The ArgR-Regulated ADI Pathway Facilitates the Survival of Vibrio fluvialis under Acidic Conditions
Abstract
1. Introduction
2. Results
2.1. Bioinformatic Analysis of ADI Gene Clusters and Regulatory Genes in the Genus Vibrio
2.2. L-Arginine Enhances the Growth of V. fluvialis at Low pH
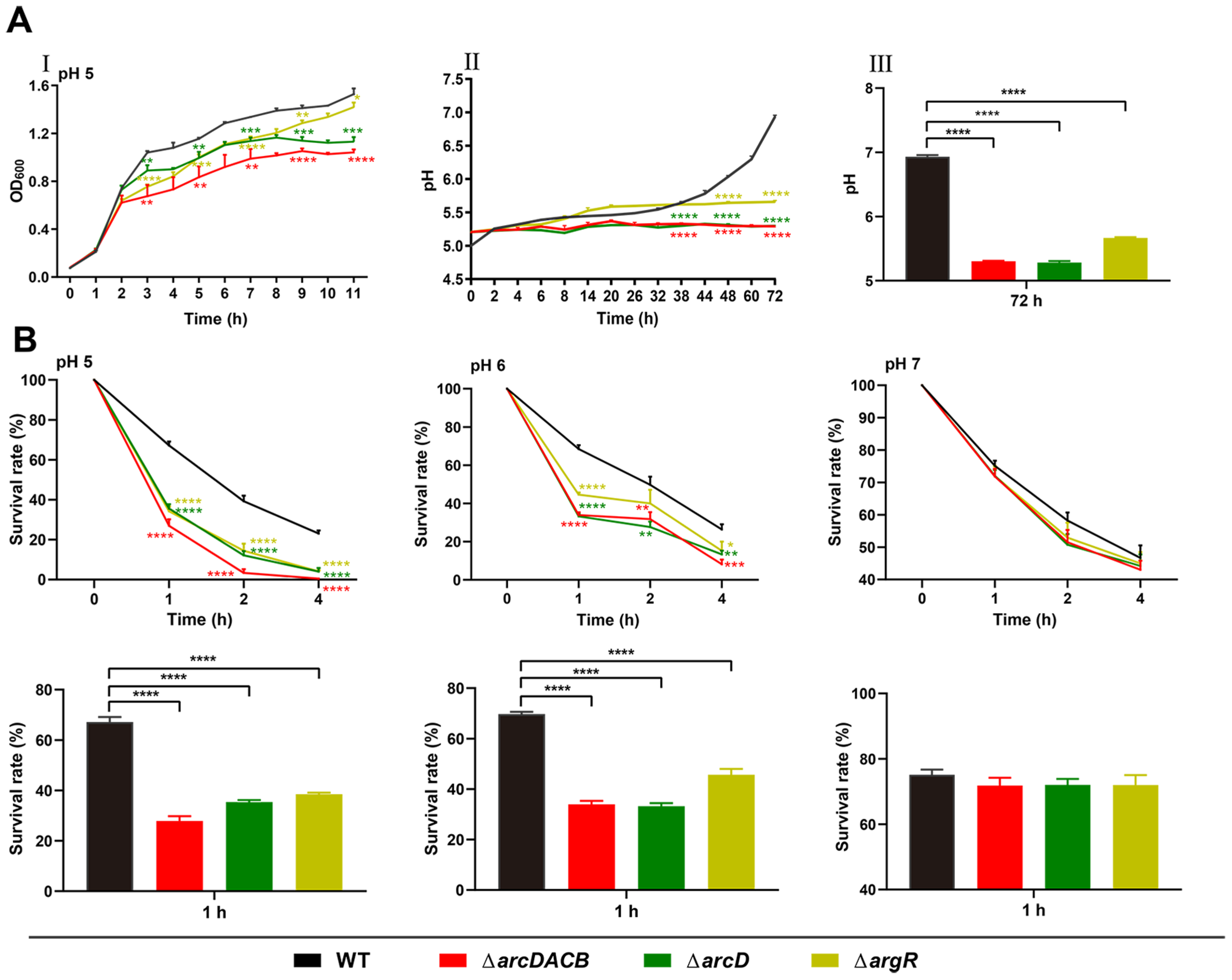
2.3. Role of the ADI Gene Cluster and argR in Acid Resistance in V. fluvialis
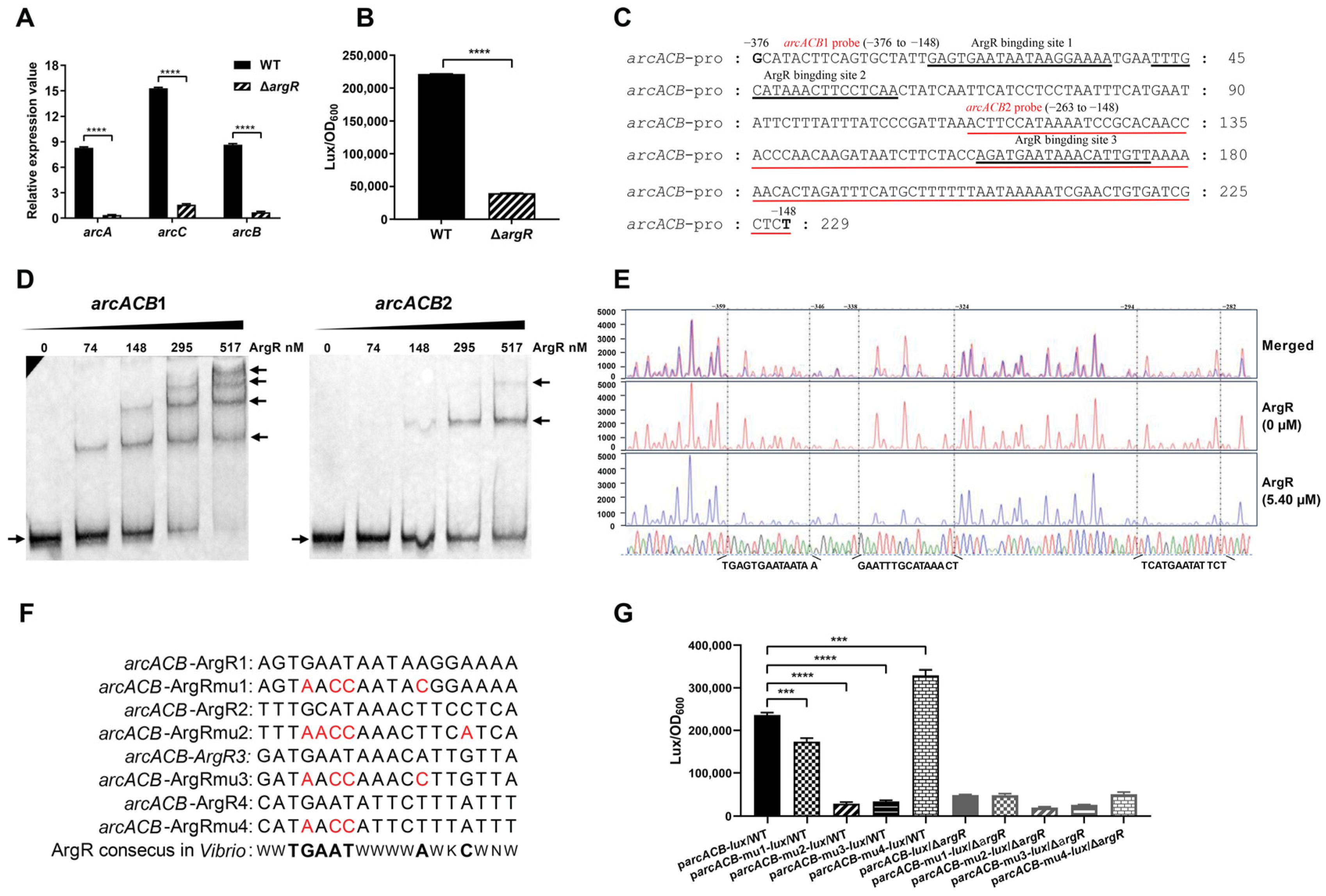
2.4. ArgR Activates arcD at the Transcriptional Level by Directly Binding to Its Promoter Region
2.5. ArgR Activates the arcACB Operon by Physically Binding to the Promoter Region
2.6. Effects of the ADI Gene Cluster and argR on In Vivo Colonization of V. fluvialis
2.7. ADI Pathway Deficiency Enhances the Phagocytosis of V. fluvialis by Macrophages
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Culture Conditions, and Plasmids
4.2. Distribution and Phylogenetic Analysis of the ADI Cluster in Vibrio Species
4.3. Construction of in-Frame Deletion Mutants
4.4. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
4.5. 5′-Rapid Amplification of cDNA End (5′-RACE)
4.6. Growth Analysis
4.7. Acid Resistance of V. fluvialis Strains
4.8. Analysis of the Transcriptional Activity of Promoters with a Luciferase Reporter Gene Assay
4.9. Cloning, Expression, and Purification of ArgR-His6
4.10. Electrophoretic Mobility Shift Assay (EMSA)
4.11. DNase I Footprinting Assay
4.12. Mouse Competition Assay
4.13. Adhesion, Invasion, and Intracellular Survival of V. fluvialis in RAW 264.7 Cells
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hornef, M.W.; Wick, M.J.; Rhen, M.; Normark, S. Bacterial strategies for overcoming host innate and adaptive immune responses. Nat. Immunol. 2002, 3, 1033–1040. [Google Scholar] [CrossRef]
- Tlaskalová-Hogenová, H.; Štěpánková, R.; Hudcovic, T.; Tučková, L.; Cukrowska, B.; Lodinová-Žádníková, R.; Kozáková, H.; Rossmann, P.; Bártová, J.; Sokol, D.; et al. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol. Lett. 2004, 93, 97–108. [Google Scholar] [CrossRef]
- Donnenberg, M.S.; Whittam, T.S. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Investig. 2001, 107, 539–548. [Google Scholar] [CrossRef]
- Hooper, L.V.; Gordon, J.I. Commensal Host-Bacterial Relationships in the Gut. Science 2001, 292, 1115–1118. [Google Scholar] [CrossRef]
- Lynch, J.P.; Lesser, C.F. A host metabolite promotes Salmonella survival. Science 2021, 371, 344–345. [Google Scholar] [CrossRef]
- Manchester, M.; Das, P.; Lahiri, A.; Lahiri, A.; Chakravortty, D. Modulation of the Arginase Pathway in the Context of Microbial Pathogenesis: A Metabolic Enzyme Moonlighting as an Immune Modulator. PLoS Pathog. 2010, 6, e1000899. [Google Scholar] [CrossRef]
- Gogoi, M.; Datey, A.; Wilson, K.T.; Chakravortty, D. Dual role of arginine metabolism in establishing pathogenesis. Curr. Opin. Microbiol. 2016, 29, 43–48. [Google Scholar] [CrossRef]
- Abdelal, A.T. Arginine Catabolism by Microorganisms. Annu. Rev. Microbiol. 1979, 33, 139–168. [Google Scholar] [CrossRef]
- Stadelmann, B.; Hanevik, K.; Andersson, M.K.; Bruserud, O.; Svärd, S.G. The role of arginine and arginine-metabolizing enzymes during Giardia—Host cell interactions in vitro. BMC Microbiol. 2013, 13, 256. [Google Scholar] [CrossRef]
- Xiong, L.F.; Teng, J.L.L.; Botelho, M.G.; Lo, R.C.; Lau, S.K.P.; Woo, P.C.Y. Arginine Metabolism in Bacterial Pathogenesis and Cancer Therapy. Int. J. Mol. Sci. 2016, 17, 363. [Google Scholar] [CrossRef]
- Hernández, V.M.; Arteaga, A.; Dunn, M.F. Diversity, properties and functions of bacterial arginases. Fems Microbiol. Rev. 2021, 45, fuab034. [Google Scholar] [CrossRef]
- Xiong, L.; Teng, J.L.L.; Watt, R.M.; Kan, B.; Lau, S.K.P.; Woo, P.C.Y. Arginine deiminase pathway is far more important than urease for acid resistance and intracellular survival in Laribacter hongkongensis: A possible result of arc gene cassette duplication. BMC Microbiol. 2014, 14, 42. [Google Scholar] [CrossRef]
- Weathers, P.J.; Chee, H.L.; Allen, M.M. Arginine catabolism in Aphanocapsa 6308. Arch. Microbiol. 1978, 118, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-D.; Winteler, H.; Abdelal, A.; Haas, D. The ArgR Regulatory Protein, a Helper to the Anaerobic Regulator ANR during Transcriptional Activation of the arcD Promoter in Pseudomonas aeruginosa. J. Bacteriol. 1999, 181, 2459–2464. [Google Scholar] [CrossRef]
- Minic, Z.; Hervé, G. Arginine Metabolism in the Deep Sea Tube Worm Riftia pachyptila and Its Bacterial Endosymbiont. J. Biol. Chem. 2003, 278, 40527–40533. [Google Scholar] [CrossRef]
- Caldara, M.; Charlier, D.; Cunin, R. The arginine regulon of Escherichia coli: Whole-system transcriptome analysis discovers new genes and provides an integrated view of arginine regulation. Microbiology 2006, 152, 3343–3354. [Google Scholar] [CrossRef]
- Liu, Y.; Dong, Y.; Chen, Y.-Y.M.; Burne, R.A. Environmental and Growth Phase Regulation of the Streptococcus gordonii Arginine Deiminase Genes. Appl. Environ. Microbiol. 2008, 74, 5023–5030. [Google Scholar] [CrossRef]
- Schulz, C.; Gierok, P.; Petruschka, L.; Lalk, M.; Mäder, U.; Hammerschmidt, S.; Thornton, J.A.; McDaniel, L.S. Regulation of the Arginine Deiminase System by ArgR2 Interferes with Arginine Metabolism and Fitness of Streptococcus pneumoniae. mBio 2014, 5, e01858-14. [Google Scholar] [CrossRef]
- Coburn, J.; Richards, C.L.; Raffel, S.J.; Bontemps-Gallo, S.; Dulebohn, D.P.; Herbert, T.C.; Gherardini, F.C. The arginine deaminase system plays distinct roles in Borrelia burgdorferi and Borrelia hermsii. PLOS Pathog. 2022, 18, e1010370. [Google Scholar] [CrossRef]
- Ryan, S.; Begley, M.; Gahan, C.G.M.; Hill, C. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: Regulation and role in acid tolerance. Environ. Microbiol. 2009, 11, 432–445. [Google Scholar] [CrossRef]
- Lindgren, J.K.; Thomas, V.C.; Olson, M.E.; Chaudhari, S.S.; Nuxoll, A.S.; Schaeffer, C.R.; Lindgren, K.E.; Jones, J.; Zimmerman, M.C.; Dunman, P.M.; et al. Arginine Deiminase in Staphylococcus epidermidis Functions to Augment Biofilm Maturation through pH Homeostasis. J. Bacteriol. 2014, 196, 2277–2289. [Google Scholar] [CrossRef]
- Gruening, P.; Fulde, M.; Valentin-Weigand, P.; Goethe, R. Structure, Regulation, and Putative Function of the Arginine Deiminase System of Streptococcus suis. J. Bacteriol. 2006, 188, 361–369. [Google Scholar] [CrossRef]
- Xu, B.; Yang, X.Y.; Zhang, P.; Ma, Z.; Lin, H.X.; Fan, H.J. The arginine deiminase system facilitates environmental adaptability of Streptococcus equi ssp. zooepidemicus through pH adjustment. Res. Microbiol. 2016, 167, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Gahan, C.G.M.; Hill, C. Analysis of the role of the Listeria monocytogenes F0F1-ATPase operon in the acid tolerance response. Int. J. Food Microbiol. 2000, 60, 137–146. [Google Scholar] [CrossRef]
- Choi, Y.; Choi, J.; Groisman, E.A.; Kang, D.-H.; Shin, D.; Ryu, S.; Camilli, A. Expression of STM4467-Encoded Arginine Deiminase Controlled by the STM4463 Regulator Contributes to Salmonella enterica Serovar Typhimurium Virulence. Infect. Immun. 2012, 80, 4291–4297. [Google Scholar] [CrossRef]
- Chakraborty, B.; Burne, R.A. Effects of Arginine on Streptococcus mutans Growth, Virulence Gene Expression, and Stress Tolerance. Appl. Environ. Microbiol. 2017, 83, e00496-17. [Google Scholar] [CrossRef] [PubMed]
- Makhlin, J.; Kofman, T.; Borovok, I.; Kohler, C.; Engelmann, S.; Cohen, G.; Aharonowitz, Y. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J. Bacteriol. 2007, 189, 5976–5986. [Google Scholar] [CrossRef]
- Hitzmann, A.; Bergmann, S.; Rohde, M.; Chhatwal, G.S.; Fulde, M. Identification and characterization of the arginine deiminase system of Streptococcus canis. Vet. Microbiol. 2013, 162, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Chantratita, N.; Tandhavanant, S.; Wikraiphat, C.; Trunck, L.A.; Rholl, D.A.; Thanwisai, A.; Saiprom, N.; Limmathurotsakul, D.; Korbsrisate, S.; Day, N.P.J.; et al. Proteomic analysis of colony morphology variants of Burkholderia pseudomallei defines a role for the arginine deiminase system in bacterial survival. J. Proteom. 2012, 75, 1031–1042. [Google Scholar] [CrossRef]
- Maghnouj, A.; Abu-Bakr, A.A.W.; Baumberg, S.; Stalon, V.; Wauven, C. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol. Lett. 2000, 191, 227–234. [Google Scholar] [CrossRef]
- Winterhoff, N.; Goethe, R.; Gruening, P.; Rohde, M.; Kalisz, H.; Smith, H.E.; Valentin-Weigand, P. Identification and Characterization of Two Temperature-Induced Surface-Associated Proteins of Streptococcus suis with High Homologies to Members of the Arginine Deiminase System of Streptococcus pyogenes. J. Bacteriol. 2002, 184, 6768–6776. [Google Scholar] [CrossRef] [PubMed]
- Fulde, M.; Willenborg, J.; de Greeff, A.; Benga, L.; Smith, H.E.; Valentin-Weigand, P.; Goethe, R. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology 2011, 157, 572–582. [Google Scholar] [CrossRef]
- Grandori, R.; Lavoie, T.A.; Pflumm, M.; Tian, G.; Niersbach, H.; Maas, W.K.; Fairman, R.; Carey, J. The DNA-binding Domain of the Hexameric Arginine Repressor. J. Mol. Biol. 1995, 254, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Garnett, J.A.; Marincs, F.; Baumberg, S.; Stockley, P.G.; Phillips, S.E.V. Structure and Function of the Arginine Repressor-Operator Complex from Bacillus subtilis. J. Mol. Biol. 2008, 379, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Cherney, L.T.; Cherney, M.M.; Garen, C.R.; James, M.N.G. Crystal Structure of the Intermediate Complex of the Arginine Repressor from Mycobacterium tuberculosis Bound with Its DNA Operator Reveals Detailed Mechanism of Arginine Repression. J. Mol. Biol. 2010, 399, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Maghnouj, A.; de Sousa Cabral, T.F.; Stalon, V.; Vander Wauven, C. The arcABDC Gene Cluster, Encoding the Arginine Deiminase Pathway of Bacillus licheniformis, and Its Activation by the Arginine Repressor ArgR. J. Bacteriol. 1998, 180, 6468–6475. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, B.; Thawani, G.; Sanyal, S. Etiology of acute childhood diarrhoea in Calcutta. Trop Gastroenterol 1989, 10, 158–166. [Google Scholar] [PubMed]
- Chowdhury, G.; Pazhani, G.P.; Dutta, D.; Guin, S.; Dutta, S.; Ghosh, S.; Izumiya, H.; Asakura, M.; Yamasaki, S.; Takeda, Y.; et al. Vibrio fluvialisin Patients with Diarrhea, Kolkata, India. Emerg. Infect. Dis. 2012, 18, 1868–1871. [Google Scholar] [CrossRef]
- Kobayashi, K.; Taguchi, M.; Shimada, T.; Sakazaki, R. Ten cases of gastroenteritis possibly caused by Vibrio fluvialis and its enterotoxigenicity. J. Jpn. Assoc. Infect. Dis. 1983, 57, 375–382. [Google Scholar] [CrossRef][Green Version]
- Igbinosa, E.O.; Obi, L.C.; Okoh, A.I. Occurrence of potentially pathogenic Vibrios in final effluents of a wastewater treatment facility in a rural community of the Eastern Cape Province of South Africa. Res. Microbiol. 2009, 160, 531–537. [Google Scholar] [CrossRef]
- Huq, M.I.; Alam, A.K.; Brenner, D.J.; Morris, G.K. Isolation of Vibrio-like group, EF-6, from patients with diarrhea. J. Clin. Microbiol. 1980, 11, 621–624. [Google Scholar] [CrossRef]
- Lu, X.; Liang, W.; Wang, Y.; Xu, J.; Zhu, J.; Kan, B. Identification of Genetic Bases of Vibrio fluvialis Species-Specific Biochemical Pathways and Potential Virulence Factors by Comparative Genomic Analysis. Appl. Environ. Microbiol. 2014, 80, 2029–2037. [Google Scholar] [CrossRef]
- Xiao, D.; Yang, W.; Wang, H.; Bi, Z.; Lu, L.; Kan, B. Vibrio Cholera Prevention and Control Manual, 6th ed.; Wang, L., Wang, D., Wang, M., Wang, G., Feng, Z., Bi, Z., Su, H., Yang, W., Xiao, D., Wang, H., et al., Eds.; People’s Health Publishing House: Beijing, China, 2013; p. 91. (in Chinese) [Google Scholar]
- Brenner, D.J.; Hickman-Brenner, F.W.; Lee, J.V.; Steigerwalt, A.G.; Fanning, G.R.; Hollis, D.G.; Farmer, J.J.; Weaver, R.E.; Joseph, S.W.; Seidler, R.J. Vibrio furnissii (formerly aerogenic biogroup of Vibrio fluvialis), a new species isolated from human feces and the environment. J. Clin. Microbiol. 1983, 18, 816–824. [Google Scholar] [CrossRef]
- Liang, P.; Cui, X.; Du, X.; Kan, B.; Liang, W. The virulence phenotypes and molecular epidemiological characteristics of Vibrio fluvialis in China. Gut Pathog. 2013, 5, 6. [Google Scholar] [CrossRef]
- Kala, K.; Shahi, N.; Singh, S.; Rawat, S.; Patiyal, R.S.; Pande, V.; Mallik, S.K. New host record of Vibrio anguillarum associated with haemorrhagic septicaemia in golden mahseer, Tor putitora (Hamilton, 1822) from India. Indian J. Comp. Microbiol. Immunol. Infect. Dis. 2021, 42, 71–83. [Google Scholar] [CrossRef]
- Makarova, K.S.; Mironov, A.A.; Gelfand, M.S. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2001, 2, RESEARCH0013. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Morgan, X.C.; Segata, N.; Waldron, L.; Reyes, J.; Earl, A.M.; Giannoukos, G.; Boylan, M.R.; Ciulla, D.; Gevers, D.; et al. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. USA 2014, 111, E2329–E2338. [Google Scholar] [CrossRef]
- Degnan, B.A.; Tuomanen, E.I.; Fontaine, M.C.; Doebereiner, A.H.; Lee, J.J.; Mastroeni, P.; Dougan, G.; Goodacre, J.A.; Kehoe, M.A. Characterization of an Isogenic Mutant of Streptococcus pyogenes Manfredo Lacking the Ability To Make Streptococcal Acid Glycoprotein. Infect. Immun. 2000, 68, 2441–2448. [Google Scholar] [CrossRef]
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric oxide synthases: Structure, function and inhibition. Biochem. J. 2001, 357, 593–615. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Gotoh, T. Arginine Metabolic Enzymes, Nitric Oxide and Infection. J. Nutr. 2004, 134, 2820S–2825S. [Google Scholar] [CrossRef] [PubMed]
- Cusumano, Z.T.; Watson, M.E.; Caparon, M.G.; Camilli, A. Streptococcus pyogenes Arginine and Citrulline Catabolism Promotes Infection and Modulates Innate Immunity. Infect. Immun. 2014, 82, 233–242. [Google Scholar] [CrossRef]
- Krysenko, S.; Emani, C.S.; Bäuerle, M.; Oswald, M.; Kulik, A.; Meyners, C.; Hillemann, D.; Merker, M.; Wohlers, I.; Hausch, F.; et al. GlnA3Mt is able to glutamylate spermine but it is not essential for the detoxification of spermine in Mycobacterium tuberculosis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Gamper, M.; Zimmermann, A.; Haas, D. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 1991, 173, 4742–4750. [Google Scholar] [CrossRef]
- Maas, W.K. The arginine repressor of Escherichia coli. Microbiol. Rev. 1994, 58, 631–640. [Google Scholar] [CrossRef]
- Klingel, U.; Miller, C.M.; North, A.K.; Stockley, P.G.; Baumberg, S. A binding site for activation by the Bacillus subtilis AhrC protein, a repressor/activator of arginine metabolism. Mol. Gen. Genet. MGG 1995, 248, 329–340. [Google Scholar] [CrossRef]
- Rodríguez-García, A.; Ludovice, M.; Martín, J.F.; Liras, P. Arginine boxes and the argR gene in Streptomyces clavuligerus: Evidence for a clear regulation of the arginine pathway. Mol. Microbiol. 2003, 25, 219–228. [Google Scholar] [CrossRef]
- Charlier, D.; Roovers, M.; Van Vliet, F.; Boyen, A.; Cunin, R.; Nakamura, Y.; Glansdorff, N.; Piérard, A. Arginine regulon of Escherichia coli K-12. J. Mol. Biol. 1992, 226, 367–386. [Google Scholar] [CrossRef]
- Tian, G.; Lim, D.; Carey, J.; Maas, W.K. Binding of the arginine repressor of Escherichia coli K12 to its operator sites. J. Mol. Biol. 1992, 226, 387–397. [Google Scholar] [CrossRef]
- Miller, C.M.; Baumberg, S.; Stockley, P.G. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: Novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol. Microbiol. 2003, 26, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Dong, Y.; Burne, R.A. Characterization of cis-acting sites controlling arginine deiminase gene expression in Streptococcus gordonii. J. Bacteriol. 2006, 188, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Marquis, R.E.; Bender, G.R.; Murray, D.R.; Wong, A. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 1987, 53, 198–200. [Google Scholar] [CrossRef] [PubMed]
- Bassoe, C.F.; Bjerknes, R. Phagocytosis by Human Leukocytes, Phagosomal pH and Degradation of Seven Species of Bacteria Measured by Flow Cytometry. J. Med. Microbiol. 1985, 19, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Fulde, M.; Willenborg, J.; Huber, C.; Hitzmann, A.; Willms, D.; Seitz, M.; Eisenreich, W.; Valentin-Weigand, P.; Goethe, R. The arginine-ornithine antiporter ArcD contributes to biological fitness of Streptococcus suis. Front. Cell. Infect. Microbiol. 2014, 4, 107. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Zhao, M.; Li, J.; Gao, H.; Kan, B.; Liang, W. Direct regulation of the natural competence regulator gene tfoX by cyclic AMP (cAMP) and cAMP receptor protein (CRP) in Vibrios. Sci. Rep. 2015, 5, 14921. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.W.; Xing, R.X.; Zhang, W.H.; Li, L.; Wu, Y.; Hu, J.; Wang, C.; Luo, Q.L.; Shen, J.L.; Chen, X. Toxoplasma ROP16I/III ameliorated inflammatory bowel diseases via inducing M2 phenotype of macrophages. World J. Gastroenterol. 2019, 25, 6634–6635. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.J.; Zhao, M.; Huang, Y.M.; Li, J.; Liu, X.S.; Ren, Z.H.; Kan, B.; Liang, W.L. Integration Host Factor Modulates the Expression and Function of T6SS2 in Vibrio fluvialis. Front. Microbiol. 2018, 9, 00962. [Google Scholar] [CrossRef]
- Zianni, M.; Tessanne, K.; Merighi, M.; Laguna, R.; Tabita, F.R. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 2006, 17, 103–113. [Google Scholar]
- Monack, D.M.; Raupach, B.; Hromockyj, A.E.; Falkow, S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 1996, 93, 9833–9838. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Q.; Han, Y.; Xiao, Y.; Li, Z.; Qin, A.; Ji, S.; Kan, B.; Liang, W. The ArgR-Regulated ADI Pathway Facilitates the Survival of Vibrio fluvialis under Acidic Conditions. Int. J. Mol. Sci. 2024, 25, 5679. https://doi.org/10.3390/ijms25115679
Cheng Q, Han Y, Xiao Y, Li Z, Qin A, Ji S, Kan B, Liang W. The ArgR-Regulated ADI Pathway Facilitates the Survival of Vibrio fluvialis under Acidic Conditions. International Journal of Molecular Sciences. 2024; 25(11):5679. https://doi.org/10.3390/ijms25115679
Chicago/Turabian StyleCheng, Qian, Yu Han, Yue Xiao, Zhe Li, Aiping Qin, Saisen Ji, Biao Kan, and Weili Liang. 2024. "The ArgR-Regulated ADI Pathway Facilitates the Survival of Vibrio fluvialis under Acidic Conditions" International Journal of Molecular Sciences 25, no. 11: 5679. https://doi.org/10.3390/ijms25115679
APA StyleCheng, Q., Han, Y., Xiao, Y., Li, Z., Qin, A., Ji, S., Kan, B., & Liang, W. (2024). The ArgR-Regulated ADI Pathway Facilitates the Survival of Vibrio fluvialis under Acidic Conditions. International Journal of Molecular Sciences, 25(11), 5679. https://doi.org/10.3390/ijms25115679





