Participation of miR165a in the Phytochrome Signal Transduction in Maize (Zea mays L.) Leaves under Changing Light Conditions
Abstract
:1. Introduction
2. Results
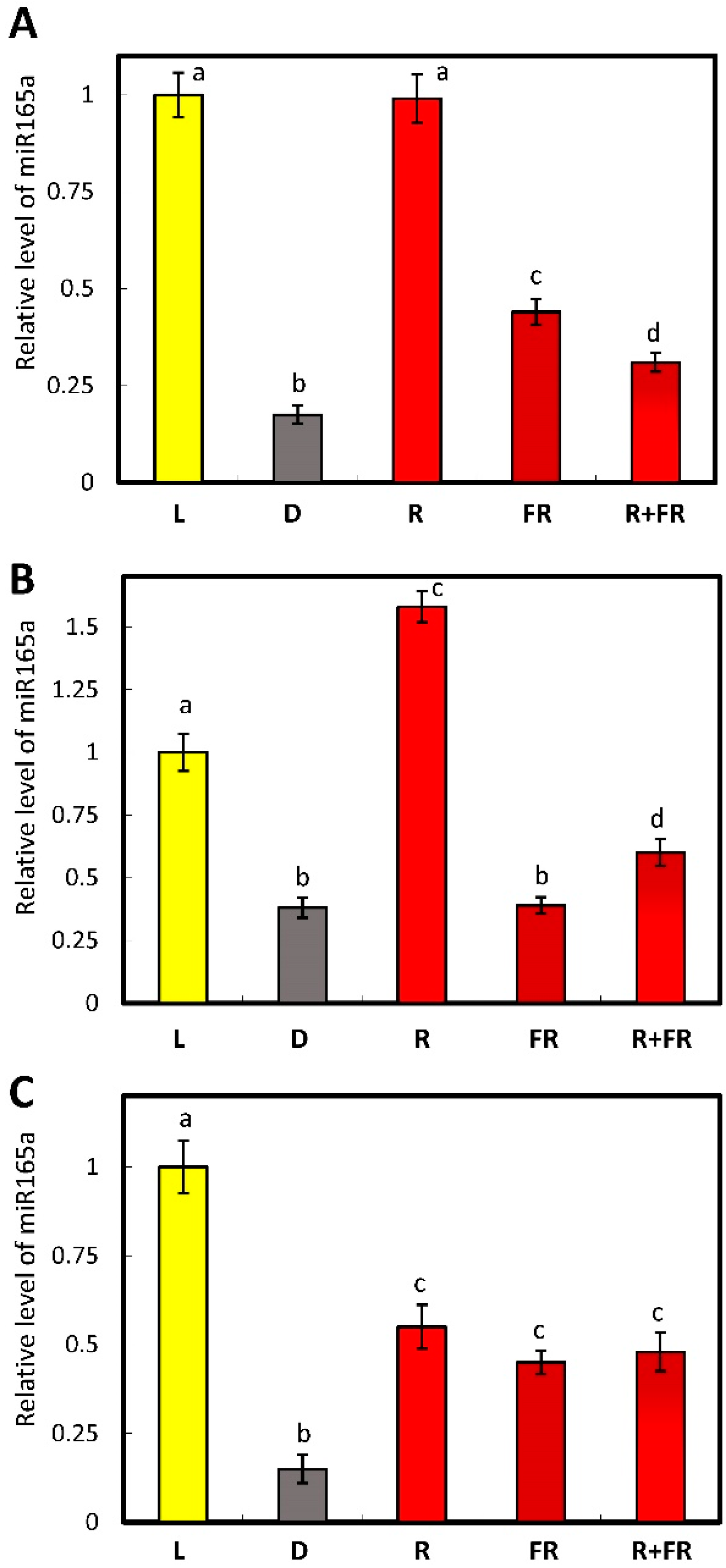
2.1. Estimation of the Amount of miR165a and of the Expression of the Genes Encoding the Proteins Regulating the Level of Micrornas in Maize Leaves upon Irradiation with Light of Different Wavelengths
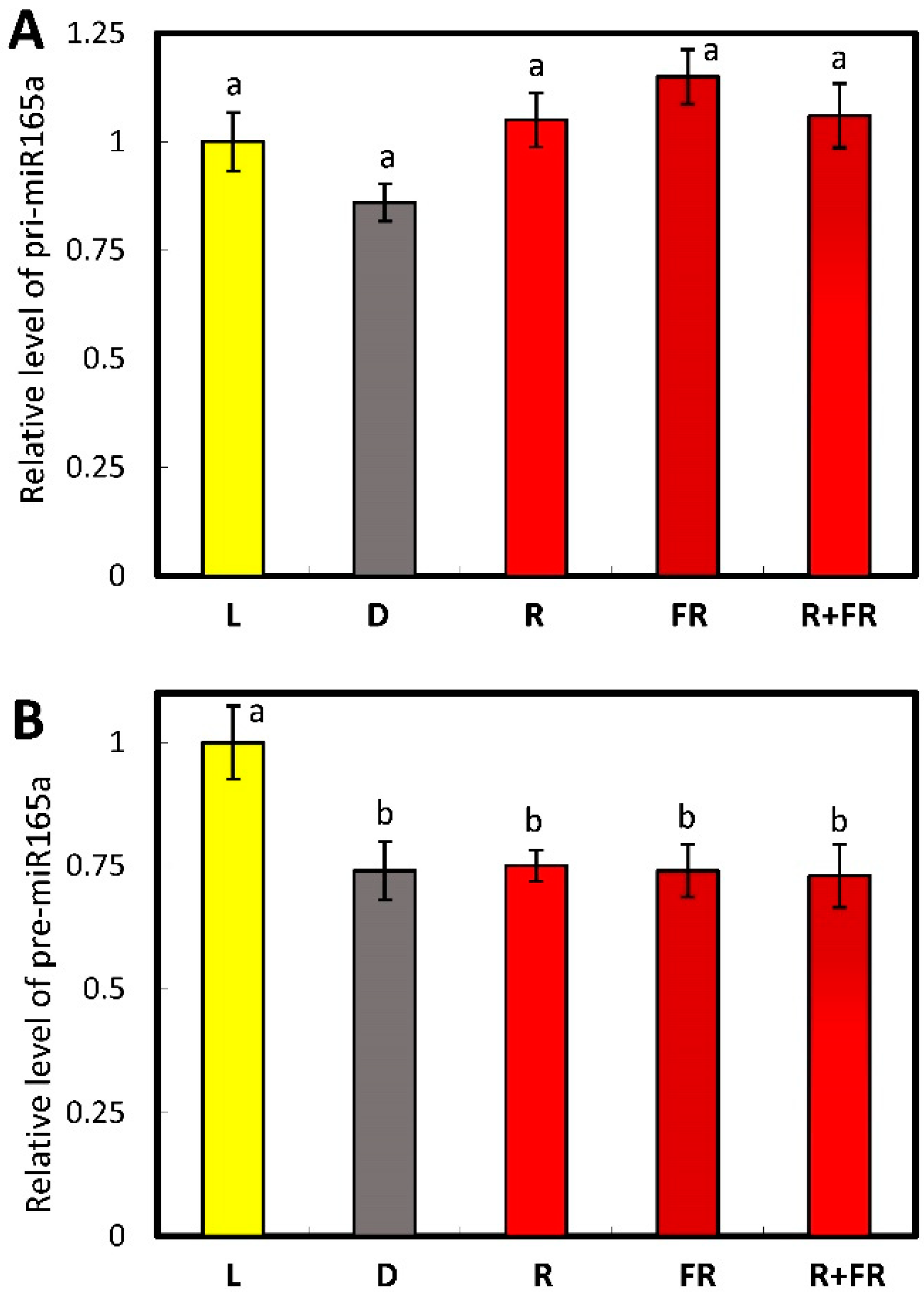
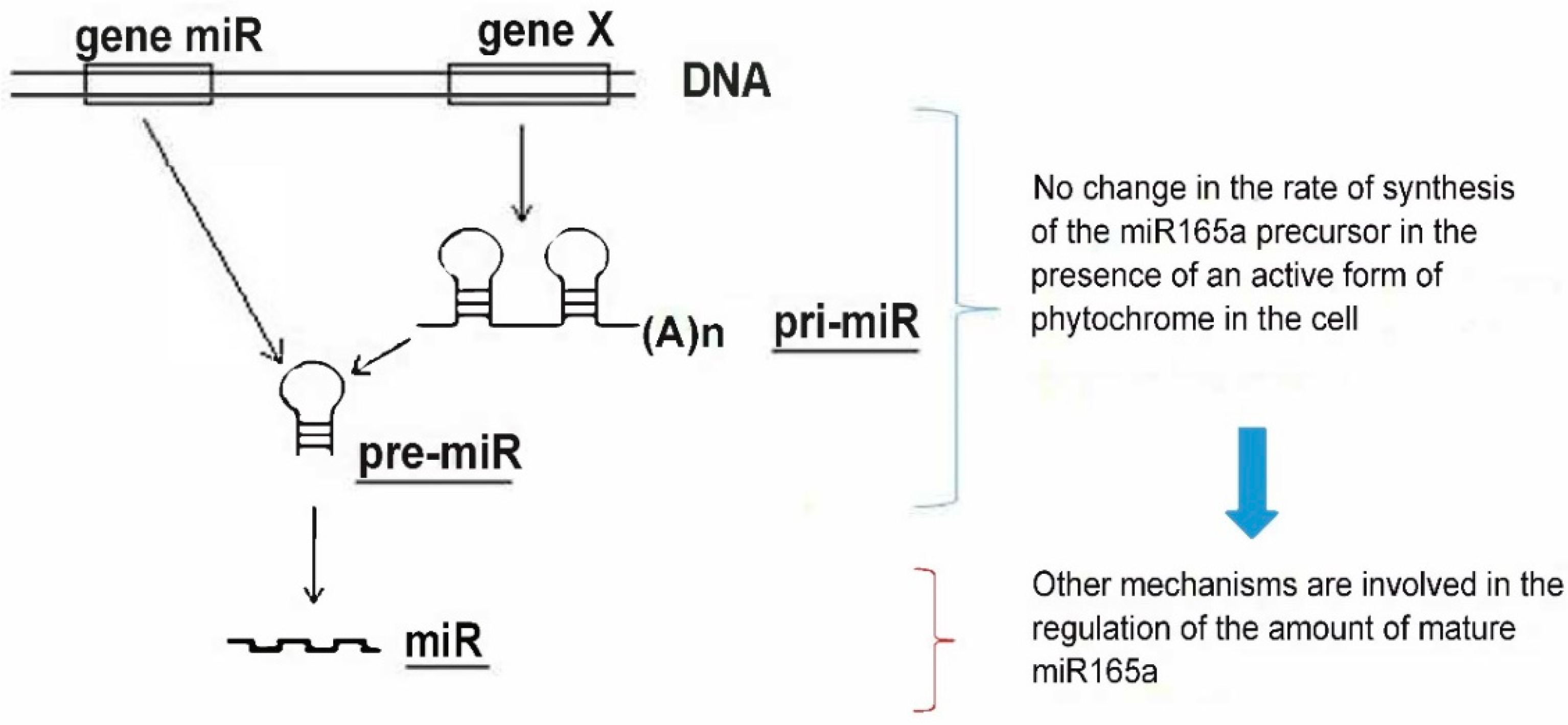
2.2. Evaluation of miR165a Precursors in Maize Leaves upon Irradiation of Plants with Light of Different Wavelengths
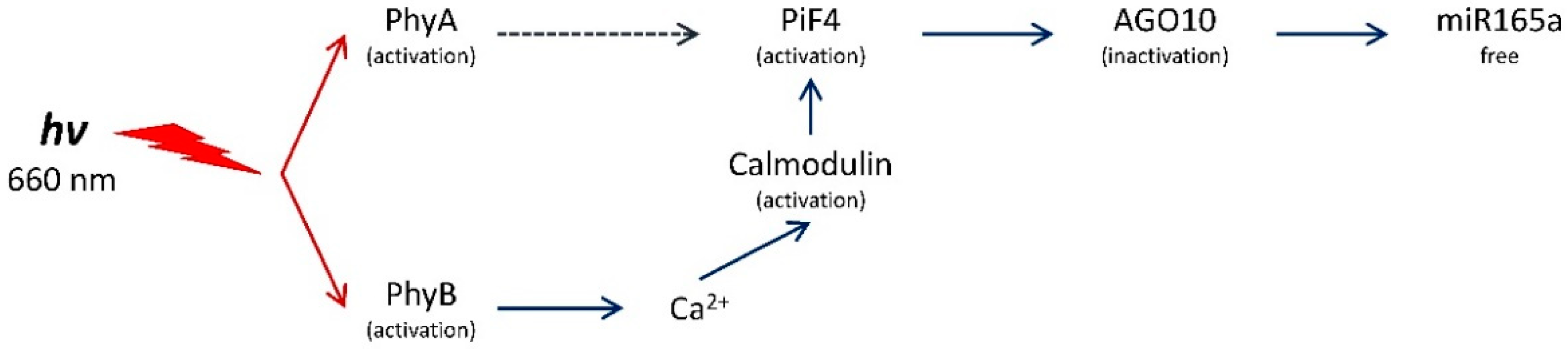
2.3. Determination of the Role of Phytochromes A and B in the Regulation of the Content of Free miR165a in Arabidopsis Leaves
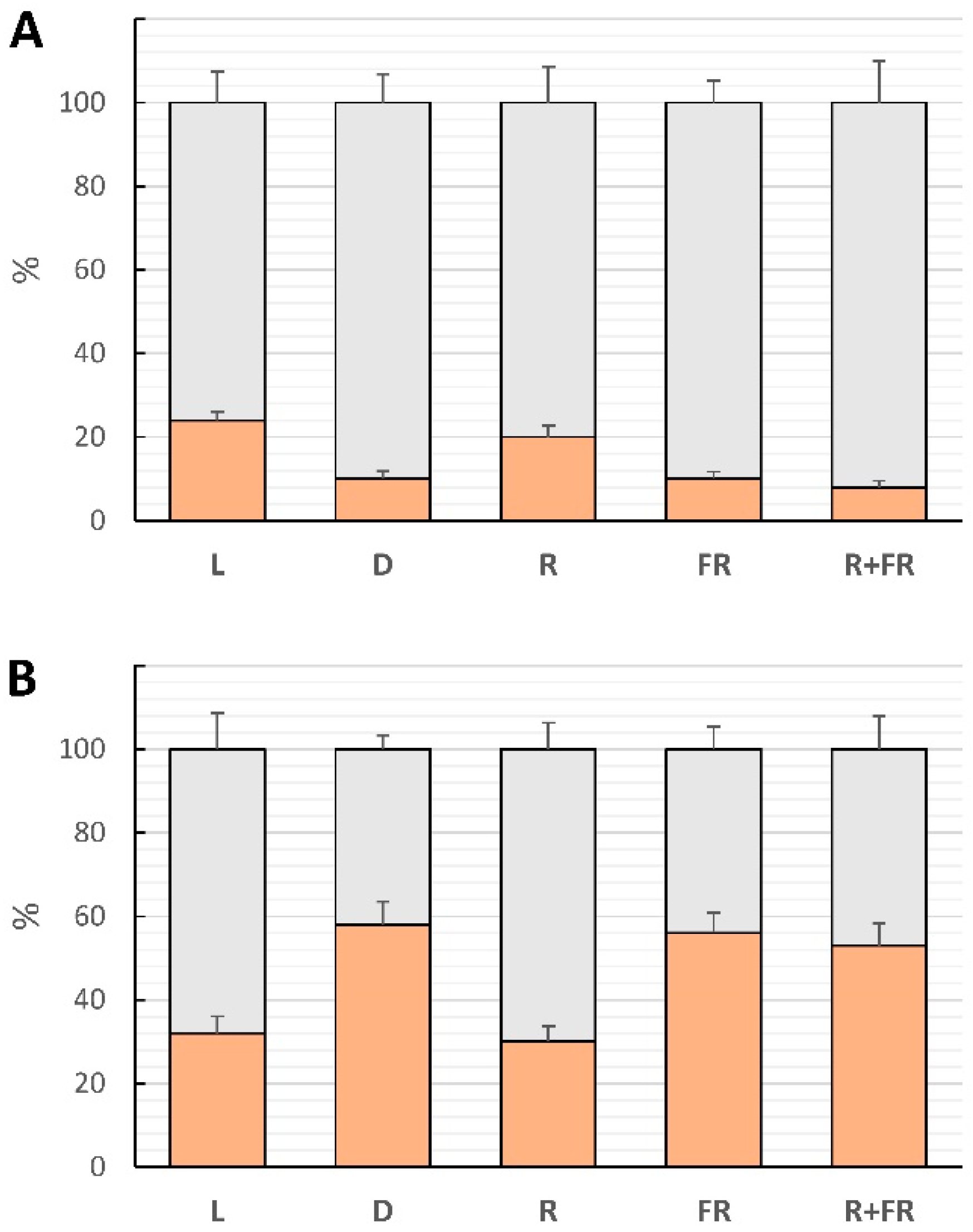
2.4. Involvement of miR165a in Phytochrome-Dependent RNA Interference
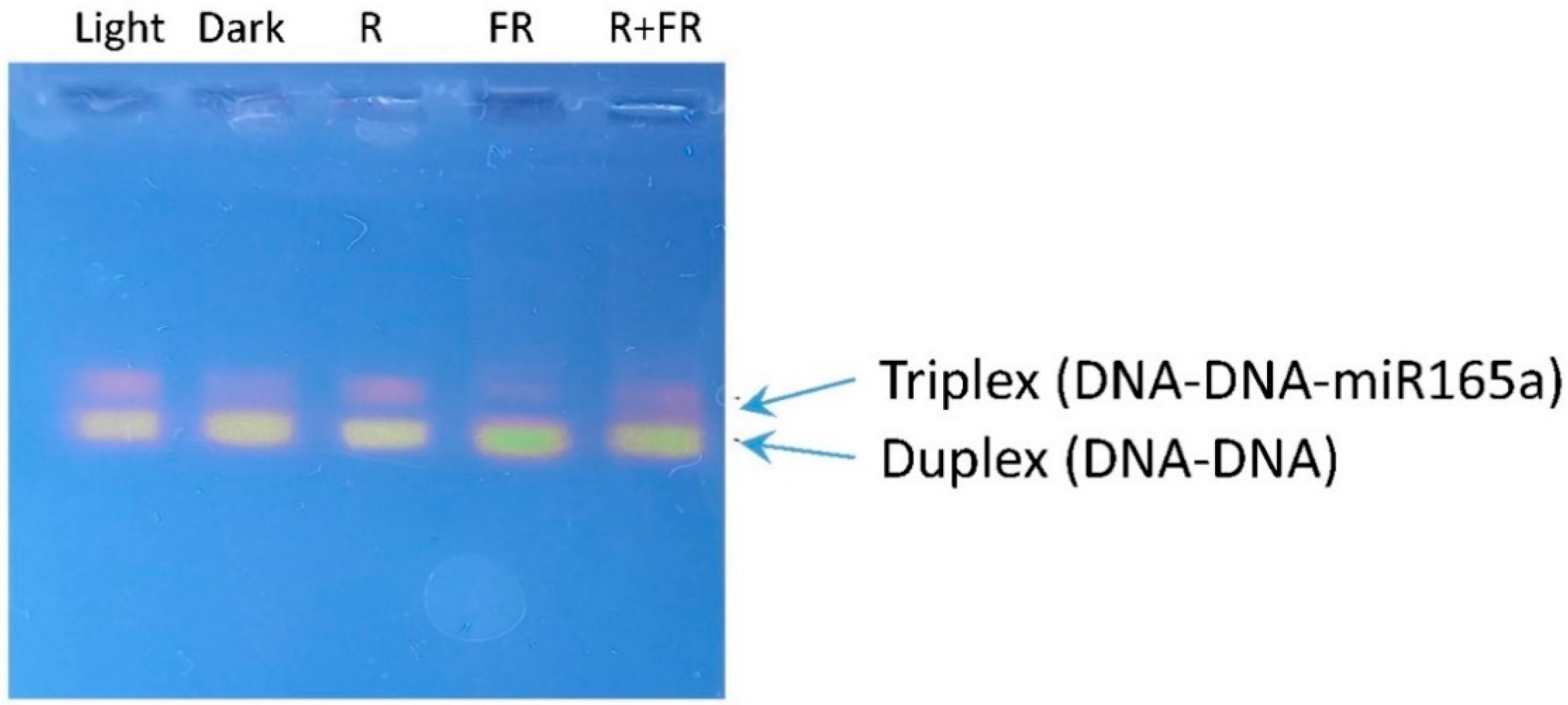
2.5. Involvement of miR165a in RNA-Dependent DNA Methylation
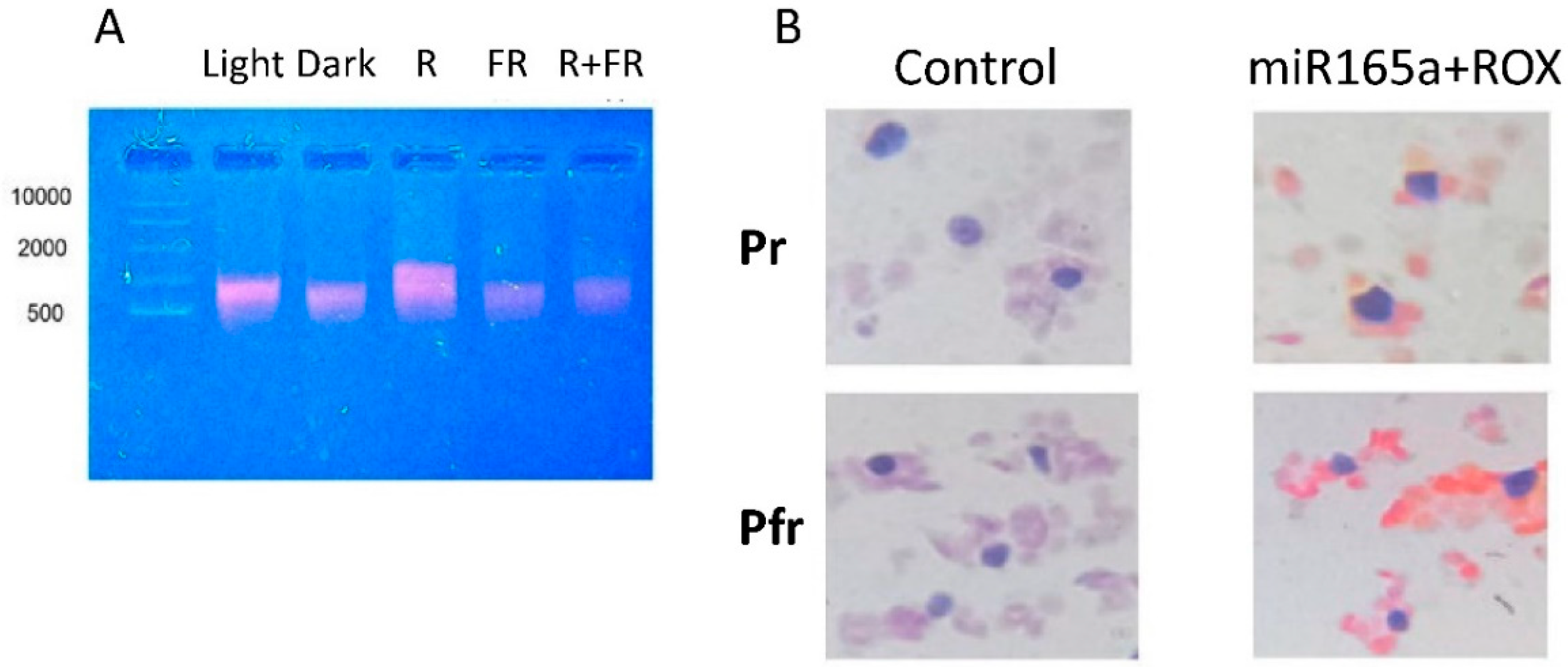
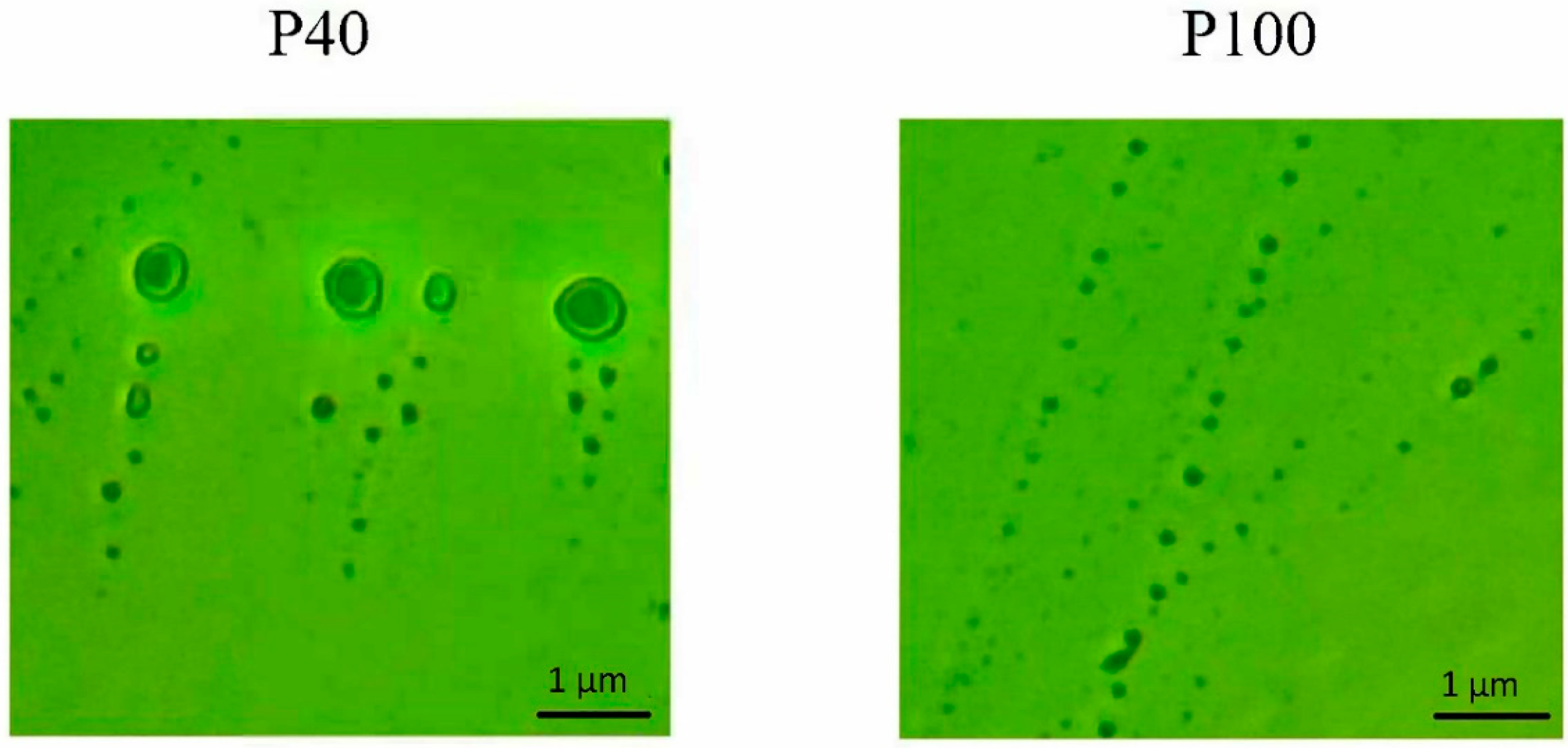
2.6. Participation of Extracellular Vesicles in miR165a Transport
3. Discussion
4. Material and Methods
4.1. Object of Investigation
4.2. RNA Isolation and Reverse Transcription
4.3. Polymerase Chain Reaction
4.4. Analysis of RNA Interference and DNA Methylation
4.5. Subcellular Distribution of miRNA
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AGO1 | ARGONAUTE1 |
| AGO10 | ARGONAUTE10 |
| miRNA | microRNA |
| PhyA | phytochrome A |
| PhyB | phytochrome B |
| PIF | phytochrome-interacting factor |
| pri-miR | initially transcribed long primary transcripts from the precursor gene of the corresponding mi-croRNA |
| pre-miR | hairpin-shaped precursor processed into microRNA |
| RdDM | RNA-directed DNA methylation |
| TF | transcription factor |
| TCA cycle | tricarboxylic acid cycle |
References
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.C.; Jones, P.A. Epigenetics and microRNAs. Pediatr. Res. 2007, 61, 24R–29R. [Google Scholar] [CrossRef] [PubMed]
- Pashkovskiy, P.; Kreslavski, V.; Khudyakova, A.; Pojidaeva, E.S.; Kosobryukhov, A.; Kuznetsov, V.; Allakhverdiev, S.I. Independent Responses of Photosynthesis and Plant Morphology to Alterations of PIF Proteins and Light-Dependent MicroRNA Contents in Arabidopsis thaliana pif Mutants Grown under Lights of Different Spectral Compositions. Cells 2022, 11, 3981. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Li, M.; Zhou, Y.; Guo, T.; Liu, Y.; Zhang, H.; Fang, Y. Coordinated regulation of Arabidopsis microRNA biogenesis and red light signaling through Dicer-like 1 and phytochrome-interacting factor 4. PLoS Genet. 2018, 14, e1007247. [Google Scholar] [CrossRef] [PubMed]
- Huq, E.; Quail, P.H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002, 21, 2441–2450. [Google Scholar] [CrossRef] [PubMed]
- Jang, I.C.; Henriques, R.; Seo, H.S.; Nagatani, A.; Chua, N.H. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 2010, 22, 2370–2383. [Google Scholar] [CrossRef]
- Zhang, H.; He, H.; Wang, X.; Wang, X.; Yang, X.; Li, L.; Deng, X.W. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011, 65, 346–358. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Kibet, N.K.; Song, C.; Zhang, C.; Li, X.; Han, J.; Fang, J. Deep sequencing of grapevine flower and berry short RNA library for discovery of novel microRNAs and validation of precise sequences of grapevine microRNAs deposited in miRBase. Physiol. Plant. 2011, 143, 64–81. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xu, X.H.; Wu, X.; Wang, Y.; Lu, X.; Sun, H.; Xie, X. Genome-wide identification of microRNAs and their targets in wild type and phyB mutant provides a key link between microRNAs and the phyB-mediated light signaling pathway in rice. Front. Plant Sci. 2015, 6, 372. [Google Scholar] [CrossRef]
- Gulyás, Z.; Székely, A.; Kulman, K.; Kocsy, G. Light-Dependent Regulatory Interactions between the Redox System and miRNAs and Their Biochemical and Physiological Effects in Plants. Int. J. Mol. Sci. 2023, 24, 8323. [Google Scholar] [CrossRef] [PubMed]
- Skopelitis, D.S.; Hill, K.; Klesen, S.; Marco, C.F.; von Born, P.; Chitwood, D.H.; Timmermans, M.C.P. Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat. Commun. 2018, 9, 3107. [Google Scholar] [CrossRef] [PubMed]
- López de Las Hazas, M.C.; Tomé-Carneiro, J.; Del Pozo-Acebo, L.; Del Saz-Lara, A.; Chapado, L.A.; Balaguer, L.; Rojo, E.; Espín, J.C.; Crespo, C.; Moreno, D.A.; et al. Therapeutic potential of plant-derived extracellular vesicles as nanocarriers for exogenous miRNAs. Pharmacol. Res. 2023, 198, 106999. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Pontes, O.; Searle, I.; Yelina, N.; Yousafzai, F.K.; Herr, A.J.; Pikaard, C.S.; Baulcombe, D.C. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 2007, 19, 1507–1521. [Google Scholar] [CrossRef] [PubMed]
- Levine, A. Regulation of stress responses by intracellular vesicle trafficking? Plant Physiol. Biochem. 2002, 40, 531–535. [Google Scholar] [CrossRef]
- Yao, X.; Wang, H.; Li, H.; Yuan, Z.; Li, F.; Yang, L.; Huang, H. Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes. FEBS Lett. 2009, 583, 3711–3717. [Google Scholar] [CrossRef]
- McConnell, J.R.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001, 411, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.J.; Valdés, A.E.; Wang, G.; Ramachandran, P.; Beste, L.; Uddenberg, D.; Carlsbecker, A. PHABULOSA Mediates an Auxin Signaling Loop to Regulate Vascular Patterning in Arabidopsis. Plant Physiol. 2016, 170, 956–970. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, Y.; Zhang, J.; Zhu, C.; Tang, G.; Yan, J. The miR165/166-PHABULOSA module promotes thermotolerance by transcriptionally and posttranslationally regulating HSFA1. Plant Cell 2023, 35, 2952–2971. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, J.; Ryu, K.H.; Zhou, J.; Tarkowská, D.; Tarkowski, P.; Cho, Y.H.; Yoo, S.D.; Kim, E.S.; Lee, J.Y. PHABULOSA controls the quiescent center-independent root meristem activities in Arabidopsis thaliana. PLoS Genet. 2015, 11, e1004973. [Google Scholar] [CrossRef] [PubMed]
- Kidner, C.A.; Martienssen, R.A. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 2004, 428, 81–84. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.H. Regulation of HD-ZIP III Genes by MicroRNA 165. Plant Signal. Behav. 2007, 2, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.K.; Kubo, M.; Zhong, R.; Demura, T.; Ye, Z.H. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007, 48, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, H.; Srivastava, A.K.; Pan, Y.; Bai, J.; Fang, J.; Shi, H.; Zhu, J.K. Knockdown of Rice MicroRNA166 Confers Drought Resistance by Causing Leaf Rolling and Altering Stem Xylem Development. Plant Physiol. 2018, 176, 2082–2094. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yang, T.; Guo, Z.; Wang, Q.; Chai, M.; Wu, M.; Li, X.; Li, W.; Li, G.; Tang, J.; et al. Maize microRNA166 Inactivation Confers Plant Development and Abiotic Stress Resistance. Int. J. Mol. Sci. 2020, 21, 9506. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, J.; Watanabe, Y. miR165⁄166 and the development of land plants. Dev. Growth Differ. 2012, 54, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, J.; Cheng, J.; Jiang, Z.; Xu, N.; An, X.; Chen, Z.; Hao, J.; Yang, S.; Xu, Z.; et al. Identification of UV-B radiation responsive microRNAs and their target genes in chrysanthemum (Chrysanthemum morifolium Ramat) using high-throughput sequencing. Industr. Crop. Prod. 2020, 151, 112484. [Google Scholar] [CrossRef]
- Malitsky, S.; Blum, E.; Less, H.; Venger, I.; Elbaz, M.; Morin, S.; Eshed, Y.; Aharoni, A. The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 2008, 148, 2021–2049. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhao, C.; Zhou, J.; Yang, Y.; Wang, P.; Zhu, X.; Tang, G.; Bressan, R.A.; Zhu, J.K. The miR165/166 Mediated Regulatory Module Plays Critical Roles in ABA Homeostasis and Response in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006416. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Ding, N.; Fan, W.; Yan, J.; Gu, Y.; Tang, X.; Li, R.; Tang, G. Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. 2015, 233, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Wang, Y.; Teotia, S.; Wang, Z.; Shi, C.; Sun, H.; Gu, Y.; Zhang, Z.; Tang, G. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci. Rep. 2019, 9, 2832. [Google Scholar] [CrossRef] [PubMed]
- Popov, V.N.; Eprintsev, A.T.; Fedorin, D.N.; Igamberdiev, A.U. Succinate dehydrogenase in Arabidopsis thaliana is regulated by light via phytochrome A. FEBS Lett. 2010, 584, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Eprintsev, A.T.; Fedorin, D.N.; Igamberdiev, A.U. Ca²⁺ is involved in phytochrome A-dependent regulation of the succinate dehydrogenase gene sdh1-2 in Arabidopsis. J. Plant Physiol. 2013, 170, 1349–1352. [Google Scholar] [CrossRef] [PubMed]
- Eprintsev, A.T.; Fedorin, D.N.; Sazonova, O.V. Phytochrome-dependent regulation of fumarate hydratase activity in maize green leaves. Russ. J. Plant Physiol. 2015, 62, 441–447. [Google Scholar] [CrossRef]
- Eprintsev, A.T.; Fedorin, D.N.; Sazonova, O.V.; Igamberdiev, A.U. Light inhibition of fumarase in Arabidopsis leaves is phytochrome A-dependent and mediated by calcium. Plant Physiol. Biochem. 2016, 102, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Eprintsev, A.T.; Fedorin, D.N.; Igamberdiev, A.U. Light-Dependent Expression and Promoter Methylation of the Genes Encoding Succinate Dehydrogenase, Fumarase, and NAD-Malate Dehydrogenase in Maize (Zea mays L.) Leaves. Int. J. Mol. Sci. 2023, 24, 10211. [Google Scholar] [CrossRef] [PubMed]
- Eprintsev, A.T.; Fedorin, D.N.; Karabutova, L.A.; Igamberdiev, A.U. Expression of genes encoding subunits A and B of succinate dehydrogenase in germinating maize seeds is regulated by methylation of their promoters. J. Plant Physiol. 2016, 205, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Eprintsev, A.T.; Fedorin, D.N.; Igamberdiev, A.U. Light Dependent Changes in Adenylate Methylation of the Promoter of the Mitochondrial Citrate Synthase Gene in Maize (Zea mays L.) Leaves. Int. J. Mol. Sci. 2022, 23, 13495. [Google Scholar] [CrossRef] [PubMed]
- Fedorin, D.N.; Eprintsev, A.T.; Igamberdiev, A.U. The role of promoter methylation of the genes encoding the enzymes metabolizing di- and tricarboxylic acids in the regulation of plant respiration by light. J. Plant Physiol. 2024, 294, 154195. [Google Scholar] [CrossRef]
- Quail, P.H. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 2002, 3, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Eprintsev, A.T.; Fedorin, D.N.; Popov, V.N. Phytochrome-mediated regulation of plant respiration and photorespiration. Plant Cell Environ. 2014, 37, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Hoang, Q.T.N.; Han, Y.J.; Kim, J.I. Regulation of Photomorphogenic Development by Plant Phytochromes. Int. J. Mol. Sci. 2019, 20, 6165. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Sun, G.; Liu, F.; Hu, W. Functions of Plant Phytochrome Signaling Pathways in Adaptation to Diverse Stresses. Int. J. Mol. Sci. 2023, 24, 13201. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.C.; Kathare, P.K.; Paik, I.; Huq, E. Phytochrome Signaling Networks. Annu. Rev. Plant Biol. 2021, 72, 217–244. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Ro, S.; Yan, W. In situ hybridization detection of microRNAs. Methods Mol. Biol. 2010, 629, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.N.; Srivastava, D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 2010, 7, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell. Pharmacol. 2011, 3, 83–92. [Google Scholar] [PubMed]
- Xu, Y.; Zhu, Z. PIF4 and PIF4-Interacting Proteins: At the Nexus of Plant Light, Temperature and Hormone Signal Integrations. Int. J. Mol. Sci. 2021, 22, 10304. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ji, L.; Le, B.H.; Zhai, J.; Chen, J.; Luscher, E.; Gao, L.; Liu, C.; Cao, X.; Mo, B.; et al. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 2017, 15, e2001272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Fan, L.; Le, B.H.; Ye, P.; Mo, B.; Chen, X. Regulation of ARGONAUTE10 Expression Enables Temporal and Spatial Precision in Axillary Meristem Initiation in Arabidopsis. Dev. Cell 2020, 55, 603–616.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.H.; Liou, L.W.; Barefoot, A.; Dickman, M.; Zhang, X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, A.; Nagel, M.K.; Popp, C.; Wüst, F.; Bindics, J.; Viczián, A.; Hiltbrunner, A.; Nagy, F.; Kunkel, T.; Schäfer, E. Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl Acad. Sci. USA 2012, 109, 5892–5897. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shi, H.; Pan, Y.; Lyu, M.; Yang, Z.; Kou, X.; Deng, X.W.; Zhong, S. Sensory circuitry controls cytosolic calcium-mediated phytochrome B phototransduction. Cell 2023, 186, 1230–1243.e14. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef] [PubMed]
- Bernardo-García, S.; de Lucas, M.; Martínez, C.; Espinosa-Ruiz, A.; Davière, J.M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Swulius, M.T.; Waxham, M.N. Ca2+/calmodulin-dependent protein kinases. Cell. Mol. Life Sci. 2008, 65, 2637–2657. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, A.; Kunkel, T.; Hiltbrunner, A.; Neuhaus, G.; Wolf, I.; Speth, V.; Adam, E.; Nagy, F.; Schäfer, E. A cell-free system for light-dependent nuclear import of phytochrome. Plant J. 2009, 57, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Hoang, Q.T.N.; Han, Y.J.; Kim, J.I. Plant Phytochromes and their Phosphorylation. Int. J. Mol. Sci. 2019, 20, 3450. [Google Scholar] [CrossRef]
- Choi, D.M.; Kim, S.H.; Han, Y.J.; Kim, J.I. Regulation of Plant Photoresponses by Protein Kinase Activity of Phytochrome A. Int. J. Mol. Sci. 2023, 24, 2110. [Google Scholar] [CrossRef]
- Sun, Q.; Wang, S.; Xu, G.; Kang, X.; Zhang, M.; Ni, M. SHB1 and CCA1 interaction desensitizes light responses and enhances thermomorphogenesis. Nat. Commun. 2019, 10, 3110. [Google Scholar] [CrossRef] [PubMed]
- Schalch, T.; Job, G.; Shanker, S.; Partridge, J.F.; Joshua-Tor, L. The Chp1-Tas3 core is a multifunctional platform critical for gene silencing by RITS. Nat. Struct. Mol. Biol. 2011, 18, 1351–1357. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Paugh, S.W.; Coss, D.R.; Bao, J.; Laudermilk, L.T.; Grace, C.R.; Ferreira, A.M.; Waddell, M.B.; Ridout, G.; Naeve, D.; Leuze, M.; et al. MicroRNAs Form Triplexes with Double Stranded DNA at Sequence-Specific Binding Sites; a Eukaryotic Mechanism via which microRNAs Could Directly Alter Gene Expression. PLoS Comput. Biol. 2016, 12, e1004744. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Liu, C.; Qi, Y. DNA methylation mediated by a microRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; He, B.; Wang, S.; Fletcher, S.; Niu, D.; Mitter, N.; Birch, P.R.J.; Jin, H. Message in a Bubble: Shuttling Small RNAs and Proteins Between Cells and Interacting Organisms Using Extracellular Vesicles. Annu. Rev. Plant Biol. 2021, 72, 497–524. [Google Scholar] [CrossRef]
- Kameli, N.; Dragojlovic-Kerkache, A.; Savelkoul, P.; Stassen, F.R. Plant-Derived Extracellular Vesicles: Current Findings, Challenges, and Future Applications. Membranes 2021, 11, 411. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 2021, 7, 342–352. [Google Scholar] [CrossRef]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [PubMed]
- Fedorin, D.N.; Chuykova, V.O.; Eprintsev, A.T. Modification of the Method for Isolating MicroRNA from Plants by Phenol–Chloroform Extraction Using Polyethylene Glycol 1500. Biochem. Suppl. Ser. B 2023, 17, 26–30. [Google Scholar] [CrossRef]
- Kramer, M.F. Stem-loop RT-qPCR for miRNAs. Curr. Protoc. Mol. Biol. 2011, 95, 15.10.1–15.10.15. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, S.; Cai, Q.; Jin, H. Effective methods for isolation and purification of extracellular vesicles from plants. J. Integr. Plant Biol. 2021, 63, 2020–2030. [Google Scholar] [CrossRef] [PubMed]
- Bao, N.; Lye, K.W.; Barton, M.K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 2004, 7, 653–662. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedorin, D.N.; Eprintsev, A.T.; Chuykova, V.O.; Igamberdiev, A.U. Participation of miR165a in the Phytochrome Signal Transduction in Maize (Zea mays L.) Leaves under Changing Light Conditions. Int. J. Mol. Sci. 2024, 25, 5733. https://doi.org/10.3390/ijms25115733
Fedorin DN, Eprintsev AT, Chuykova VO, Igamberdiev AU. Participation of miR165a in the Phytochrome Signal Transduction in Maize (Zea mays L.) Leaves under Changing Light Conditions. International Journal of Molecular Sciences. 2024; 25(11):5733. https://doi.org/10.3390/ijms25115733
Chicago/Turabian StyleFedorin, Dmitry N., Alexander T. Eprintsev, Victoria O. Chuykova, and Abir U. Igamberdiev. 2024. "Participation of miR165a in the Phytochrome Signal Transduction in Maize (Zea mays L.) Leaves under Changing Light Conditions" International Journal of Molecular Sciences 25, no. 11: 5733. https://doi.org/10.3390/ijms25115733




