Novel Proteins of the High-Affinity Nitrate Transporter Family NRT2, SaNRT2.1 and SaNRT2.5, from the Euhalophyte Suaeda altissima: Molecular Cloning and Expression Analysis
Abstract
1. Introduction
2. Results
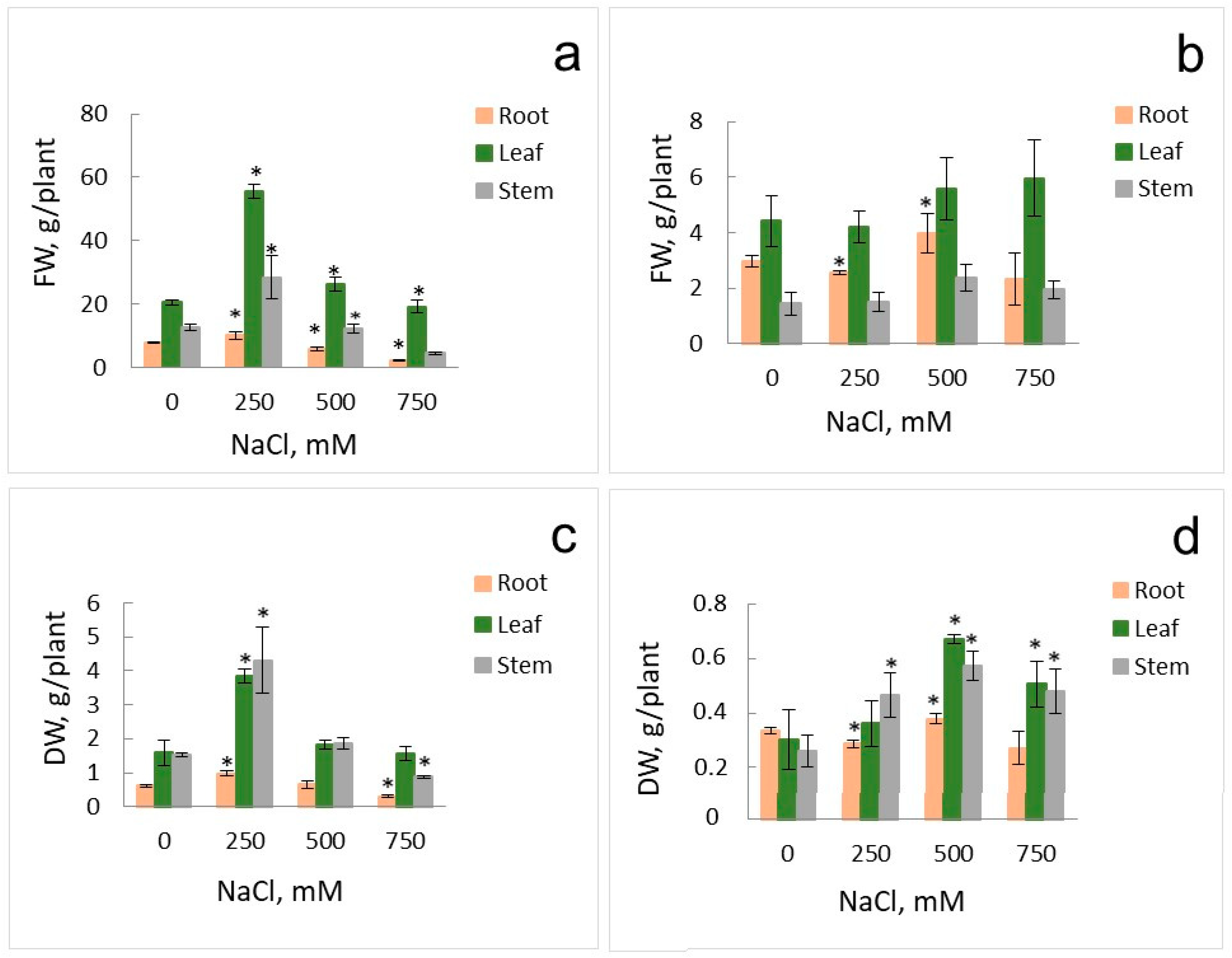
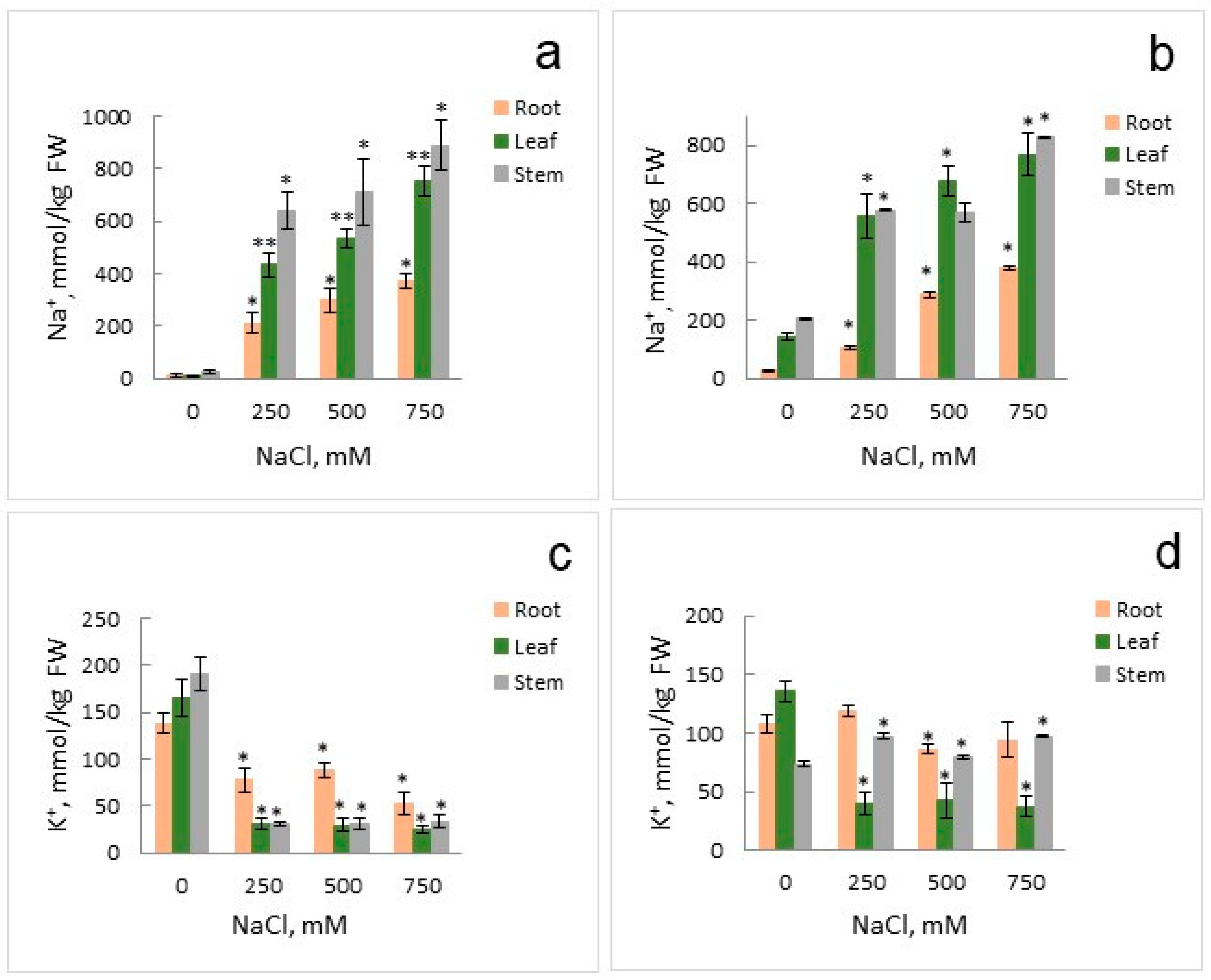
2.1. Growth Parameters and Anion Accumulation in S. altissima Organs
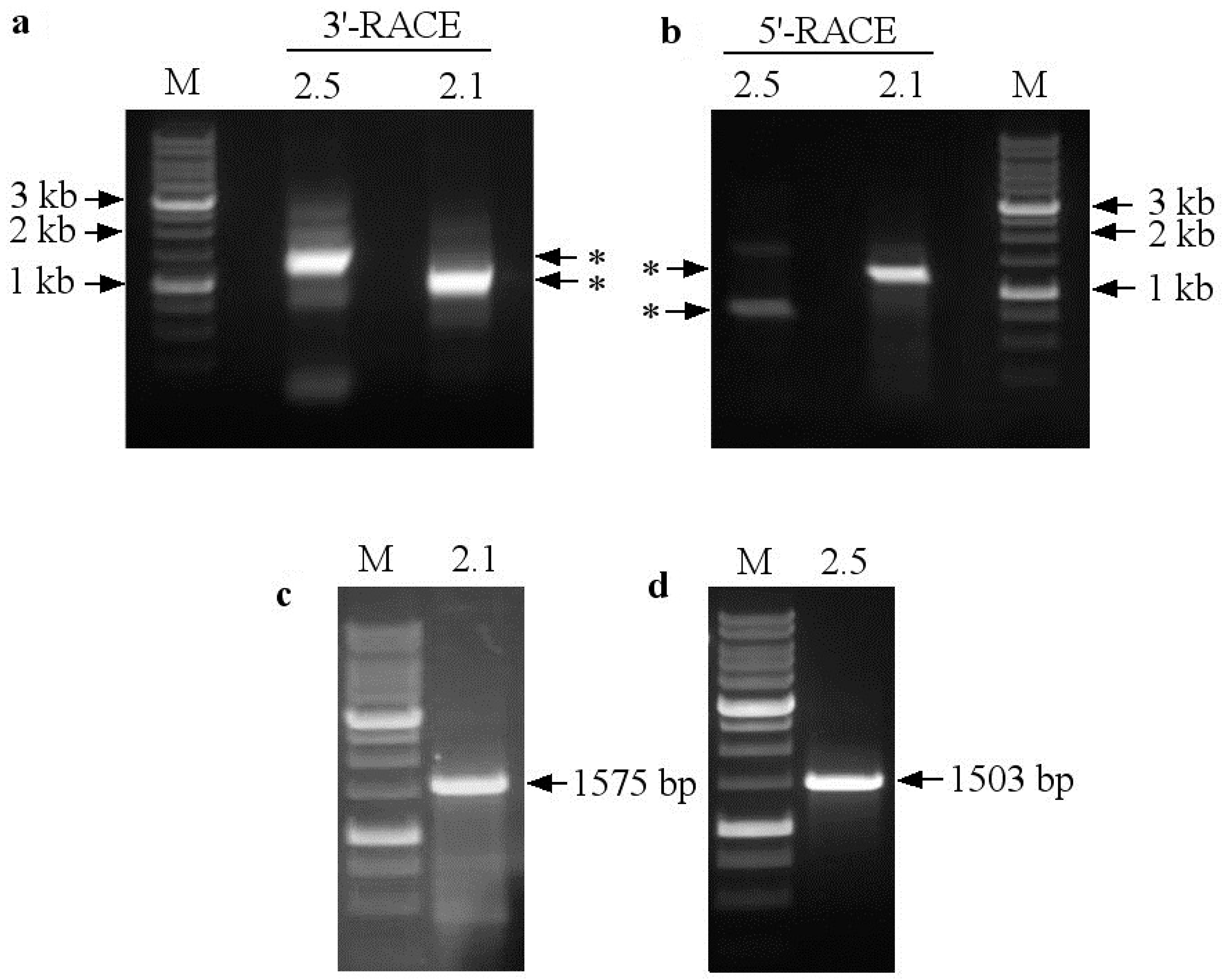
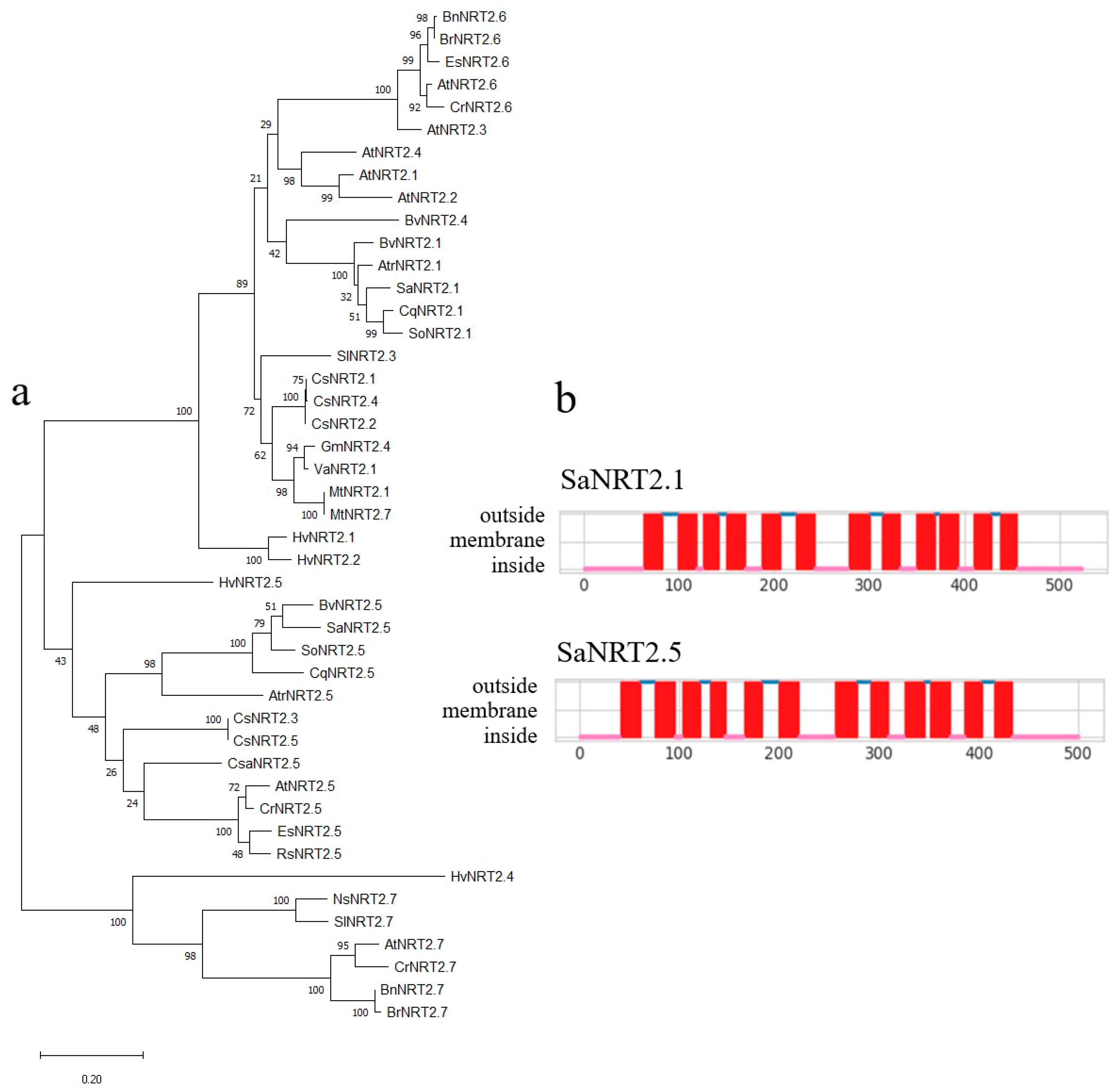
2.2. Identification of the Full-Length Coding Sequences SaNRT2.1 and SaNRT2.5 for High-Affinity Nitrate Transporters and In Silico Analysis of the Protein Structures
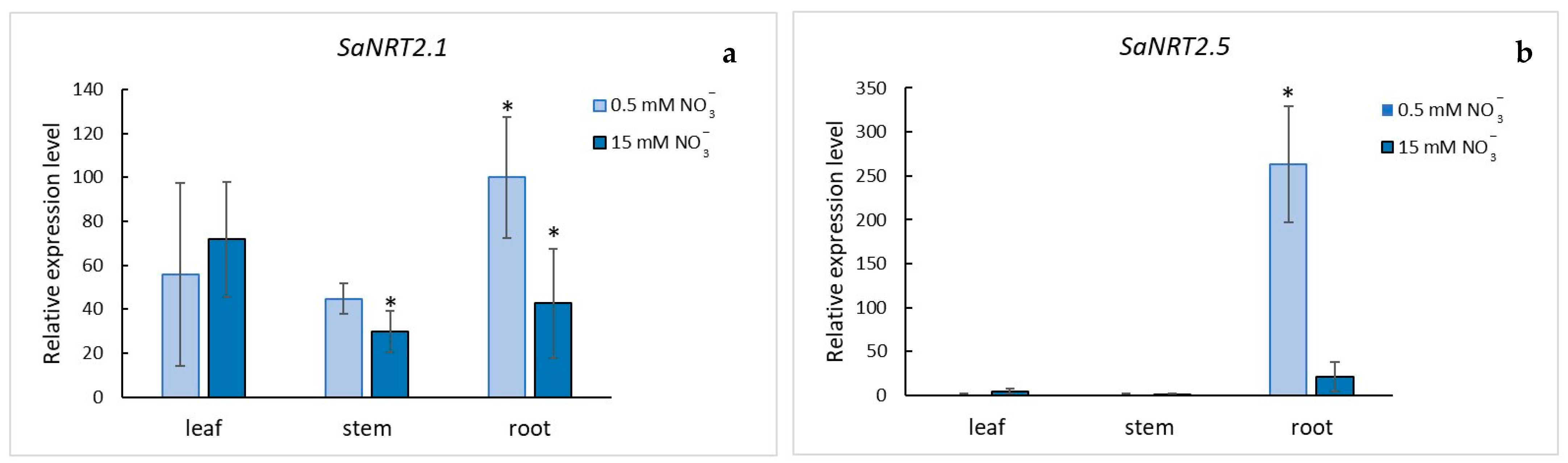
2.3. Quantitative Analysis of SaNRT2.1 and SaNRT2.5 Transcripts in S. altissima Organs
2.4. Experiments on Functional Complementation of Yeast Mutant Δynt1 by SaNRT2.1 and SaNRT2.5 Expression in H. polymorpha Cells
3. Discussion
4. Material and Methods
4.1. Plant Material
4.2. Yeast Strain and Vectors Used in This Study
4.3. Plant Organ Fresh and Dry Weight Analysis
4.4. Determination of NO3− and Cl− Contents in S. altissima Organs
4.5. Extraction of Total RNA from Plant Material and First-Strand cDNA Synthesis
4.6. Primer Design
4.7. Identification of the Full-Length SaNRT2.1 and SaNRT2.5 Coding Sequences
4.8. Quantitative Analysis of SaNRT2.1 and SaNRT2.5 Transcripts in S. altissima Organs
4.9. Cultivation of H. polymorpha WT Strain and Δynt1 Transformants
4.10. Bioinformatic Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A. Nitrate Transport, Sensing, and Responses in Plants. Mol. Plant. 2016, 9, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Dechorgnat, J.; Nguyen, C.T.; Armengaud, P.; Jossier, M.; Diatloff, E.; Filleur, S.; Daniel-Vedele, F. From the soil to the seeds: The long journey of nitrate in plants. J. Exp. Bot. 2011, 62, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Carillo, P.; Rouphael, Y. Nitrate Uptake and Use Efficiency: Pros and Cons of Chloride Interference in the Vegetable Crops. Front. Plant Sci. 2022, 13, 899522. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.Y.; Glass, A.D.M.; Ruth, T.J.; Rufty, T.W. Studies of the uptake of nitrate in barley: I. Kinetics of 13NO3− influx. Plant Physiol. 1990, 93, 1426–1432. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.J.; Fan, X.; Orsel, M.; Smith, S.J.; Wells, D.M. Nitrate transport and signalling. J. Exp. Bot. 2007, 58, 2297–2306. [Google Scholar] [CrossRef] [PubMed]
- Lezhneva, L.; Kiba, T.; Feria-Bourrellier, A.B.; Lafouge, F.; Boutet-Mercey, S.; Zoufan, P.; Sakakibara, H.; Daniel-Vedele, F.; Krapp, A. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 2014, 80, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Orsel, M.; Krapp, A.; Daniel-Vedele, F. Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 2002, 129, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Filleur, S.; Daniel-Vedele, F. Expression analysis of a high affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 1999, 207, 461–469. [Google Scholar] [CrossRef]
- Cerezo, M.; Tillard, P.; Filleur, S.; Munos, S.; Daniel-Vedele, F.; Gojon, A. Major alterations of the regulation of root NO3-uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 2001, 127, 262–271. [Google Scholar] [CrossRef]
- Orsel, M.; Eulenburg, K.; Krapp, A.; Daniel-Vedele, F. Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 2004, 219, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Feria-Bourrellier, A.B.; Lafouge, F.; Lezhneva, L.; Boutet-Mercey, S.; Orsel, M.; Brehaut, V.; Miller, A.; Daniel-Vedele, F.; Sakakibara, H.; et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 2012, 24, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Chopin, F.; Orsel, M.; Dorbe, M.F.; Chardon, F.; Truong, H.N.; Miller, A.J.; Krapp, A.; Daniel-Vedele, F. The Arabidopsis AtNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 2007, 19, 1590–1602. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Gong, X.; Yang, K.; Huang, Y.; Zhang, S.; Shen, L.; Sun, Y.; Wu, D.; Ye, C.; Zhu, Q.H.; et al. Technology-enabled great leap in deciphering plant genomes. Nat. Plants 2024, 10, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.M.; Yan, M.; Fan, X.R.; Li, B.Z.; Shen, Q.R.; Miller, A.J.; Xu, G.H. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J. Exp. Bot. 2011, 62, 2319–2332. [Google Scholar] [CrossRef] [PubMed]
- Plett, D.; Toubia, J.; Garnett, T.; Tester, M.; Kaiser, B.N.; Baumann, U. Dichotomy in the NRT gene families of dicots and grass species. PLoS ONE 2010, 5, e15289. [Google Scholar] [CrossRef]
- Yin, L.P.; Li, P.; Wen, B.; Wen, B.; Taylor, D.; Berry, J.O. Characterization and expression of a high-affinity nitrate system transporter gene (TaNRT2.1) from wheat roots, and its evolutionary relationship to other NTR2 genes. Plant Sci. 2007, 172, 621–631. [Google Scholar] [CrossRef]
- Guo, T.C.; Xuan, H.M.; Yang, Y.Y.; Wang, L.N.; Wei, L.T.; Wang, Y.H.; Kang, G.Z. Transcription analysis of genes encoding the wheat root transporter NRT1 and NRT2 families during nitrogen starvation. J. Plant Growth Regul. 2014, 33, 837–848. [Google Scholar] [CrossRef]
- Guo, B.; Li, Y.; Wang, S.; Li, D.; Lv, C.; Xu, R. Characterization of the Nitrate Transporter gene family and functional identification of HvNRT2.1 in barley (Hordeum vulgare L.). PLoS ONE 2020, 15, e0232056. [Google Scholar] [CrossRef]
- You, H.; Liu, Y.; Minh, T.N.; Lu, H.; Zhang, P.; Li, W.; Xiao, J.; Ding, X.; Li, Q. Genome-wide identification and expression analyses of nitrate transporter family genes in wild soybean (Glycine soja). J. Appl. Genet. 2020, 61, 489–501. [Google Scholar] [CrossRef]
- Tong, J.; Walk, T.C.; Han, P.; Chen, L.; Shen, X.; Li, Y.; Gu, C.; Xie, L.; Hu, X.; Liao, X.; et al. Genome-wide identification and analysis of high-affinity nitrate transporter 2 (NRT2) family genes in rapeseed (Brassica napus L.) and their responses to various stresses. BMC Plant Biol. 2020, 20, 464. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q. Identification and characterization of the NPF, NRT2 and NRT3 in spinach. Plant Physiol. Biochem. 2021, 158, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Li, M.; Xu, W.; Liu, J.-H.; Li, C. Genome-Wide Identification of NRT Gene Family and Expression Analysis of Nitrate Transporters in Response to Salt Stress in Poncirus trifoliata. Genes 2022, 13, 1115. [Google Scholar] [CrossRef] [PubMed]
- Rubinigg, M.; Posthumus, F.; Ferschke, M.; Elzenga, J.T.M.; Stulen, I. Effects of NaCl salinity on 15N-nitrate fluxes and specific root length in the halophyte Plantago maritima L. Plant Soil. 2003, 250, 201–213. [Google Scholar] [CrossRef]
- Debouba, M.; Maâroufi-Dghimi, H.; Suzuki, A.; Ghorbel, M.H.; Gouia, H. Changes in growth and activity of enzymes involved in nitrate reduction and ammonium assimilation in tomato seedlings in response to NaCl stress. Ann. Bot. 2007, 99, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.X.; Zhu, Y.X.; Fang, X.Z.; Ye, J.Y.; Du, W.X.; Zhu, Q.Y.; Lin, X.Y.; Jin, C.W. Ammonium aggravates salt stress in plants by entrapping them in a chloride over-accumulation state in an NRT1.1-dependent manner. Sci. Total Environ. 2020, 746, 141244. [Google Scholar] [CrossRef]
- Abdelgadir, E.M.; Oka, M.; Fujiyama, H. Characteristics of Nitrate Uptake by Plants Under Salinity. J. Plant Nutr. 2005, 28, 33–46. [Google Scholar] [CrossRef]
- Kudo, N.; Fujiyama, H. Responses of halophyte Salicornia bigelovii to different forms of nitrogen source. Pedosphere 2010, 20, 311–317. [Google Scholar] [CrossRef]
- Yuan, J.F.; Tian, C.Y.; Feng, G. Effects of sodium on nitrate uptake and osmotic adjustment of Suaeda physophora. J. Arid Land. 2010, 2, 190–196. [Google Scholar]
- Flowers, T.J. Salt tolerance in Suaeda maritima (L.) Dum: A comparison of mitochondria isolated from green tissues of Suaeda and Pisum. J. Exp. Bot. 1974, 25, 101–110. [Google Scholar] [CrossRef]
- Schütze, P.; Freitag, H.; Weising, K. An integrated molecular and morphological study of the subfamily Suaedoideae Ulbr. (Chenopodiaceae). Plant Syst. Evol. 2003, 239, 257–286. [Google Scholar] [CrossRef]
- Wang, X.; Shao, X.; Zhang, W.; Sun, T.; Ding, Y.; Lin, Z.; Li, Y. Genus Suaeda: Advances in Phytology, Chemistry, Pharmacology and Clinical Application (1895–2021). Pharmacol. Res. 2022, 179, 106203. [Google Scholar] [CrossRef] [PubMed]
- Balnokin, Y.V.; Kotov, A.A.; Myasoedov, N.A.; Khailova, G.F.; Kurkova, E.B.; Lun’kov, R.V.; Kotova, L.M. Involvement of long-distance Na+ transport in maintaining water potential gradient in the medium-root-leaf system of a halophyte Suaeda altissima. Russ. J. Plant Physiol. 2005, 52, 489–496. [Google Scholar] [CrossRef]
- Perdomo, G.; Navarro, F.J.; Medina, B.; Machín, F.; Tejera, P.; Siverio, J.M. Tobacco Nia2 cDNA functionally complements a Hansenula polymorpha yeast mutant lacking nitrate reductase. A new expression system for the study of plant proteins involved in nitrate assimilation. Plant Mol. Biol. 2002, 50, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Martín, Y.; Navarro, F.J.; Siverio, J.M. Functional characterization of the Arabidopsis thaliana nitrate transporter CHL1 in the yeast Hansenula polymorpha. Plant Mol. Biol. 2008, 68, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Machín, F.; Medina, B.; Navarro, F.J.; Pérez, M.D.; Veenhuis, M.; Tejera, P.; Lorenzo, H.; Lancha, A.; Siverio, J.M. The role of Ynt1 in nitrate and nitrite transport in the yeast Hansenula polymorpha. Yeast 2004, 21, 265–276. [Google Scholar] [CrossRef]
- Shuvalov, A.V.; Yurchenko, A.A.; Nedelyaeva, O.I.; Myasoedov, N.A.; Karpichev, I.V.; Khalilova, L.A.; Popova, L.G.; Balnokin, Y.V. Identification of Some Anion Transporter Genes in the Halophyte Suaeda altissima (L.) Pall. and Their Expression under Nitrate Deficiency and Salinity. Russ. J. Plant Physiol. 2021, 68, 873–882. [Google Scholar] [CrossRef]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G. Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 2000, 1465, 219–235. [Google Scholar] [CrossRef]
- Liu, R.; Cui, B.; Lu, X.; Song, J. The positive effect of salinity on nitrate uptake in Suaeda salsa. Plant Physiol. Biochem. 2021, 166, 958–963. [Google Scholar] [CrossRef]
- Maathuis, F.J.M.; Amtmann, A. K+ Nutrition and Na+ Toxicity: The Basis of Cellular K+/Na+ Ratios. Ann. Bot. 1999, 84, 123–133. [Google Scholar] [CrossRef]
- Shabala, S.; Cuin, T.A. Potassium transport and plant salt tolerance. Physiol. Plant. 2008, 133, 651–669. [Google Scholar] [CrossRef]
- Volkov, V.; Wang, B.; Dominy, P.J.; Fricke, W.; Amtmann, A. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell Environ. 2004, 27, 1–14. [Google Scholar] [CrossRef]
- Bojórquez-Quintal, E.; Velarde-Buendía, A.; Ku-González, A.; Carillo-Pech, M.; Ortega-Camacho, D.; Echevarría-Machado, I.; Pottosin, I.; Martínez-Estévez, M. Mechanisms of salt tolerance in habanero pepper plants (Capsicum chinense Jacq.): Proline accumulation, ions dynamics and sodium root-shoot partition and compartmentation. Front. Plant Sci. 2014, 5, 605. [Google Scholar]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Jia, T.; Cui, B.; Song, J. The expression patterns and putative function of nitrate transporter 2.5 in plants. Plant Signal. Behav. 2020, 15, e1815980. [Google Scholar] [CrossRef] [PubMed]
- Orsel, M.; Chopin, F.; Leleu, O.; Smith, S.J.; Krapp, A.; Daniel-Vedele, F.; Miller, A.J. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 2006, 142, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Nedelyaeva, O.I.; Khramov, D.E.; Khalilova, L.A.; Konoshenkova, A.O.; Ryabova, A.V.; Popova, L.G.; Volkov, V.S.; Balnokin, Y.V. Molecular Cloning, Expression and Transport Activity of SaNPF6.3/SaNRT1.1, a Novel Protein of the Low-Affinity Nitrate Transporter Family from the Euhalophyte Suaeda altissima (L.) Pall. Membranes 2023, 13, 845. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-J.; Fernandez, E.; Galvan, A.; Miller, A.J. A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Lett. 2000, 466, 225–227. [Google Scholar] [CrossRef]
- Feng, H.; Fan, X.; Yan, M.; Liu, X.; Miller, A.J.; Xu, G. Multiple roles of nitrate transport accessory protein NAR2 in plants. Plant Signal. Behav. 2011, 6, 1286–1289. [Google Scholar] [CrossRef]
- Yong, Z.; Kotur, Z.; Glass, A.D. Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J. 2010, 63, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Zhang, X.; Jian, J.; Guan, Z.; Zhao, S.; Fang, W.; Liao, Y.; Chen, S.; Chen, F. Chrysanthemum CmNAR2 interacts with CmNRT2 in the control of nitrate uptake. Sci. Rep. 2014, 4, 5833. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.P.; Downton, W.J.S. Potassium, sodium and chloride ion concentrations in leaves and isolated chloroplasts of the halophyte Suaeda australis R. Br. Aust. J. Plant Physiol. 1985, 12, 471–479. [Google Scholar] [CrossRef]
- Bogdanova, A.I.; Agaphonov, M.O.; Ter-Avanesyan, M.D. Plasmid reorganization during integrative transformation in Hansenula polymorpha. Yeast 1995, 11, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Agaphonov, M.O.; Romanova, N.V.; Trushkina, P.M.; Smirnov, V.N.; Ter-Avanesyan, M.D. Aggregation and retention of human urokinase type plasminogen activator in the yeast endoplasmic reticulum. BMC Mol. Biol. 2002, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Nedelyaeva, O.I.; Popova, L.G.; Khramov, D.E.; Volkov, V.S.; Balnokin, Y.V. Chloride Channel Family in the Euhalophyte Suaeda altissima (L.) Pall.: Cloning of Novel Members SaCLCa2 and SaCLCc2, General Characterization of the Family. Int. J. Mol. Sci. 2023, 24, 941. [Google Scholar] [CrossRef] [PubMed]
- Sohn, J.H.; Choi, E.S.; Kim, C.H.; Agaphonov, M.O.; Ter-Avanesyan, M.D.; Rhee, J.S.; Rhee, S.K. A novel autonomously replicating sequence (ARS) for multiple integration in the yeast Hansenula polymorpha DL-1. J. Bacteriol. 1996, 178, 4420–4428. [Google Scholar] [CrossRef] [PubMed]
- Khramov, D.E.; Nedelyaeva, O.I.; Konoshenkova, A.O.; Volkov, V.S.; Balnokin, Y.V. Identification and selection of reference genes for analysis of gene expression by quantitative real time PCR in the euhalophyte Suaeda altissima (L.) Pall. Commun. Integr. Biol. 2024, in press. [Google Scholar]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv 2022. [CrossRef]




| SaNRT2.1 | AtNRT2.1 | AtNRT2.2 | AtNRT2.3 | AtNRT2.4 | AtNRT2.5 | AtNRT2.6 | AtNRT2.7 | |
|---|---|---|---|---|---|---|---|---|
| SaNRT2.5 | 53.37 | 53.46 | 53.27 | 54.38 | 54.21 | 65.36 | 55.47 | 49.79 |
| AtNRT2.1 | 74.48 | 87.26 | 69.04 | 84.23 | 59.75 | 68.28 | 46.52 | |
| AtNRT2.2 | 71.76 | 87.26 | 66.8 | 80.98 | 58.58 | 66.93 | 46.53 | |
| AtNRT2.3 | 68.68 | 69.04 | 66.8 | 71.76 | 57.02 | 89.3 | 47.98 | |
| AtNRT2.4 | 73.9 | 83.24 | 80.98 | 71.76 | 58.14 | 71.16 | 47.17 | |
| AtNRT2.5 | 58.21 | 59.75 | 58.58 | 57.02 | 58.14 | 57.76 | 49.9 | |
| AtNRT2.6 | 69.19 | 68.28 | 66.93 | 89.3 | 71.16 | 57.76 | 47.75 | |
| AtNRT2.7 | 46.86 | 46.52 | 46.53 | 47.98 | 47.17 | 49.9 | 47.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khramov, D.E.; Rostovtseva, E.I.; Matalin, D.A.; Konoshenkova, A.O.; Nedelyaeva, O.I.; Volkov, V.S.; Balnokin, Y.V.; Popova, L.G. Novel Proteins of the High-Affinity Nitrate Transporter Family NRT2, SaNRT2.1 and SaNRT2.5, from the Euhalophyte Suaeda altissima: Molecular Cloning and Expression Analysis. Int. J. Mol. Sci. 2024, 25, 5648. https://doi.org/10.3390/ijms25115648
Khramov DE, Rostovtseva EI, Matalin DA, Konoshenkova AO, Nedelyaeva OI, Volkov VS, Balnokin YV, Popova LG. Novel Proteins of the High-Affinity Nitrate Transporter Family NRT2, SaNRT2.1 and SaNRT2.5, from the Euhalophyte Suaeda altissima: Molecular Cloning and Expression Analysis. International Journal of Molecular Sciences. 2024; 25(11):5648. https://doi.org/10.3390/ijms25115648
Chicago/Turabian StyleKhramov, Dmitrii E., Elena I. Rostovtseva, Dmitrii A. Matalin, Alena O. Konoshenkova, Olga I. Nedelyaeva, Vadim S. Volkov, Yurii V. Balnokin, and Larissa G. Popova. 2024. "Novel Proteins of the High-Affinity Nitrate Transporter Family NRT2, SaNRT2.1 and SaNRT2.5, from the Euhalophyte Suaeda altissima: Molecular Cloning and Expression Analysis" International Journal of Molecular Sciences 25, no. 11: 5648. https://doi.org/10.3390/ijms25115648
APA StyleKhramov, D. E., Rostovtseva, E. I., Matalin, D. A., Konoshenkova, A. O., Nedelyaeva, O. I., Volkov, V. S., Balnokin, Y. V., & Popova, L. G. (2024). Novel Proteins of the High-Affinity Nitrate Transporter Family NRT2, SaNRT2.1 and SaNRT2.5, from the Euhalophyte Suaeda altissima: Molecular Cloning and Expression Analysis. International Journal of Molecular Sciences, 25(11), 5648. https://doi.org/10.3390/ijms25115648







