.; Lebiecka, Z.; Misiak, B.; Samochowiec, J. The Role of Cytokines in the Pathogenesis of Schizophrenia. J. Clin. Med. 2021, 10, 3849. [Google Scholar] [CrossRef]
- Choubey, L.; Collette, J.C.; Smith, K.M. Quantitative assessment of fibroblast growth factor receptor 1 expression in neurons and glia. PeerJ 2017, 5, e3173. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, G.; Pattison, M.B.; Midkiff, C.C. The FGF/FGFR system in the microglial neuroinflammation with Borrelia burgdorferi: Likely intersectionality with other neurological conditions. J. Neuroinflammation 2023, 20, 10. [Google Scholar] [CrossRef]
- Benson, C.A.; Powell, H.R.; Liput, M.; Dinham, S.; Freedman, D.A.; Ignatowski, T.A.; Stachowiak, E.K.; Stachowiak, M.K. Immune Factor, TNFalpha, Disrupts Human Brain Organoid Development Similar to Schizophrenia-Schizophrenia Increases Developmental Vulnerability to TNFalpha. Front. Cell. Neurosci. 2020, 14, 233. [Google Scholar] [CrossRef]
- Sporns, O.; Betzel, R.F. Modular Brain Networks. Annu. Rev. Psychol. 2016, 67, 613–640. [Google Scholar] [CrossRef]
- Coleman, S.J.; Watt, J.; Arumugam, P.; Solaini, L.; Carapuca, E.; Ghallab, M.; Grose, R.P.; Kocher, H.M. Pancreatic cancer organotypics: High throughput, preclinical models for pharmacological agent evaluation. World J. Gastroenterol. 2014, 20, 8471–8481. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhiman, S.; Manoj, N.; Liput, M.; Sangwan, A.; Diehl, J.; Balcerak, A.; Sudhakar, S.; Augustyniak, J.; Jornet, J.M.; Bae, Y.; et al. Systems Genome: Coordinated Gene Activity Networks, Recurring Coordination Modules, and Genome Homeostasis in Developing Neurons. Int. J. Mol. Sci. 2024, 25, 5647. https://doi.org/10.3390/ijms25115647
Dhiman S, Manoj N, Liput M, Sangwan A, Diehl J, Balcerak A, Sudhakar S, Augustyniak J, Jornet JM, Bae Y, et al. Systems Genome: Coordinated Gene Activity Networks, Recurring Coordination Modules, and Genome Homeostasis in Developing Neurons. International Journal of Molecular Sciences. 2024; 25(11):5647. https://doi.org/10.3390/ijms25115647
Chicago/Turabian StyleDhiman, Siddhartha, Namya Manoj, Michal Liput, Amit Sangwan, Justin Diehl, Anna Balcerak, Sneha Sudhakar, Justyna Augustyniak, Josep M. Jornet, Yongho Bae, and et al. 2024. "Systems Genome: Coordinated Gene Activity Networks, Recurring Coordination Modules, and Genome Homeostasis in Developing Neurons" International Journal of Molecular Sciences 25, no. 11: 5647. https://doi.org/10.3390/ijms25115647
APA StyleDhiman, S., Manoj, N., Liput, M., Sangwan, A., Diehl, J., Balcerak, A., Sudhakar, S., Augustyniak, J., Jornet, J. M., Bae, Y., Stachowiak, E. K., Dutta, A., & Stachowiak, M. K. (2024). Systems Genome: Coordinated Gene Activity Networks, Recurring Coordination Modules, and Genome Homeostasis in Developing Neurons. International Journal of Molecular Sciences, 25(11), 5647. https://doi.org/10.3390/ijms25115647










