Identification of an FMNL2 Interactome by Quantitative Mass Spectrometry
Abstract
1. Introduction
2. Results
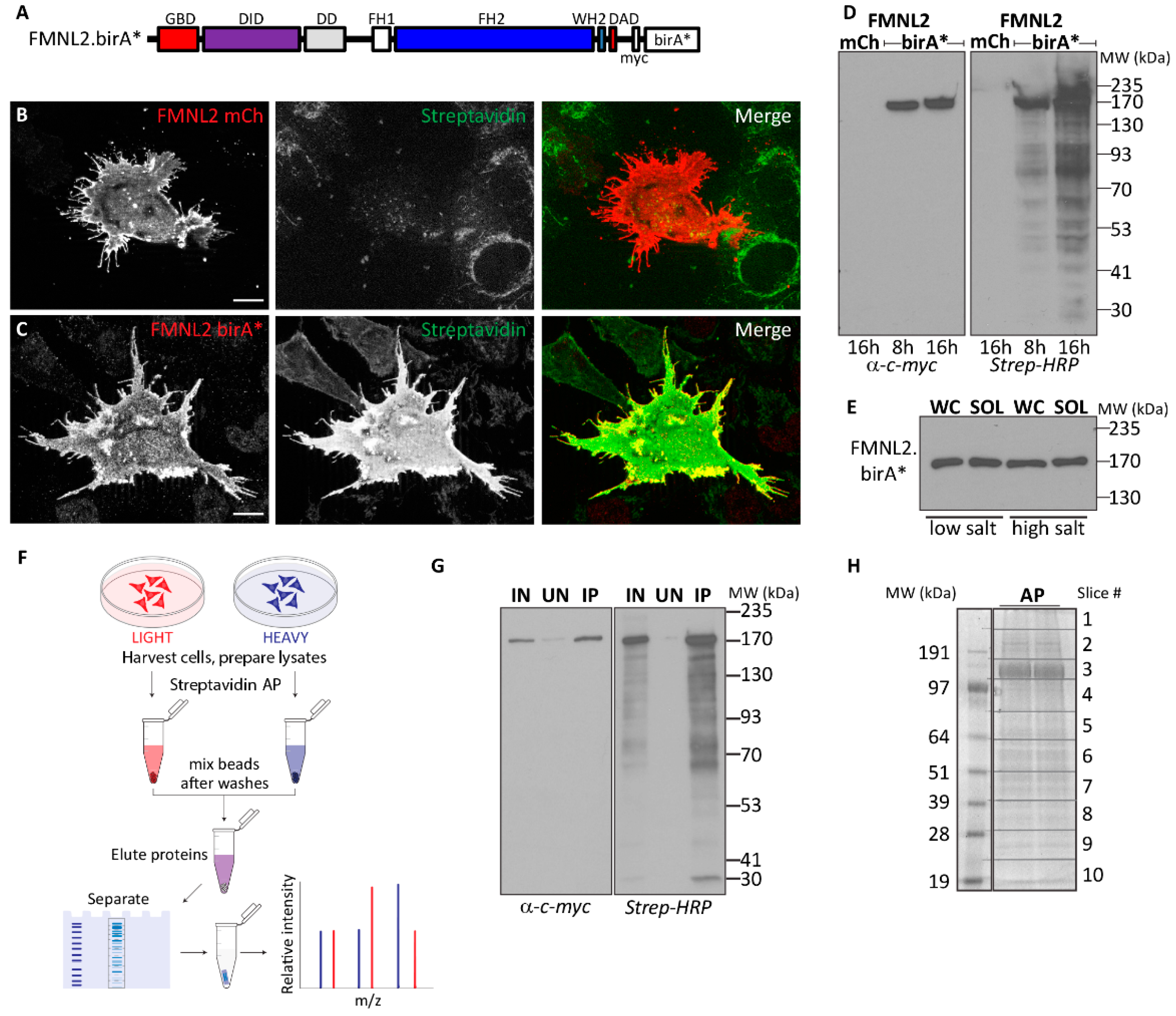
2.1. FMNL2 BioID Screen
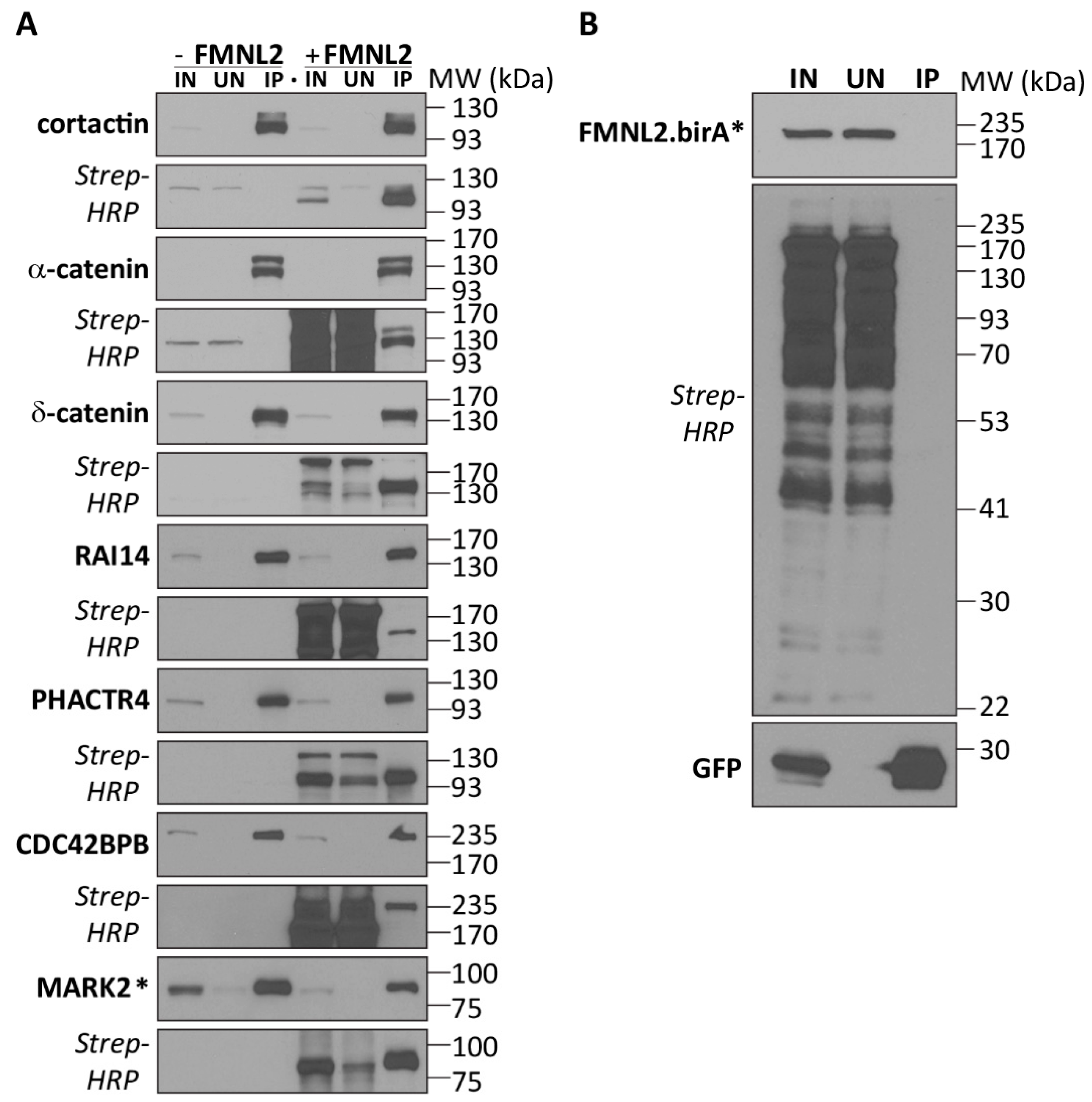
2.2. Functional FMNL2 Interacting Proteins
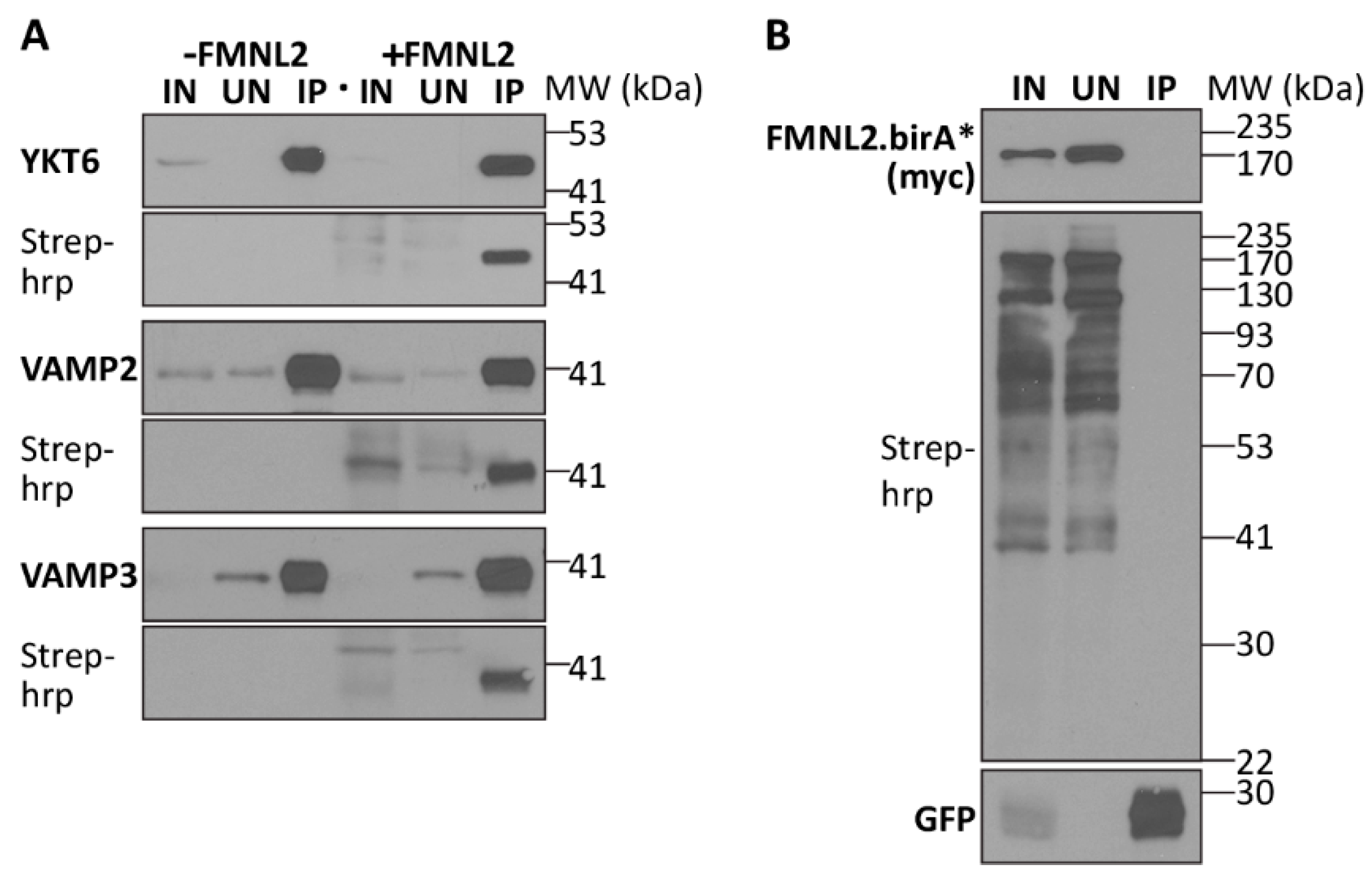
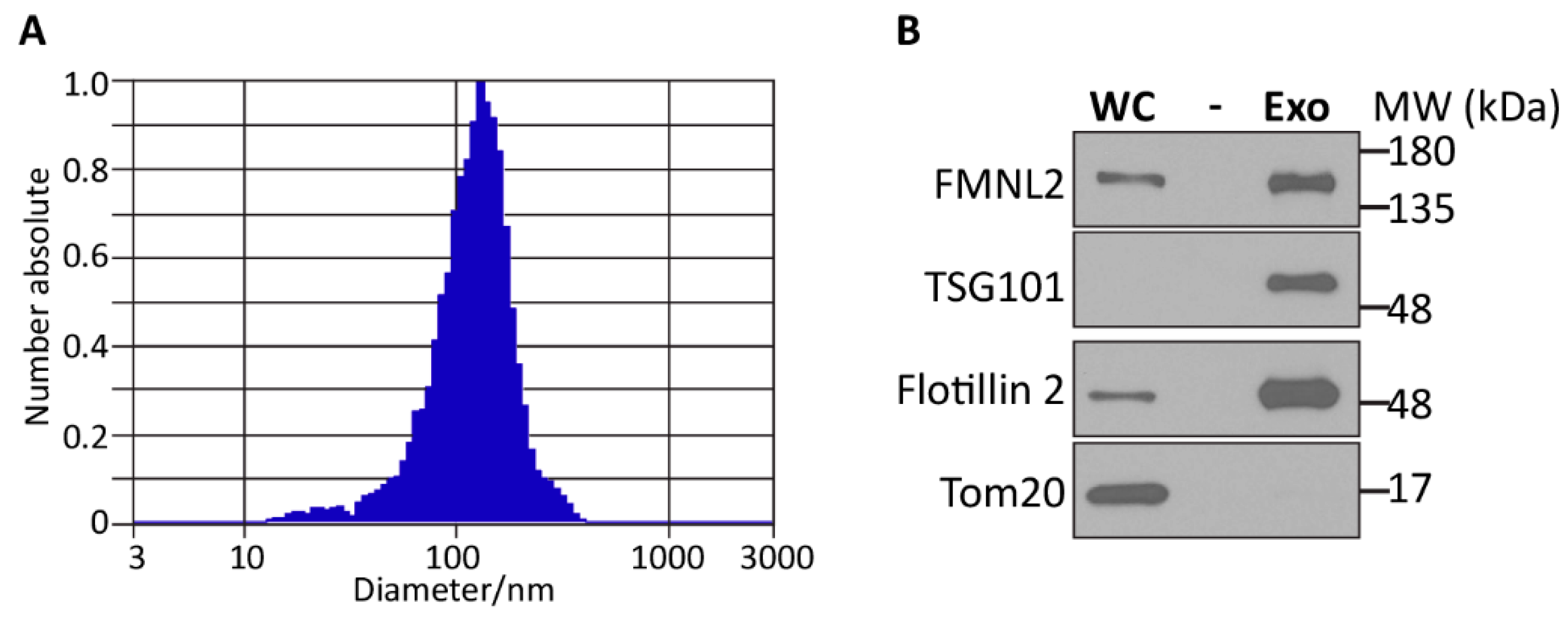
2.3. FMNL2 and Extracellular Vesicles
3. Discussion
4. Materials and Methods
4.1. Reagents and Plasmids
4.2. Cell Culture, Transfections, and Treatments
4.3. Immunofluorescence
4.4. BioID Screen
4.5. Gene Ontology
4.6. BioID Validation
4.7. Extracellular Vesicle Isolation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Young, K.G.; Copeland, J.W. Formins in cell signaling. Biochim. Biophys. Acta 2010, 1803, 183–190. [Google Scholar] [CrossRef]
- Higgs, H.N. Formin proteins: A domain-based approach. Trends Biochem. Sci. 2005, 30, 342–353. [Google Scholar] [CrossRef]
- Truong, D.; Copeland, J.W.; Brumell, J.H. Bacterial subversion of host cytoskeletal machinery: Hijacking formins and the Arp2/3 complex. Bioessays 2014, 36, 687–696. [Google Scholar] [CrossRef]
- Labat-de-Hoz, L.; Alonso, M.A. Formins in Human Disease. Cells 2021, 10, 2554. [Google Scholar] [CrossRef]
- Chen, A.; Arora, P.D.; Lai, C.C.; Copeland, J.W.; Moraes, T.F.; McCulloch, C.A.; Lavoie, B.D.; Wilde, A. The scaffold-protein IQGAP1 enhances and spatially restricts the actin-nucleating activity of Diaphanous-related formin 1 (DIAPH1). J. Biol. Chem. 2020, 295, 3134–3147. [Google Scholar] [CrossRef]
- Zweifel, M.E.; Sherer, L.A.; Mahanta, B.; Courtemanche, N. Nucleation limits the lengths of actin filaments assembled by formin. Biophys. J. 2021, 120, 4442–4456. [Google Scholar] [CrossRef]
- Courtemanche, N. Mechanisms of formin-mediated actin assembly and dynamics. Biophys. Rev. 2018, 10, 1553–1569. [Google Scholar] [CrossRef]
- Vaillant, D.C.; Copeland, S.J.; Davis, C.; Thurston, S.F.; Abdennur, N.; Copeland, J.W. Interaction of the N- and C-terminal autoregulatory domains of FRL2 does not inhibit FRL2 activity. J. Biol. Chem. 2008, 283, 33750–33762. [Google Scholar] [CrossRef]
- Esue, O.; Harris, E.S.; Higgs, H.N.; Wirtz, D. The filamentous actin cross-linking/bundling activity of mammalian formins. J. Mol. Biol. 2008, 384, 324–334. [Google Scholar] [CrossRef]
- Harris, E.S.; Rouiller, I.; Hanein, D.; Higgs, H.N. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J. Biol. Chem. 2006, 281, 14383–14392. [Google Scholar] [CrossRef]
- Bartolini, F.; Andres-Delgado, L.; Qu, X.; Nik, S.; Ramalingam, N.; Kremer, L.; Alonso, M.A.; Gundersen, G.G. An mDia1-INF2 formin activation cascade facilitated by IQGAP1 regulates stable microtubules in migrating cells. Mol. Biol. Cell 2016, 27, 1797–1808. [Google Scholar] [CrossRef]
- Bartolini, F.; Gundersen, G.G. Formins and microtubules. Biochim. Biophys. Acta 2010, 1803, 164–173. [Google Scholar] [CrossRef]
- Thurston, S.F.; Kulacz, W.A.; Shaikh, S.; Lee, J.M.; Copeland, J.W. The ability to induce microtubule acetylation is a general feature of formin proteins. PLoS ONE 2012, 7, e48041. [Google Scholar] [CrossRef]
- Young, K.G.; Thurston, S.F.; Copeland, S.; Smallwood, C.; Copeland, J.W. INF1 is a novel microtubule-associated formin. Mol. Biol. Cell 2008, 19, 5168–5180. [Google Scholar] [CrossRef]
- Fernandez-Barrera, J.; Alonso, M.A. Coordination of microtubule acetylation and the actin cytoskeleton by formins. Cell Mol. Life Sci. 2018, 75, 3181–3191. [Google Scholar] [CrossRef]
- Schonichen, A.; Geyer, M. Fifteen formins for an actin filament: A molecular view on the regulation of human formins. Biochim. Biophys. Acta 2010, 1803, 152–163. [Google Scholar] [CrossRef]
- Pruyne, D. Probing the origins of metazoan formin diversity: Evidence for evolutionary relationships between metazoan and non-metazoan formin subtypes. PLoS ONE 2017, 12, e0186081. [Google Scholar] [CrossRef]
- Hegsted, A.; Yingling, C.V.; Pruyne, D. Inverted formins: A subfamily of atypical formins. Cytoskeleton 2017, 74, 405–419. [Google Scholar] [CrossRef]
- Breitsprecher, D.; Goode, B.L. Formins at a glance. J. Cell Sci. 2013, 126 Pt 1, 1–7. [Google Scholar] [CrossRef]
- Chesarone, M.A.; DuPage, A.G.; Goode, B.L. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 2010, 11, 62–74. [Google Scholar] [CrossRef]
- Copeland, S.J.; McRae, A.; Guarguaglini, G.; Trinkle-Mulcahy, L.; Copeland, J.W. Actin-dependent regulation of cilia length by the inverted formin FHDC1. Mol. Biol. Cell 2018, 29, 1611–1627. [Google Scholar] [CrossRef] [PubMed]
- Copeland, S.J.; Thurston, S.F.; Copeland, J.W. Actin- and microtubule-dependent regulation of Golgi morphology by FHDC1. Mol. Biol. Cell 2016, 27, 260–276. [Google Scholar] [CrossRef] [PubMed]
- Fattouh, R.; Kwon, H.; Czuczman, M.A.; Copeland, J.W.; Pelletier, L.; Quinlan, M.E.; Muise, A.M.; Higgins, D.E.; Brumell, J.H. The diaphanous-related formins promote protrusion formation and cell-to-cell spread of Listeria monocytogenes. J. Infect. Dis. 2015, 211, 1185–1195. [Google Scholar] [CrossRef]
- Hetheridge, C.; Scott, A.N.; Swain, R.K.; Copeland, J.W.; Higgs, H.N.; Bicknell, R.; Mellor, H. The formin FMNL3 is a cytoskeletal regulator of angiogenesis. J. Cell Sci. 2012, 125 Pt 6, 1420–1428. [Google Scholar] [CrossRef]
- Lim, S.M.; Cruz, V.E.; Antoku, S.; Gundersen, G.G.; Schwartz, T.U. Structures of FHOD1-Nesprin1/2 complexes reveal alternate binding modes for the FH3 domain of formins. Structure 2021, 29, 540–552. [Google Scholar] [CrossRef]
- Qu, X.; Yuan, F.N.; Corona, C.; Pasini, S.; Pero, M.E.; Gundersen, G.G.; Shelanski, M.L.; Bartolini, F. Stabilization of dynamic microtubules by mDia1 drives Tau-dependent Abeta1-42 synaptotoxicity. J. Cell Biol. 2017, 216, 3161–3178. [Google Scholar] [CrossRef]
- Fox, S.; Tran, A.; Trinkle-Mulcahy, L.; Copeland, J.W. Cooperative assembly of filopodia by the formin FMNL2 and I-BAR domain protein IRTKS. J. Biol. Chem. 2022, 298, 102512. [Google Scholar] [CrossRef] [PubMed]
- Peladeau, C.; Heibein, A.; Maltez, M.T.; Copeland, S.J.; Copeland, J.W. A specific FMNL2 isoform is up-regulated in invasive cells. BMC Cell Biol. 2016, 17, 32. [Google Scholar] [CrossRef]
- Block, J.; Breitsprecher, D.; Kuhn, S.; Winterhoff, M.; Kage, F.; Geffers, R.; Duwe, P.; Rohn, J.L.; Baum, B.; Brakebusch, C.; et al. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 2012, 22, 1005–1012. [Google Scholar] [CrossRef]
- Zhu, X.L.; Zeng, Y.F.; Guan, J.; Li, Y.F.; Deng, Y.J.; Bian, X.W.; Ding, Y.Q.; Liang, L. FMNL2 is a positive regulator of cell motility and metastasis in colorectal carcinoma. J. Pathol. 2011, 224, 377–388. [Google Scholar] [CrossRef]
- Kitzing, T.M.; Wang, Y.; Pertz, O.; Copeland, J.W.; Grosse, R. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene 2010, 29, 2441–2448. [Google Scholar] [CrossRef]
- Kage, F.; Winterhoff, M.; Dimchev, V.; Mueller, J.; Thalheim, T.; Freise, A.; Bruhmann, S.; Kollasser, J.; Block, J.; Dimchev, G.; et al. FMNL formins boost lamellipodial force generation. Nat. Commun. 2017, 8, 14832. [Google Scholar] [CrossRef]
- Lorenzen, L.; Frank, D.; Schwan, C.; Grosse, R. Spatiotemporal Regulation of FMNL2 by N-Terminal Myristoylation and C-Terminal Phosphorylation Drives Rapid Filopodia Formation. Biomolecules 2023, 13, 548. [Google Scholar] [CrossRef]
- Pfisterer, K.; Levitt, J.; Lawson, C.D.; Marsh, R.J.; Heddleston, J.M.; Wait, E.; Ameer-Beg, S.M.; Cox, S.; Parsons, M. FMNL2 regulates dynamics of fascin in filopodia. J. Cell Biol. 2020, 219, e201906111. [Google Scholar] [CrossRef]
- Kuhn, S.; Erdmann, C.; Kage, F.; Block, J.; Schwenkmezger, L.; Steffen, A.; Rottner, K.; Geyer, M. The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation. Nat. Commun. 2015, 6, 7088. [Google Scholar] [CrossRef]
- Grobe, H.; Wustenhagen, A.; Baarlink, C.; Grosse, R.; Grikscheit, K. A Rac1-FMNL2 signaling module affects cell-cell contact formation independent of Cdc42 and membrane protrusions. PLoS ONE 2018, 13, e0194716. [Google Scholar] [CrossRef]
- Grikscheit, K.; Frank, T.; Wang, Y.; Grosse, R. Junctional actin assembly is mediated by Formin-like 2 downstream of Rac1. J. Cell Biol. 2015, 209, 367–376. [Google Scholar] [CrossRef]
- Wang, Y.; Arjonen, A.; Pouwels, J.; Ta, H.; Pausch, P.; Bange, G.; Engel, U.; Pan, X.; Fackler, O.T.; Ivaska, J.; et al. Formin-like 2 Promotes beta1-Integrin Trafficking and Invasive Motility Downstream of PKCalpha. Dev. Cell 2015, 34, 475–483. [Google Scholar] [CrossRef]
- Frank, D.; Moussi, C.J.; Ulferts, S.; Lorenzen, L.; Schwan, C.; Grosse, R. Vesicle-Associated Actin Assembly by Formins Promotes TGFbeta-Induced ANGPTL4 Trafficking, Secretion and Cell Invasion. Adv. Sci. 2023, 10, e2204896. [Google Scholar] [CrossRef]
- Kage, F.; Steffen, A.; Ellinger, A.; Ranftler, C.; Gehre, C.; Brakebusch, C.; Pavelka, M.; Stradal, T.; Rottner, K. FMNL2 and -3 regulate Golgi architecture and anterograde transport downstream of Cdc42. Sci. Rep. 2017, 7, 9791. [Google Scholar] [CrossRef]
- Woodham, E.F.; Paul, N.R.; Tyrrell, B.; Spence, H.J.; Swaminathan, K.; Scribner, M.R.; Giampazolias, E.; Hedley, A.; Clark, W.; Kage, F.; et al. Coordination by Cdc42 of Actin, Contractility, and Adhesion for Melanoblast Movement in Mouse Skin. Curr. Biol. 2017, 27, 624–637. [Google Scholar] [CrossRef]
- Moriya, K.; Yamamoto, T.; Takamitsu, E.; Matsunaga, Y.; Kimoto, M.; Fukushige, D.; Kimoto, C.; Suzuki, T.; Utsumi, T. Protein N-myristoylation is required for cellular morphological changes induced by two formin family proteins, FMNL2 and FMNL3. Biosci. Biotechnol. Biochem. 2012, 76, 1201–1209. [Google Scholar] [CrossRef]
- Isogai, T.; van der Kammen, R.; Goerdayal, S.S.; Heck, A.J.; Altelaar, A.F.; Innocenti, M. Proteomic analyses uncover a new function and mode of action for mouse homolog of Diaphanous 2 (mDia2). Mol. Cell Proteomics 2015, 14, 1064–1078. [Google Scholar] [CrossRef]
- Saleh, A.; Subramaniam, G.; Raychaudhuri, S.; Dhawan, J. Cytoplasmic sequestration of the RhoA effector mDiaphanous1 by Prohibitin2 promotes muscle differentiation. Sci. Rep. 2019, 9, 8302. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yu, G.; Sarioglu, H.; Caballero-Martinez, A.; Schlott, F.; Ueffing, M.; Haase, H.; Peschel, C.; Krackhardt, A.M. Proteomic investigation of the interactome of FMNL1 in hematopoietic cells unveils a role in calcium-dependent membrane plasticity. J. Proteomics 2013, 78, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Rollason, R.; Wherlock, M.; Heath, J.A.; Heesom, K.J.; Saleem, M.A.; Welsh, G.I. Disease causing mutations in inverted formin 2 regulate its binding to G-actin, F-actin capping protein (CapZ alpha-1) and profilin 2. Biosci. Rep. 2016, 36, e00302. [Google Scholar] [CrossRef]
- Lambert, J.P.; Tucholska, M.; Go, C.; Knight, J.D.; Gingras, A.C. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J. Proteomics 2015, 118, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Ren XL, Qiao YD, Li JY, Li XM, Zhang D, Zhang XJ, Zhu XH, Zhou WJ, Shi J, Wang W et al: Cortactin recruits FMNL2 to promote actin polymerization and endosome motility in invadopodia formation. Cancer Lett. 2018, 419, 245–256. [CrossRef]
- Schackmann, R.C.; Tenhagen, M.; van de Ven, R.A.; Derksen, P.W. p120-catenin in cancer—Mechanisms, models and opportunities for intervention. J. Cell Sci. 2013, 126 Pt 16, 3515–3525. [Google Scholar] [CrossRef]
- Pieters, T.; van Roy, F.; van Hengel, J. Functions of p120ctn isoforms in cell-cell adhesion and intracellular signaling. Front. Biosci. 2012, 17, 1669–1694. [Google Scholar] [CrossRef]
- Kowalczyk, A.P.; Reynolds, A.B. Protecting your tail: Regulation of cadherin degradation by p120-catenin. Curr. Opin. Cell Biol. 2004, 16, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Hofbrucker-MacKenzie, S.A.; Izadi, M.; Seemann, E.; Steiniger, F.; Schwintzer, L.; Koch, D.; Kessels, M.M.; Qualmann, B. Ankyrin repeat-containing N-Ank proteins shape cellular membranes. Nat. Cell Biol. 2019, 21, 1191–1205. [Google Scholar] [CrossRef]
- Izadi, M.; Wolf, D.; Seemann, E.; Ori, A.; Schwintzer, L.; Steiniger, F.; Kessels, M.M.; Qualmann, B. Membrane shapers from two distinct superfamilies cooperate in the development of neuronal morphology. J. Cell Biol. 2023, 222, e202211032. [Google Scholar] [CrossRef] [PubMed]
- Huet, G.; Rajakyla, E.K.; Viita, T.; Skarp, K.P.; Crivaro, M.; Dopie, J.; Vartiainen, M.K. Actin-regulated feedback loop based on Phactr4, PP1 and cofilin maintains the actin monomer pool. J. Cell Sci. 2013, 126 Pt 2, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Unbekandt, M.; Belshaw, S.; Bower, J.; Clarke, M.; Cordes, J.; Crighton, D.; Croft, D.R.; Drysdale, M.J.; Garnett, M.J.; Gill, K.; et al. Discovery of Potent and Selective MRCK Inhibitors with Therapeutic Effect on Skin Cancer. Cancer Res. 2018, 78, 2096–2114. [Google Scholar] [CrossRef] [PubMed]
- Unbekandt, M.; Croft, D.R.; Crighton, D.; Mezna, M.; McArthur, D.; McConnell, P.; Schuttelkopf, A.W.; Belshaw, S.; Pannifer, A.; Sime, M.; et al. A novel small-molecule MRCK inhibitor blocks cancer cell invasion. Cell Commun. Signal 2014, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.I.; Cheng, C.Y. MARK2 and MARK4 Regulate Sertoli Cell BTB Dynamics Through Microtubule and Actin Cytoskeletons. Endocrinology 2022, 163, bqac130. [Google Scholar]
- Pasapera, A.M.; Heissler, S.M.; Eto, M.; Nishimura, Y.; Fischer, R.S.; Thiam, H.R.; Waterman, C.M. MARK2 regulates directed cell migration through modulation of myosin II contractility and focal adhesion organization. Curr. Biol. 2022, 32, 2704–2718 e2706. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Shi, M.; Stewart, T.; Fernagut, P.O.; Huang, Y.; Tian, C.; Dehay, B.; Atik, A.; Yang, D.; De Giorgi, F.; et al. Reduced oligodendrocyte exosome secretion in multiple system atrophy involves SNARE dysfunction. Brain 2020, 143, 1780–1797. [Google Scholar]
- Somasundaram, A.; Taraska, J.W. Local protein dynamics during microvesicle exocytosis in neuroendocrine cells. Mol. Biol. Cell 2018, 29, 1891–1903. [Google Scholar]
- de Wit, H.; Lichtenstein, Y.; Kelly, R.B.; Geuze, H.J.; Klumperman, J.; van der Sluijs, P. Rab4 regulates formation of synaptic-like microvesicles from early endosomes in PC12 cells. Mol. Biol. Cell 2001, 12, 3703–3715. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, P.; Dong, W.; Liu, H.; Sun, J.; Zhao, L. LncRNA PVT1 promotes exosome secretion through YKT6, RAB7, and VAMP3 in pancreatic cancer. Aging 2020, 12, 10427–10440. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, S.N.; Rider, M.A.; Bundy, J.L.; Liu, X.; Singh, R.K.; Meckes, D.G., Jr. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget 2016, 7, 86999–87015. [Google Scholar] [CrossRef] [PubMed]
- Clancy, J.W.; Sedgwick, A.; Rosse, C.; Muralidharan-Chari, V.; Raposo, G.; Method, M.; Chavrier, P.; D’Souza-Schorey, C. Regulated delivery of molecular cargo to invasive tumour-derived microvesicles. Nat. Commun. 2015, 6, 6919. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, S.M.; Zhang, F.; Ding, C.; Montoya-Durango, D.E.; Hu, X.; Yang, C.; Wang, Z.; Yuan, F.; Fox, M.; Zhang, H.G.; et al. Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming. Cell Metab. 2021, 33, 2040–2058 e2010. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, R.; Kemper, S.; Brigstock, D.R. Pathways of production and delivery of hepatocyte exosomes. J. Cell Commun. Signal 2018, 12, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Martinez, M.; Navarro, A.; Marrades, R.M.; Vinolas, N.; Santasusagna, S.; Munoz, C.; Ramirez, J.; Molins, L.; Monzo, M. YKT6 expression, exosome release, and survival in non-small cell lung cancer. Oncotarget 2016, 7, 51515–51524. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Popovic, A.; Miihkinen, M.; Ghimire, S.; Saup, R.; Gronloh, M.L.B.; Ball, N.J.; Goult, B.T.; Ivaska, J.; Jacquemet, G. Myosin-X recruits lamellipodin to filopodia tips. J. Cell Sci. 2023, 136, jcs260574. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Cannon, K.S.; Curtis, B.N.; Edelmaier, C.; Gladfelter, A.S.; Nazockdast, E. Curvature sensing as an emergent property of multiscale assembly of septins. Proc. Natl. Acad. Sci. USA 2023, 120, e2208253120. [Google Scholar] [CrossRef]
- Zhao, H.; Pykalainen, A.; Lappalainen, P. I-BAR domain proteins: Linking actin and plasma membrane dynamics. Curr. Opin. Cell Biol. 2011, 23, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Faix, J.; Rottner, K. Ena/VASP proteins in cell edge protrusion, migration and adhesion. J. Cell Sci. 2022, 135, jcs259226. [Google Scholar] [CrossRef] [PubMed]
- Gronloh, M.L.B.; Arts, J.J.G.; Mahlandt, E.K.; Nolte, M.A.; Goedhart, J.; van Buul, J.D. Primary adhered neutrophils increase JNK1-MARCKSL1-mediated filopodia to promote secondary neutrophil transmigration. iScience 2023, 26, 107406. [Google Scholar] [CrossRef] [PubMed]
- Krause, M.; Gautreau, A. Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 2014, 15, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Horton ER, Byron A, Askari JA, Ng DH, Millon-Fremillon A, Robertson J, Koper EJ, Paul NR, Warwood S, Knight D et al: Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 2015, 17, 1577–1587. [CrossRef] [PubMed]
- Meves, A.; Stremmel, C.; Gottschalk, K.; Fassler, R. The Kindlin protein family: New members to the club of focal adhesion proteins. Trends Cell Biol. 2009, 19, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Shortill, S.P.; Frier, M.S.; Conibear, E. You can go your own way: SNX-BAR coat complexes direct traffic at late endosomes. Curr. Opin. Cell Biol. 2022, 76, 102087. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Saarloos, I.; Kooistra, R.; van de Bospoort, R.; Verhage, M.; Toonen, R.F. SNAP-25 gene family members differentially support secretory vesicle fusion. J. Cell Sci. 2017, 130, 1877–1889. [Google Scholar] [CrossRef] [PubMed]
- Kugeratski, F.G.; Hodge, K.; Lilla, S.; McAndrews, K.M.; Zhou, X.; Hwang, R.F.; Zanivan, S.; Kalluri, R. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat. Cell Biol. 2021, 23, 631–641. [Google Scholar] [CrossRef]
- Vilcaes, A.A.; Chanaday, N.L.; Kavalali, E.T. Interneuronal exchange and functional integration of synaptobrevin via extracellular vesicles. Neuron 2021, 109, 971–983 e975. [Google Scholar] [CrossRef]
- He, G.; Li, W.; Zhao, W.; Men, H.; Chen, Q.; Hu, J.; Zhang, J.; Zhu, H.; Wang, W.; Deng, M.; et al. Formin-like 2 promotes angiogenesis and metastasis of colorectal cancer by regulating the EGFL6/CKAP4/ERK axis. Cancer Sci. 2023, 114, 2014–2028. [Google Scholar] [CrossRef] [PubMed]
- Lazar, I.; Clement, E.; Ducoux-Petit, M.; Denat, L.; Soldan, V.; Dauvillier, S.; Balor, S.; Burlet-Schiltz, O.; Larue, L.; Muller, C.; et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment. Cell Melanoma Res. 2015, 28, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Otasek, D.; Morris, J.H.; Boucas, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]

| Protein Class | UniProt | Gene | Peptide | Unique Peptide | SILAC Ratio | Total Intensity | Bkgd |
|---|---|---|---|---|---|---|---|
| ECVs | Q15836 | VAMP3 | 5 | 2 | 22.22 | 1.16 × 108 | - |
| Q9UQN3 | CHMP2B | 4 | 4 | Bait only | 2.28 × 107 | - | |
| P63027 | VAMP2 | 5 | 2 | Bait only | 9.39 × 106 | - | |
| O15498 | YKT6 | 2 | 2 | Bait only | 9.16 × 106 | - | |
| Q9NZZ3 | CHMP5 | 2 | 2 | 3.09 | 8.72 × 106 | - | |
| O75955 | FLOT1 | 2 | 2 | 3.12 | 2.65 × 106 | - | |
| Actin | Q14247 | CTTN * | 13 | 13 | Bait only | 1.39 × 108 | < |
| Cytoskeleton | Q14008 | CKAP5 | 20 | 20 | 3.16 | 1.16 × 108 | < |
| Q8N8S7 | ENAH | 4 | 4 | 47.86 | 3.21 × 107 | - | |
| P13797 | PLS3 | 3 | 3 | 2.08 | 2.59 × 107 | - | |
| Q9UQB8 | BAIAP2 * | 4 | 4 | 7.62 | 2.27 × 107 | - | |
| Q9UJU6 | DBNL | 4 | 4 | 22.28 | 1.53 × 107 | - | |
| P49006 | MARCKSL1 | 3 | 3 | Bait only | 1.18 × 107 | - | |
| Q7Z6K5 | ARPIN | 2 | 2 | Bait only | 7.17 × 106 | - | |
| Q9UHR4 | BAIAP2L1 * | 2 | 2 | Bait only | 6.05 × 106 | - | |
| Q05682 | CALD1 | 2 | 2 | Bait only | 2.86 × 106 | - | |
| Q3V6T2 | CCDC88A | 2 | 2 | Bait only | 2.56 × 106 | - | |
| P61586;P08134 | RHOA/C * | 2 | 2 | Bait only | 1.10 × 106 | - | |
| SNAREs | Q13596 | SNX1 | 13 | 11 | Bait only | 2.94 × 108 | - |
| and SNXs | O60749 | SNX2 | 13 | 11 | Bait only | 1.19 × 108 | < |
| Q9BSJ8 | ESYT1 | 3 | 3 | 3.51 | 2.80 × 107 | - | |
| O00161 | SNAP23 | 2 | 2 | Bait only | 1.86 × 107 | - | |
| Q9Y5X1 | SNX9 | 6 | 6 | 69.76 | 1.64 × 107 | - | |
| Q9Y5X3 | SNX5 | 5 | 5 | 3.54 | 1.60 × 107 | - | |
| O95721 | SNAP29 | 3 | 3 | Bait only | 9.11 × 106 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fox, S.; Gaudreau-LaPierre, A.; Reshke, R.; Podinic, I.; Gibbings, D.J.; Trinkle-Mulcahy, L.; Copeland, J.W. Identification of an FMNL2 Interactome by Quantitative Mass Spectrometry. Int. J. Mol. Sci. 2024, 25, 5686. https://doi.org/10.3390/ijms25115686
Fox S, Gaudreau-LaPierre A, Reshke R, Podinic I, Gibbings DJ, Trinkle-Mulcahy L, Copeland JW. Identification of an FMNL2 Interactome by Quantitative Mass Spectrometry. International Journal of Molecular Sciences. 2024; 25(11):5686. https://doi.org/10.3390/ijms25115686
Chicago/Turabian StyleFox, Sarah, Antoine Gaudreau-LaPierre, Ryan Reshke, Irina Podinic, Derrick J. Gibbings, Laura Trinkle-Mulcahy, and John W. Copeland. 2024. "Identification of an FMNL2 Interactome by Quantitative Mass Spectrometry" International Journal of Molecular Sciences 25, no. 11: 5686. https://doi.org/10.3390/ijms25115686
APA StyleFox, S., Gaudreau-LaPierre, A., Reshke, R., Podinic, I., Gibbings, D. J., Trinkle-Mulcahy, L., & Copeland, J. W. (2024). Identification of an FMNL2 Interactome by Quantitative Mass Spectrometry. International Journal of Molecular Sciences, 25(11), 5686. https://doi.org/10.3390/ijms25115686






