Trimeric and Tetrameric Cationic Styryl Dyes as Novel Fluorescence and CD Probes for ds-DNA and ds-RNA
Abstract
:1. Introduction
- (a)
- multiplicity of styryl motif through attaching three or four styryl moieties to one central aromatic core;
- (b)
- type of positive charge by comparing heterocyclic cationic species with exocyclic per-alkylated amines;
- (c)
- steric shielding of positive charge by the introduction of smaller or larger alkylating groups.
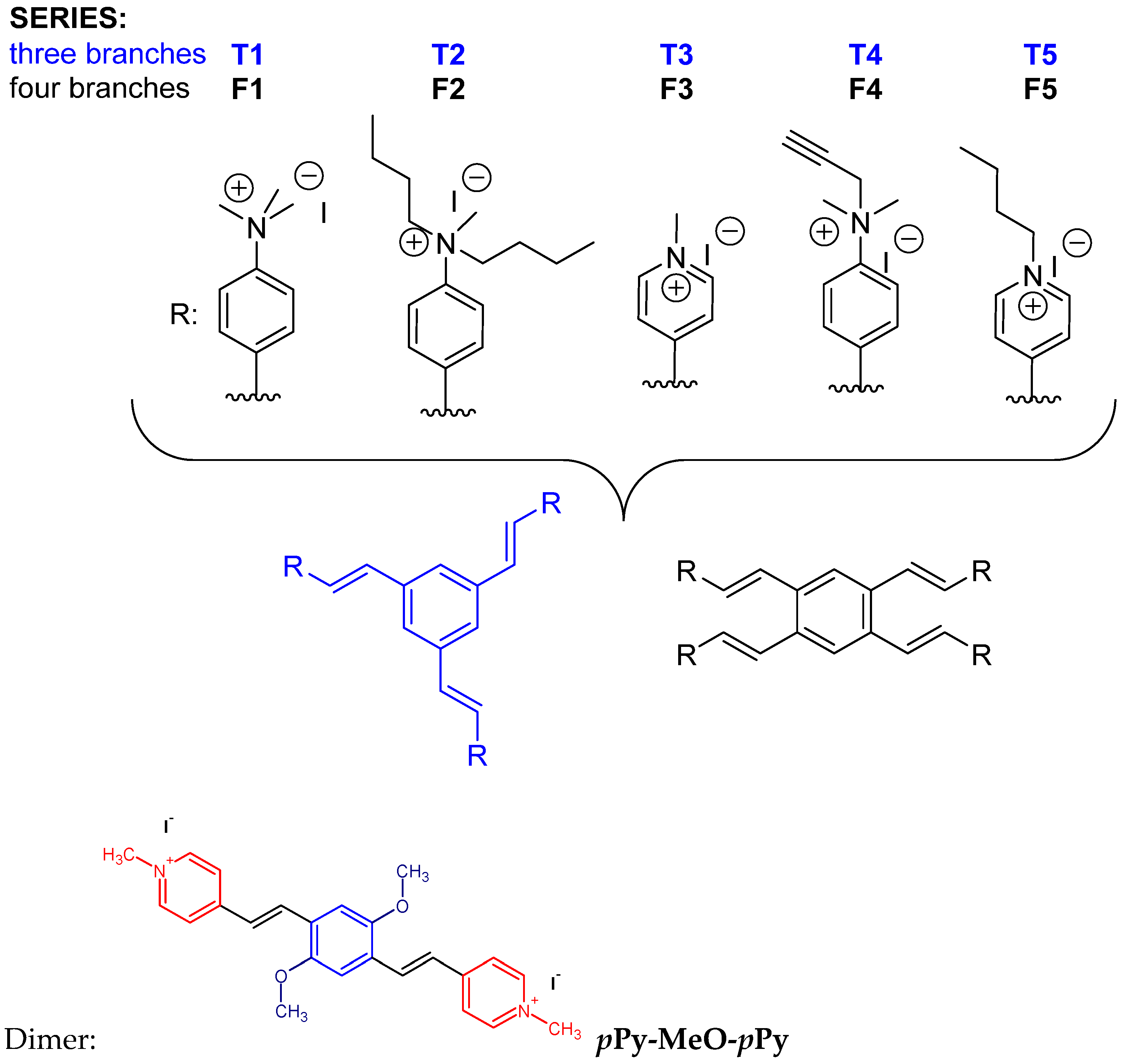
2. Results and Discussion
2.1. Synthesis
2.2. Photophysical Studies of the New Dyes in Aqueous Solutions
2.3. Interactions of New Dyes with ds-DNA and ds-RNA
2.3.1. Thermal Denaturation of ds-DNA/RNA
2.3.2. Spectrophotometric Titrations of the New Dyes with Polynucleotides
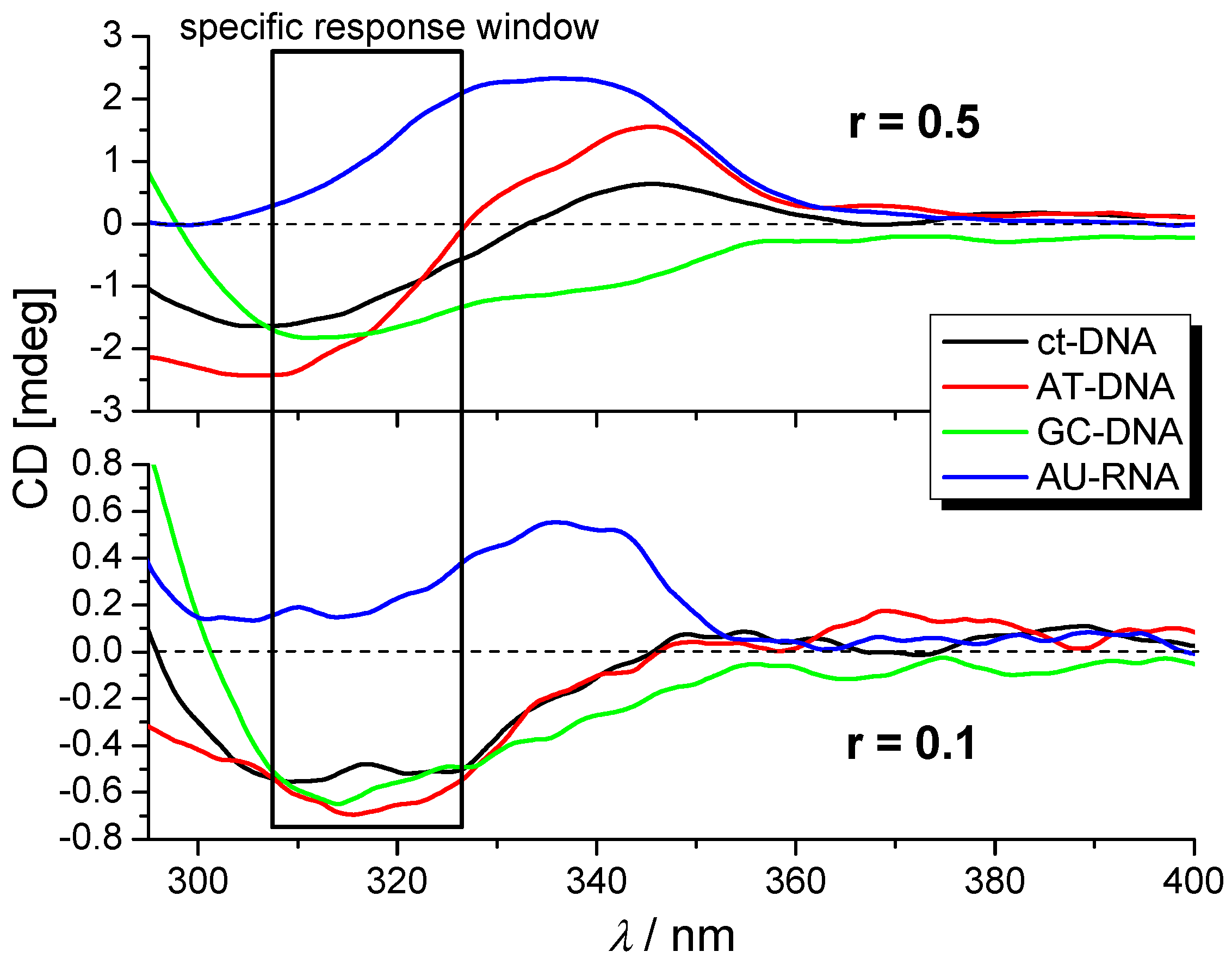
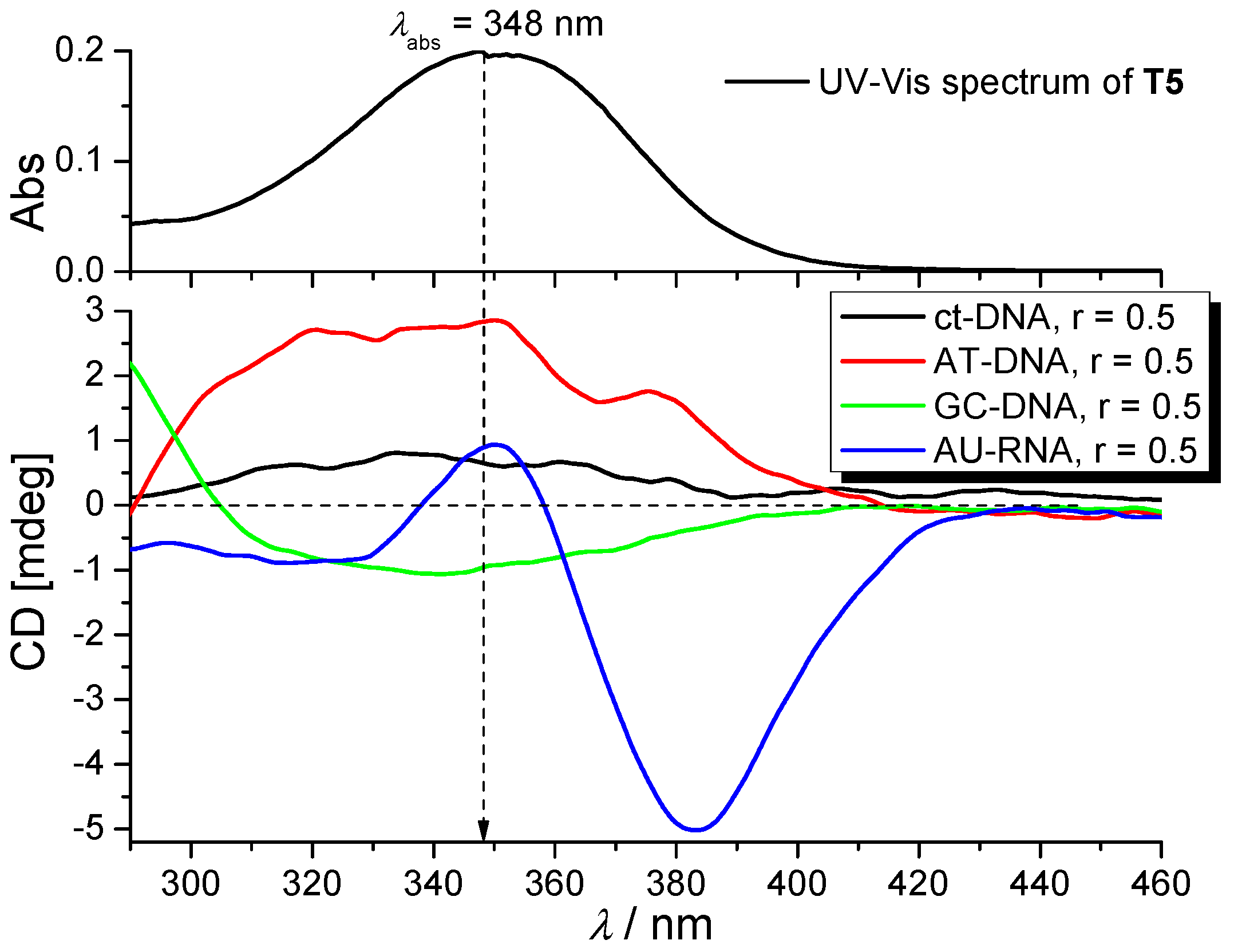
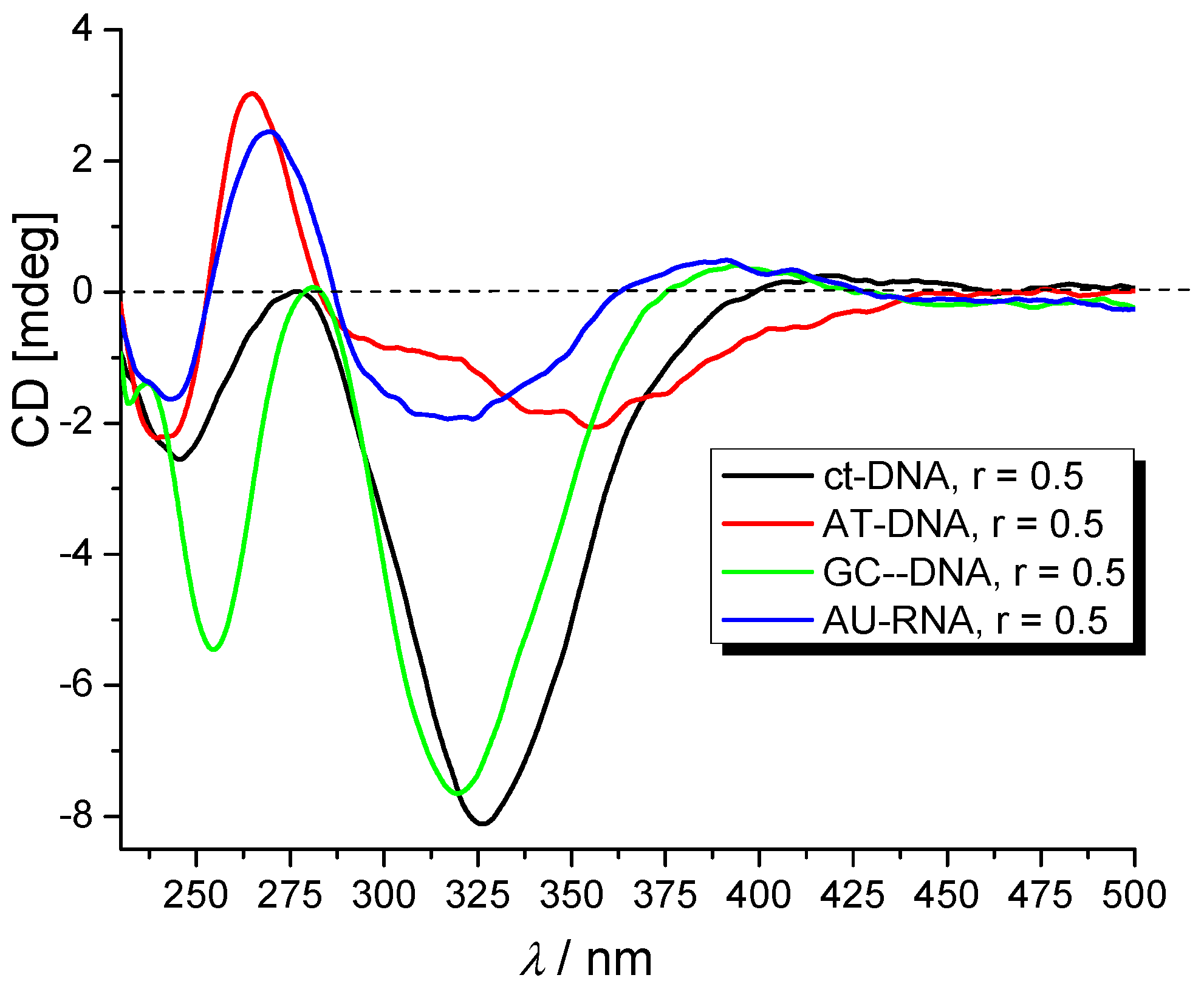
2.3.3. Circular Dichroism (CD) Experiments
2.4. Biological Experiments
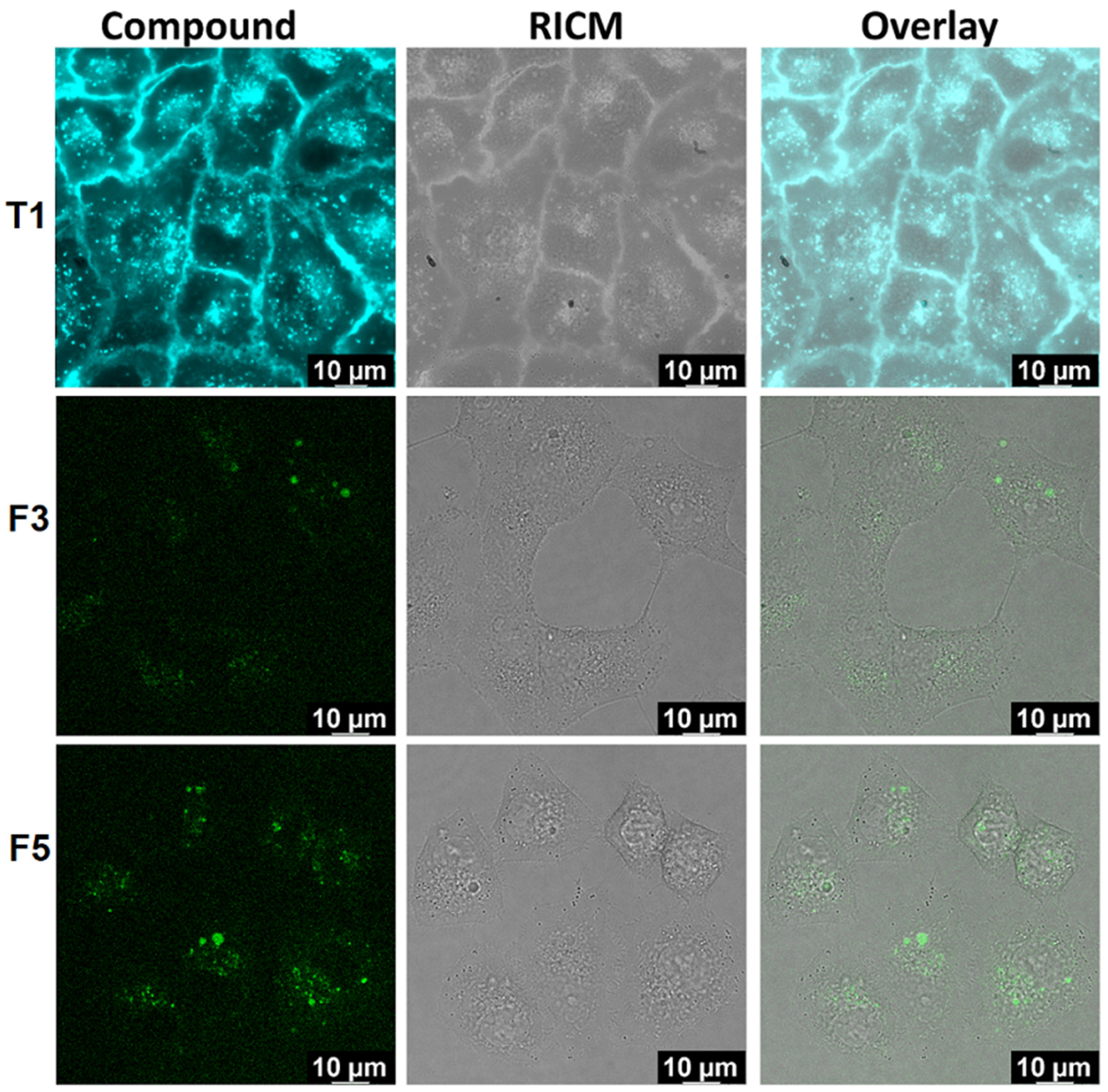
Confocal Microscopy
3. Materials and Methods
3.1. General
3.2. Synthesis
3.3. Spectrophotometric Characterization
3.4. Study of DNA/RNA Interactions
3.5. Biology
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: Boston, MA, USA, 2006. [Google Scholar]
- Yang, X.; Wang, Y.; Liu, R.; Zhang, Y.; Tang, J.; Yang, E.; Zhang, D.; Zhao, Y.; Ye, Y. A novel ICT-based two photon and NIR fluorescent probe for labile Fe2+ detection and cell imaging in living cells. Sens. Actuator B-Chem. 2019, 288, 217–224. [Google Scholar] [CrossRef]
- Wang, L.; Frei, M.S.; Salim, A.; Johnsson, K. Small-Molecule Fluorescent Probes for Live-Cell Super-Resolution Microscopy. J. Am. Chem. Soc. 2019, 141, 2770–2781. [Google Scholar] [CrossRef] [PubMed]
- Ke, C.-S.; Fang, C.-C.; Yan, J.-Y.; Tseng, P.-J.; Pyle, J.R.; Chen, C.-P.; Lin, S.-Y.; Zhang, X.; Chan, Y.-H. Molecular Engineering and Design of Semiconducting Polymer Dots with Narrow-Band, Near-Infrared Emission for in Vivo Biological Imaging. ACS Nano 2017, 11, 3166–3177. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Guo, S.; Hu, C.; Fan, J.; Peng, X. Recent Development of Chemosensors Based on Cyanine Platforms. Chem. Rev. 2016, 116, 7768–7817. [Google Scholar] [CrossRef] [PubMed]
- Nagaya, T.; Nakamura, Y.A.; Choyke, P.L.; Kobayashi, H. Fluorescence-Guided Surgery. Front. Oncol. 2017, 7, 314. [Google Scholar] [CrossRef]
- Ren, H.; Huo, F.; Wu, X.; Liu, X.; Yin, C. An ESIPT-induced NIR fluorescent probe to visualize mitochondrial sulfur dioxide during oxidative stress in vivo. Chem. Commun. 2021, 57, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, A.C.; Wu, L.; Han, H.-H.; Bull, S.D.; He, X.-P.; James, T.D.; Sessler, J.L.; Tang, B.Z.; Tian, H.; Yoon, J. Excited-state intramolecular proton-transfer (ESIPT) based fluorescence sensors and imaging agents. Chem. Soc. Rev. 2018, 47, 8842–8880. [Google Scholar] [CrossRef]
- Dahal, D.; Ojha, K.R.; Pokhrel, S.; Paruchuri, S.; Konopka, M.; Liu, Q.; Pang, Y. NIR-emitting styryl dyes with large Stokes’ shifts for imaging application: From cellular plasma membrane, mitochondria to zebrafish neuromast. Dye. Pigment. 2021, 194, 109629. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Wang, R.; Lv, C.; Chen, G.; You, J.; Yu, F. Detection of Selenocysteine with a Ratiometric near-Infrared Fluorescent Probe in Cells and in Mice Thyroid Diseases Model. Anal. Chem. 2020, 92, 1589–1597. [Google Scholar] [CrossRef]
- Song, X.; Bai, S.; He, N.; Wang, R.; Xing, Y.; Lv, C.; Yu, F. Real-Time Evaluation of Hydrogen Peroxide Injuries in Pulmonary Fibrosis Mice Models with a Mitochondria-Targeted Near-Infrared Fluorescent Probe. ACS Sens. 2021, 6, 1228–1239. [Google Scholar] [CrossRef]
- Abeywickrama, C.S.; Baumann, H.J.; Alexander, N.; Shriver, L.P.; Konopka, M.; Pang, Y. NIR-emitting benzothiazolium cyanines with an enhanced stokes shift for mitochondria imaging in live cells. Org. Biomol. Chem. 2018, 16, 3382–3388. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, L. Fluorescent probes for the selective detection of chemical species inside mitochondria. Chem. Commun. 2016, 52, 1094–1119. [Google Scholar] [CrossRef]
- Dahal, D.; McDonald, L.; Pokhrel, S.; Paruchuri, S.; Konopka, M.; Pang, Y. A NIR-emitting cyanine with large Stokes shifts for live cell imaging: Large impact of the phenol group on emission. Chem. Commun. 2019, 55, 13223–13226. [Google Scholar] [CrossRef]
- Musiol, R.; Serda, M.; Hensel-Bielowka, S.; Polanski, J. Quinoline-Based Antifungals. Curr. Med. Chem. 2010, 17, 1960–1973. [Google Scholar] [CrossRef] [PubMed]
- Quinteros, M.A.; Galera, I.L.D.; Tolosa, J.; García-Martínez, J.C.; Páez, P.L.; Paraje, M.G. Novel antifungal activity of oligostyrylbenzenes compounds on Candida tropicalis biofilms. Med. Mycol. 2021, 59, 244–252. [Google Scholar] [CrossRef]
- Tolosa, J.; De las Heras, G.S.; Carrión, B.; Segura, T.; Páez, P.L.; De Lera-Garrido, F.J.; Rodríguez-López, J.; García-Martínez, J.C. Structure-activity relationships for poly(phenylene)vinylene derivatives as antibacterial agents. ChemistrySelect 2018, 3, 7327–7332. [Google Scholar] [CrossRef]
- Solomon, V.R.; Lee, H. Quinoline as a privileged scaffold in cancer drug discovery. Curr. Med. Chem. 2011, 18, 1488–1508. [Google Scholar] [CrossRef] [PubMed]
- Musiol, R. Quinoline-based HIV integrase inhibitors. Curr. Pharm. Des. 2013, 19, 1835–1849. [Google Scholar] [CrossRef]
- Sánchez-Ruiz, A.; Sousa-Hervés, A.; Pérez-Flores, J.C.; Marín-Rueda, J.R.; Tolosa, J.; Garzón-Ruiz, A.; Rodríguez-López, J.; Canales-Vázquez, J.; García-Martínez, J.C. Inkjet printing of poly(phenylenevinylene)-based fluorophores using an environmentally friendly approach. Prog. Org. Coat. 2022, 166, 106787. [Google Scholar] [CrossRef]
- De Lera-Garrido, F.; Domínguez, R.; Fernández-Liencres, M.P.; Martín, C.; Tolosa, J.; García-Frutos, E.M.; Perles, J.; Hofkens, J.; García-Martínez, J.C.; Garzón-Ruiz, A.; et al. Effect of bulky side groups on photophysical properties and electroluminescent performance of oligo(styryl)benzenes. Dye. Pigment. 2023, 213, 111179. [Google Scholar] [CrossRef]
- Ortiz-Gómez, I.; González-Alfaro, S.; Sánchez-Ruiz, A.; De Orbe-Payá, I.; Capitán-Vallvey, L.F.; Navarro, A.; Salinas-Castillo, A.; García-Martínez, J.C. Reversal of a fluorescent fluoride chemosensor from turn-off to turn-on based on aggregation induced emission properties. ACS Sens. 2022, 7, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Cinar, R.; Nordmann, J.; Dirksen, E.; Müller, T.J.J. Domino synthesis of protochromic “ON–OFF–ON” luminescent 2-styryl quinolines. Org. Biomol. Chem. 2013, 11, 2597–2604. [Google Scholar] [CrossRef] [PubMed]
- Nosova, E.V.; Trashakhova, T.V.; Ustyugov, V.S.; Mochul’skaya, N.N.; Valova, M.S.; Lipunova, G.N.; Charushin, V.N. Fluorine-containing quinoline and quinoxaline styryl derivatives: Synthesis and photophysical properties. Russ. Chem. Bull. 2011, 60, 942–947. [Google Scholar] [CrossRef]
- Gómez-González, J.; Martínez-Castro, L.; Tolosa-Barrilero, J.; Alcalde-Ordóñez, A.; Learte-Aymamí, S.; Mascareñas, J.L.; García-Martínez, J.C.; Martínez-Costas, J.; Maréchal, J.-D.; Vázquez López, M.; et al. Selective recognition of A/T-rich DNA 3-way junctions with a three-fold symmetric tripeptide. Chem. Commun. 2022, 58, 7769–7772. [Google Scholar] [CrossRef] [PubMed]
- Staderini, M.; Aulić, S.; Bartolini, M.; Tran, H.N.A.; González-Ruiz, V.; Pérez, D.I.; Cabezas, N.; Martínez, A.; Martín, M.A.; Andrisano, V.; et al. A Fluorescent styrylquinoline with combined therapeutic and diagnostic activities against Alzheimer’s and prion diseases. ACS Med. Chem. Lett. 2013, 4, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, Y.; Ichimura, C.; Sumiya, S.; Hirai, T. Multicolor fluorescence of a styrylquinoline dye tuned by metal cations. Chem.-Eur. J. 2011, 17, 8324–8332. [Google Scholar] [CrossRef] [PubMed]
- Botti, V.; Cesaretti, A.; Ban, Ž.; Crnolatac, I.; Consiglio, G.; Elisei, F.; Piantanida, I. Fine structural tuning of styryl-based dyes for fluorescence and CD-based sensing of various ds-DNA/RNA sequences. Org. Biomol. Chem. 2019, 17, 8243–8258. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Xu, D.; Sun, A.Y.; Ho, S.L.; Poon, C.Y.; Chan, H.N.; Ng, O.T.W.; Yung, K.K.L.; Yan, H.; Li, H.W.; et al. Fluoro-substituted cyanine for reliable in vivo labelling of amyloid-beta oligomers and neuroprotection against amyloid-beta induced toxicity. Chem. Sci. 2017, 8, 8279–8284. [Google Scholar] [CrossRef] [PubMed]
- Čipor, I.; Kurutos, A.; Dobrikov, G.M.; Kamounah, F.S.; Majhen, D.; Nestić, D.; Piantanida, I. Structure-dependent mitochondria or lysosome-targeting styryl fluorophores bearing remarkable Stokes shift. Dye. Pigment. 2022, 206, 110626. [Google Scholar] [CrossRef]
- Cantor, C.R.; Schimmel, P.R. Biophysical Chemistry; WH Freeman and Co.: San Francisco, CA, USA, 1980; Volume 3. [Google Scholar]
- Egli, M.; Saenger, W. Principles of Nucleic Acid Structure; Springer: New York, NY, USA, 1983. [Google Scholar]
- Sánchez-Ruiz, A.; Rodríguez-López, J.; Garzón-Ruiz, A.; Jiménez-Pulido, S.B.; Illán-Cabeza, N.A.; Navarro, A.; García-Martínez, J.C. Shedding light on the origin of solid-state luminescence enhancementin butterfly Molecules. Chem.-Eur. J. 2020, 26, 13990–14001. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Y.; Ma, C.; Liu, W.; Li, Y.; Li, L. Naphthalene-fused BODIPY with large Stokes shift as saturated-red fluorescent dye for living cell imaging. Dye. Pigment. 2015, 122, 1–5. [Google Scholar] [CrossRef]
- Zheng, Q.; Xu, G.; Prasad, P.N. Conformationally restricted dipyrromethene boron difluoride (BODIPY) dyes: Highly fluorescent, multicolored probes for cellular imaging. Chem.-Eur. J. 2008, 14, 5812–5819. [Google Scholar] [CrossRef]
- Wilson, W.D.; Ratmeyer, L.; Zhao, M.; Strekowski, L.; Boykin, D. The search for structure-specific nucleic acid-interactive drugs: Effects of compound structure on RNA versus DNA interaction strength. Biochemistry 1993, 32, 4098–4104. [Google Scholar] [CrossRef] [PubMed]
- McGhee, J.D.; von Hippel, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef] [PubMed]
- Scatchard, G. The attractions of proteins for small molecules an ions. Ann. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- Gore, M.G. Spectrophotometry and Spectrofluorimetry. A Practical Approach; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Rodger, A.; Norden, B. Circular Dichroism and Linear Dichroism; Oxford University Press: New York, NY, USA, 1997. [Google Scholar]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids—Tutorial. Beilstein J. Org. Chem. 2018, 14, 84–105. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.; Norden, B. Linear and Circular Dichroism of Drug-Nucleic Acid Complexes. Methods Enzymol. 2001, 340, 68–98. [Google Scholar] [CrossRef] [PubMed]
- Demeunynck, M.; Bailly, C.; Wilson, W.D. Small Molecule DNA and RNA Binders: From Synthesis to Nucleic Acid Complexes; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2004. [Google Scholar]
- Winter, A.; Friebe, C.; Hager, M.D.; Schubert, U.S. Synthesis of Rigid π-Conjugated Mono-, Bis-, Tris-, and Tetrakis(terpyridine)s: Influence of the Degree and Pattern of Substitution on the Photophysical Properties. Eur. J. Org. Chem. 2009, 2009, 801–809. [Google Scholar] [CrossRef]
- Mergny, J.L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Ranjbar, B.; Gill, P. Circular Dichroism Techniques: Biomolecular and Nanostructural Analyses—A Review. Chem. Biol. Drug Des. 2009, 74, 101–120. [Google Scholar] [CrossRef] [PubMed]
- Górecki, M.; Zinna, F.; Biver, T.; Di Bari, L.J. Induced circularly polarized luminescence for revealing DNA binding with fluorescent dyes. Pharm. Biomed. Anal. 2017, 144, 6–11. [Google Scholar] [CrossRef] [PubMed]




| Compd. | maxλabs/[nm] (ε × 103/[M−1 cm−1]) | b Φf | a λexc [nm] | λem [nm] | a τ [ns] | χ2 |
|---|---|---|---|---|---|---|
| T1 | 314 (91.5) | 0.27 ± 0.05 | 340 | 412 | 10.68 (14.04%) 20.33 (85.96%) | 0.960 |
| T2 | 315 (80.0) | 0.41 ± 0.05 | 340 | 480 | 13.05 (33.74%) 21.50 (66.26%) | 0.918 |
| T3 | 348 (88.8) | 0.04 ± 0.05 | 340 | 454 | 0.78 (90.42%) 5.03 (9.58%) | 1.055 |
| T4 | 314 (86.3) | 0.37 ± 0.05 | 340 | 411 | 10.67 (18.28%) 20.24 (81.72%) | 0.931 |
| T5 | 348 (79.5) | 0.29 ± 0.05 | 340 | 448 | 0.69 (98.06%) 4.55 (1.94%) | 1.062 |
| F1 | 333 (75.4) | 0.57 ± 0.05 | 340 | 460 | 2.89 (21.82%) 7.03 (78.18%) | 0.941 |
| F2 | 335 (66.5) | 0.20 ± 0.05 | 340 | 460 | 2.55 (20.77%) 6.69 (79.23%) | 1.050 |
| F3 | 366 (83.0) | 0.07 ± 0.05 | 340 | 485 | 0.64 (59.91%) 3.56 (40.09%) | 0.963 |
| F4 | 334 (60.6) | 0.35 ± 0.05 | 340 | 460 | 2.60 (28.08%) 6.58 (71.92%) | 0.985 |
| F5 | 366 (53.8) | 0.06 ± 0.05 | 340 | 485 | 0.68 (55.54%) 3.63 (44.46%) | 0.906 |
| c dimer | 446 (35.0) | 0.21 ± 0.05 | 457 | 605 | 0.19 (21%) 1.60 (79%) | 0.98 < χ2 < 1.02 |
| Compd. | ctDNA | pApU |
|---|---|---|
| T1 | +11.9 | +23.2 |
| T2 | +6.2 | +5.1 |
| T3 | >+15.0 | +8.5 |
| T4 | +12.0 | +18.1 |
| T5 | +14.3 | +1.5 |
| F1 | >+15.0 | +12.5 |
| F2 | 0 | 0 |
| F3 | +6.6 | +6.4 |
| F4 | +4.8 | +10.4 |
| F5 | +3.2 | +1.7 |
| b dimer | +3.1 | +1.8 |
| Compd. | ctDNA | pApU |
|---|---|---|
| T1 | 8.6 (0.23) | 8.9 (0.30) |
| T2 | 8.6 (0.04) | 8.9 (0.16) |
| T3 | 8.2 (0.24) | 8.7 (0.13) |
| T4 | 8.9 (0.10) | 8.6 (0.35) |
| T5 | 8.9 (0.10) | 8.7 (0.14) |
| F1 | 9.3 (0.17) | 8.6 (0.17) |
| F2 | 8.0 (0.09) | 8.6 (0.08) |
| F3 | 7.1 (0.11) | 8.2 (0.28) |
| F4 | 8.6 (0.17) | 8.9 (0.17) |
| F5 | 8.9 (0.30) | 7.6 (0.26) |
| c dimer | 6.9 (0.80) | 7.5 (0.5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlović Saftić, D.; Krošl Knežević, I.; de Lera Garrido, F.; Tolosa, J.; Majhen, D.; Piantanida, I.; García Martínez, J.C. Trimeric and Tetrameric Cationic Styryl Dyes as Novel Fluorescence and CD Probes for ds-DNA and ds-RNA. Int. J. Mol. Sci. 2024, 25, 5724. https://doi.org/10.3390/ijms25115724
Pavlović Saftić D, Krošl Knežević I, de Lera Garrido F, Tolosa J, Majhen D, Piantanida I, García Martínez JC. Trimeric and Tetrameric Cationic Styryl Dyes as Novel Fluorescence and CD Probes for ds-DNA and ds-RNA. International Journal of Molecular Sciences. 2024; 25(11):5724. https://doi.org/10.3390/ijms25115724
Chicago/Turabian StylePavlović Saftić, Dijana, Ivona Krošl Knežević, Fernando de Lera Garrido, Juan Tolosa, Dragomira Majhen, Ivo Piantanida, and Joaquín Calixto García Martínez. 2024. "Trimeric and Tetrameric Cationic Styryl Dyes as Novel Fluorescence and CD Probes for ds-DNA and ds-RNA" International Journal of Molecular Sciences 25, no. 11: 5724. https://doi.org/10.3390/ijms25115724






