Longitudinal Neuropathological Consequences of Extracranial Radiation Therapy in Mice
Abstract
1. Introduction
2. Results
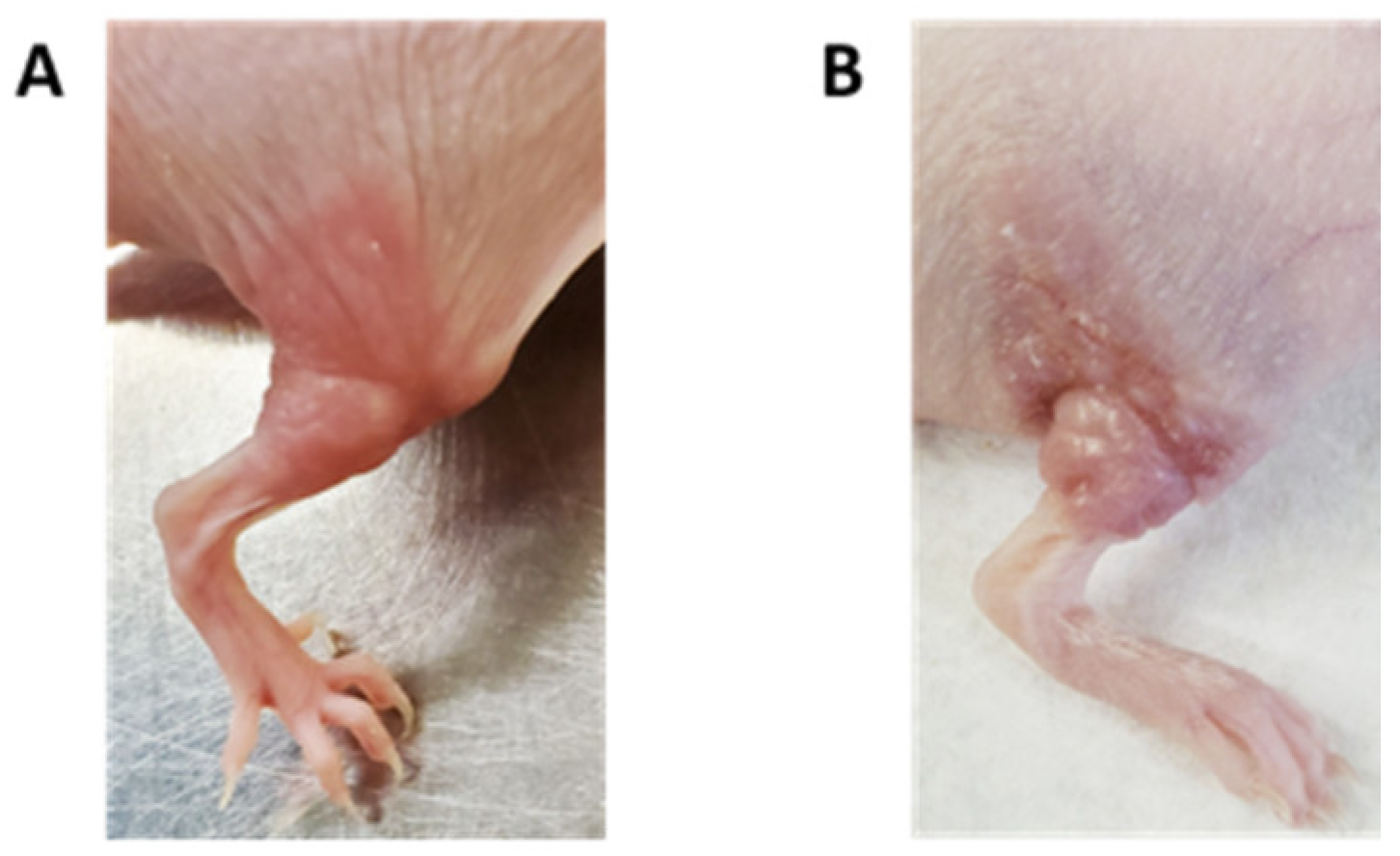
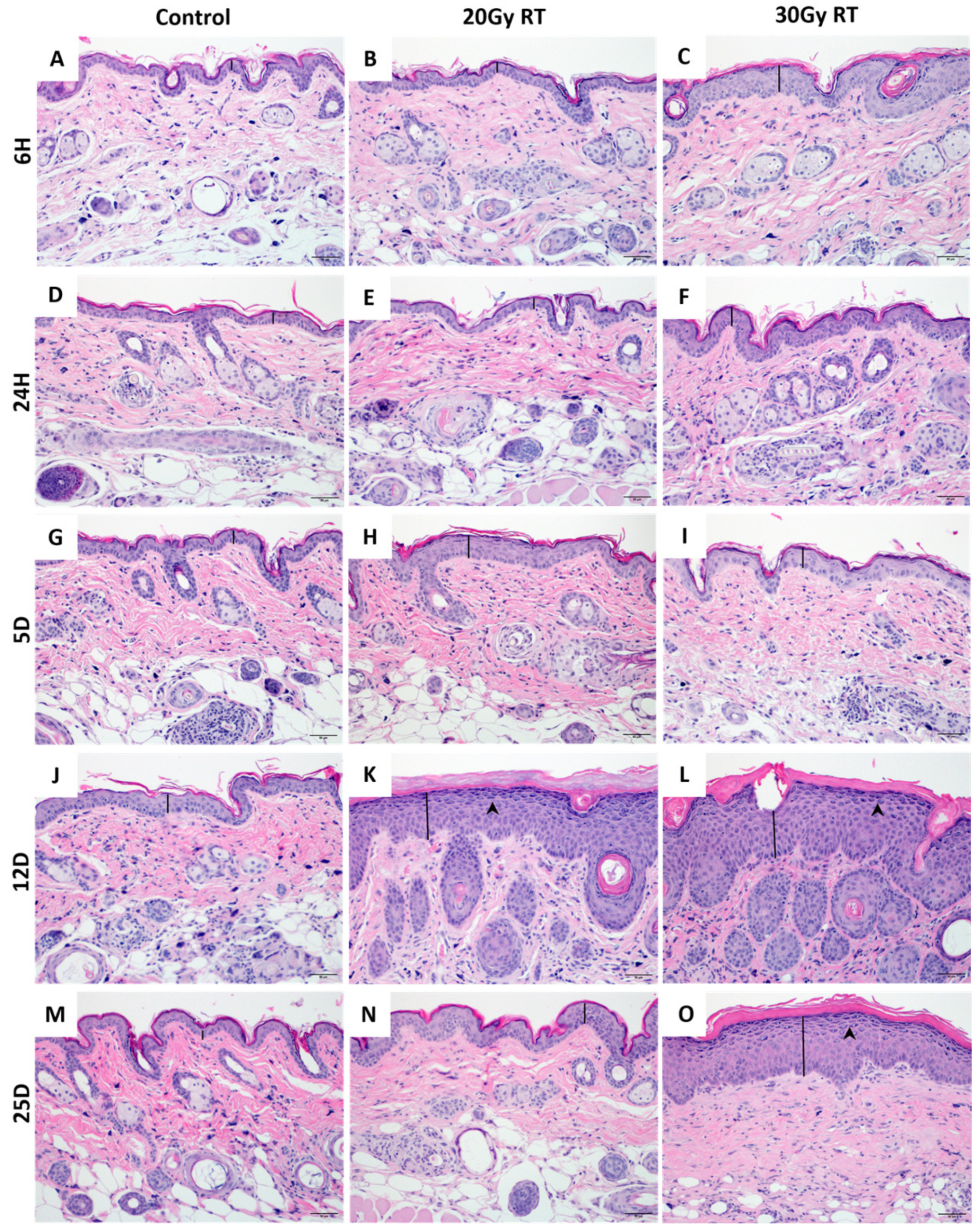
2.1. Extracranial Radiation Therapy Causes Acute Toxicity in the Skin of Mice
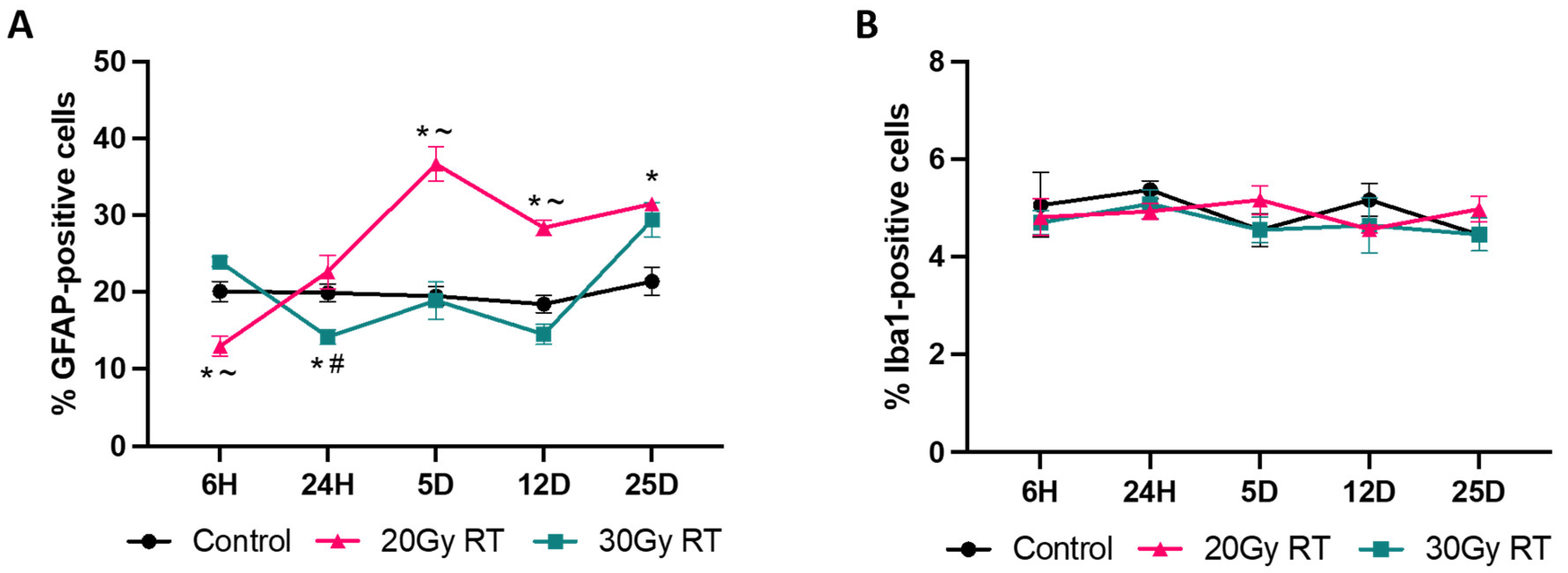
2.2. Extracranial Radiation Increases Activation of Astrocytes in the Thoracic Spinal Cord of Mice
2.3. Extracranial Causes Significant Glial Cell Activation in the Striatum
2.4. Extracranial Causes Significant Glial Cell Activation in the Retrosplenial Cortex
2.5. Extracranial Causes Significant Glial Cell Activation in the Hippocampus
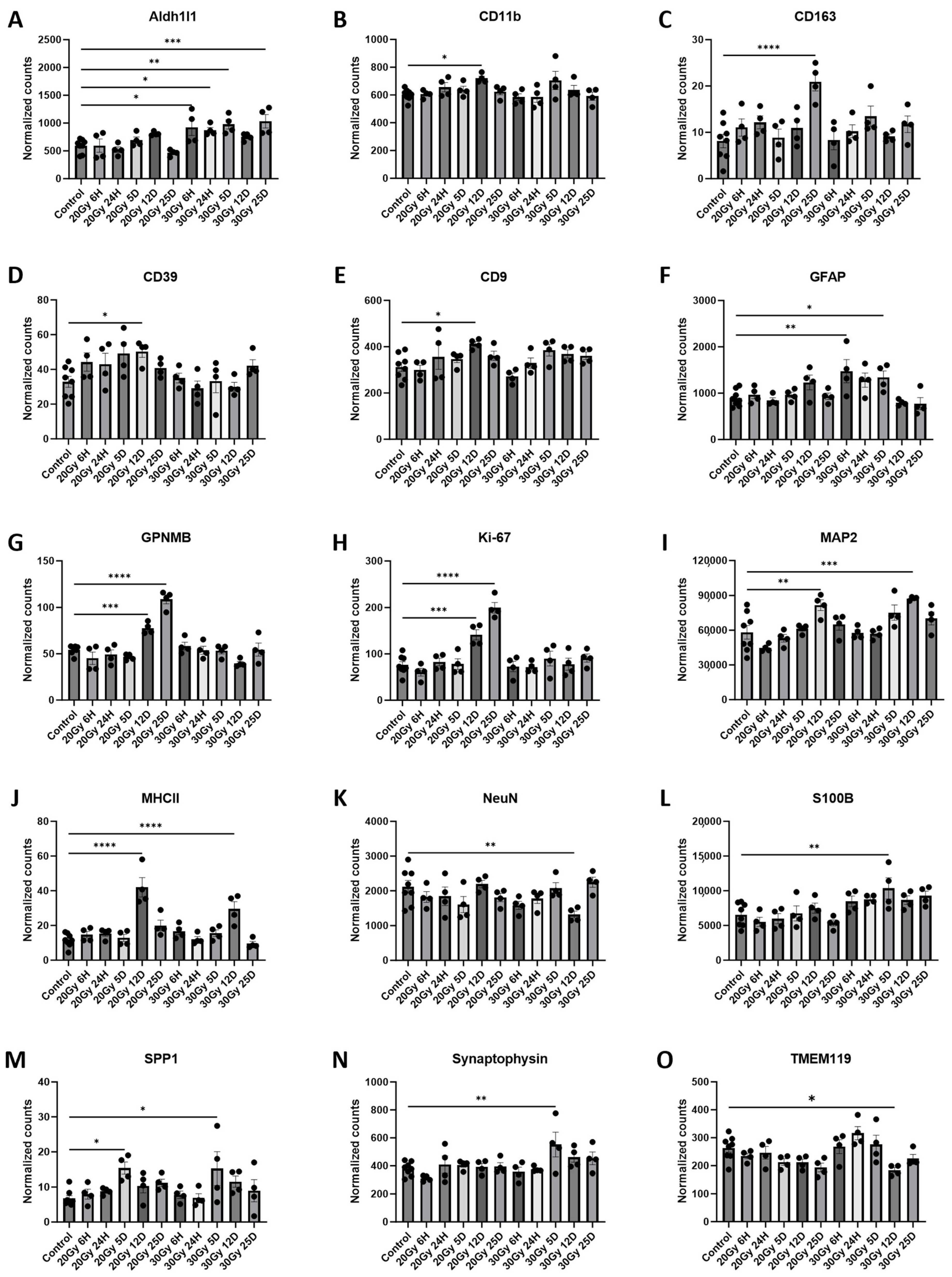
2.6. Extracranial Radiation Causes Molecular Changes in the Normal Brain
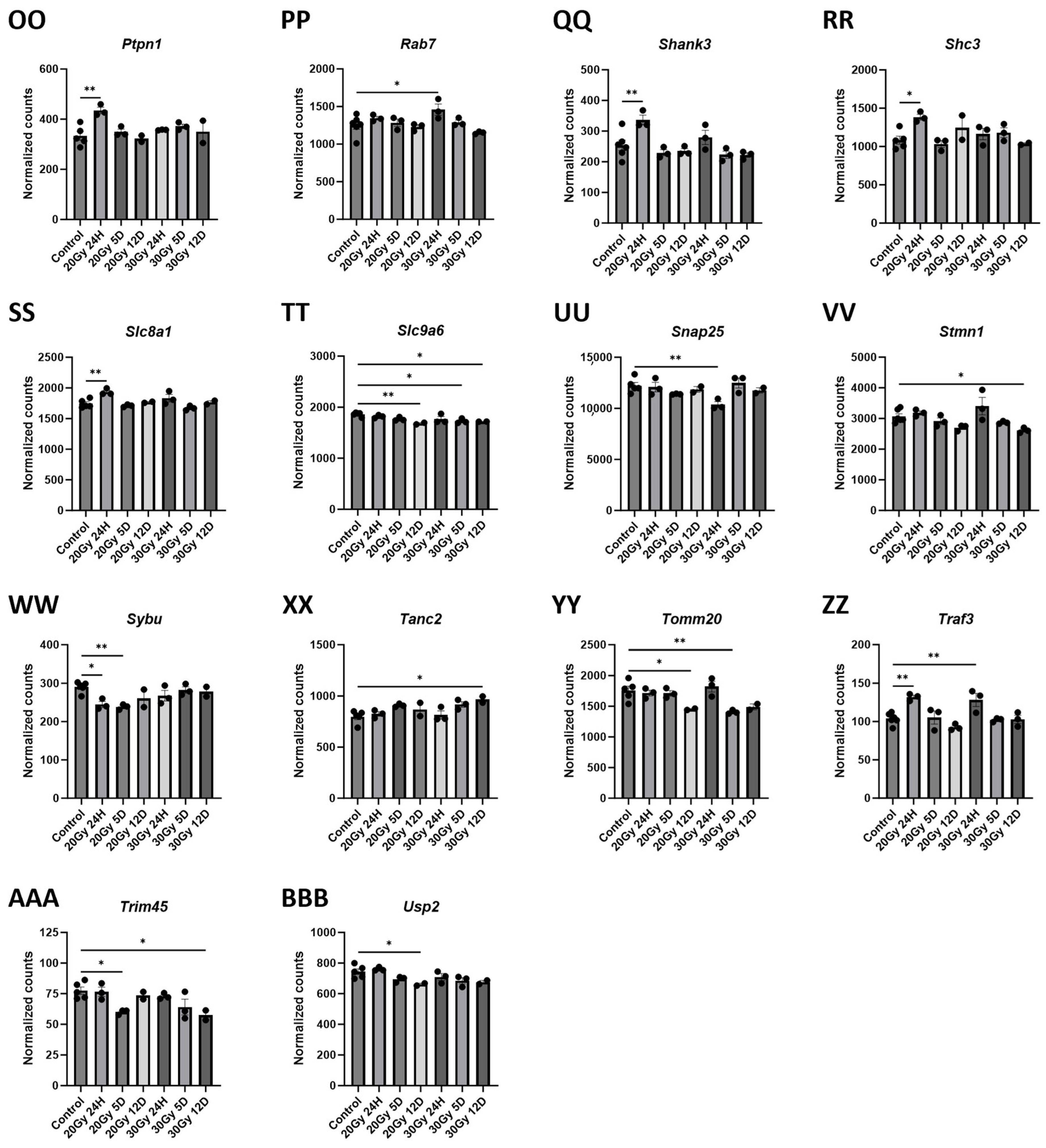
2.7. Gene Changes Caused by Extracranial Radiation Therapy Are Associated with Neuronal Transmission
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. Radiation Treatment
4.3. Hematoxylin and Eosin (H&E) Stain
4.4. Immunohistochemistry (IHC)
4.5. IHC Analysis
4.6. Tissue Preparation for Nanostring GeoMx Assay
4.7. Probe Incubation and Morphology Marker Staining
4.8. GeoMx DSP AOI Selection and Barcode Collection
4.9. GeoMX DSP Analysis
4.10. Nanostring Gene Expression Profiling Assay
4.11. Nanostring Gene Expression Profiling Analysis
4.12. Enrichr Analysis
4.13. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feiock, C.; Yagi, M.; Maidman, A.; Rendahl, A.; Hui, S.; Seelig, D. Central Nervous System Injury—A Newly Observed Bystander Effect of Radiation. PLoS ONE 2016, 11, e0163233. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.; Pyter, L.M. Neuroimmunology of Behavioral Comorbidities Associated With Cancer and Cancer Treatments. Front. Immunol. 2018, 9, 1195. [Google Scholar] [CrossRef] [PubMed]
- Demos-Davies, K.; Lawrence, J.; Rogich, A.; Lind, E.; Seelig, D. Cancer treatment induces neuroinflammation and behavioral deficits in mice. Front. Behav. Neurosci. 2022, 16, 1067298. [Google Scholar] [CrossRef] [PubMed]
- Demos-Davies, K.; Lawrence, J.; Seelig, D. Cancer related cognitive impairment: A downside of cancer treatment. Front. Oncol. 2024, 14, 1387251. [Google Scholar] [CrossRef] [PubMed]
- Janelsins, M.C.; Kohli, S.; Mohile, S.G.; Usuki, K.; Ahles, T.A.; Morrow, G.R. An update on cancer- and chemotherapy-related cognitive dysfunction: Current status. Semin. Oncol. 2011, 38, 431–438. [Google Scholar] [CrossRef] [PubMed]
- McDonald, B.C.; Conroy, S.K.; Ahles, T.A.; West, J.D.; Saykin, A.J. Gray matter reduction associated with systemic chemotherapy for breast cancer: A prospective MRI study. Breast Cancer Res. Treat. 2010, 123, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Jim, H.S.; Donovan, K.A.; Small, B.J.; Andrykowski, M.A.; Munster, P.N.; Jacobsen, P.B. Cognitive functioning in breast cancer survivors: A controlled comparison. Cancer 2009, 115, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Noal, S.; Levy, C.; Hardouin, A.; Rieux, C.; Heutte, N.; Ségura, C.; Collet, F.; Allouache, D.; Switsers, O.; Delcambre, C.; et al. One-year longitudinal study of fatigue, cognitive functions, and quality of life after adjuvant radiotherapy for breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 2011, 81, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Donovan, K.A.; Small, B.J.; Andrykowski, M.A.; Schmitt, F.A.; Munster, P.; Jacobsen, P.B. Cognitive functioning after adjuvant chemotherapy and/or radiotherapy for early-stage breast carcinoma. Cancer 2005, 104, 2499–2507. [Google Scholar] [CrossRef]
- Kohli, S.; Griggs, J.J.; Roscoe, J.A.; Jean-Pierre, P.; Bole, C.; Mustian, K.M.; Hill, R.; Smith, K.; Gross, H.; Morrow, G.R. Self-reported cognitive impairment in patients with cancer. J. Oncol. Pract. 2007, 3, 54–59. [Google Scholar] [CrossRef]
- Phillips, K.M.; Jim, H.S.; Small, B.J.; Laronga, C.; Andrykowski, M.A.; Jacobsen, P.B. Cognitive functioning after cancer treatment: A 3-year longitudinal comparison of breast cancer survivors treated with chemotherapy or radiation and noncancer controls. Cancer 2012, 118, 1925–1932. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Kim, W.D. Early diagnosis of radiodermatitis using lactate dehydrogenase isozymes in hairless mice (SKH1-hr). Lab. Anim. Res. 2012, 28, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Cuccurullo, V.; Di Stasio, G.D.; Cascini, G.L.; Gatta, G.; Bianco, C. The Molecular Effects of Ionizing Radiations on Brain Cells: Radiation Necrosis vs. Tumor Recurrence. Diagnostics 2019, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Lumniczky, K.; Szatmári, T.; Sáfrány, G. Ionizing Radiation-Induced Immune and Inflammatory Reactions in the Brain. Front. Immunol. 2017, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Gibson, E.M.; Monje, M. Microglia in Cancer Therapy-Related Cognitive Impairment. Trends Neurosci. 2021, 44, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Shibayama, O.; Yoshiuchi, K.; Inagaki, M.; Matsuoka, Y.; Yoshikawa, E.; Sugawara, Y.; Akechi, T.; Wada, N.; Imoto, S.; Murakami, K.; et al. Association between adjuvant regional radiotherapy and cognitive function in breast cancer patients treated with conservation therapy. Cancer Med. 2014, 3, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, J.; Seelig, D.; Demos-Davies, K.; Ferreira, C.; Ren, Y.; Wang, L.; Alam, S.K.; Yang, R.; Guedes, A.; Craig, A.; et al. Radiation dermatitis in the hairless mouse model mimics human radiation dermatitis. bioRxiv 2024, 5, 595074. [Google Scholar] [CrossRef]
- Demos-Davies, K.; Lawrence, J.; Ferreira, C.; Seelig, D. The Distant Molecular Effects on the Brain by Cancer Treatment. Brain Sci. 2023, 14, 22. [Google Scholar] [CrossRef]
- Hu, S.C.; Hou, M.F.; Luo, K.H.; Chuang, H.Y.; Wei, S.Y.; Chen, G.S.; Chiang, W.; Huang, C.J. Changes in biophysical properties of the skin following radiotherapy for breast cancer. J. Dermatol. 2014, 41, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Benavides, F.; Oberyszyn, T.M.; VanBuskirk, A.M.; Reeve, V.E.; Kusewitt, D.F. The hairless mouse in skin research. J. Dermatol. Sci. 2009, 53, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Juresic, E.; Barton, M.; Shafiq, J. Management of skin toxicity during radiation therapy: A review of the evidence. J. Med. Imaging Radiat. Oncol. 2010, 54, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Richardson, B.N.; Lin, J.; Buchwald, Z.S.; Bai, J. Skin Microbiome and Treatment-Related Skin Toxicities in Patients with Cancer: A Mini-Review. Front. Oncol. 2022, 12, 924849. [Google Scholar] [CrossRef] [PubMed]
- Albouy, G.; King, B.R.; Maquet, P.; Doyon, J. Hippocampus and striatum: Dynamics and interaction during acquisition and sleep-related motor sequence memory consolidation. Hippocampus 2013, 23, 985–1004. [Google Scholar] [CrossRef] [PubMed]
- Vann, S.D.; Aggleton, J.P.; Maguire, E.A. What does the retrosplenial cortex do? Nat. Rev. Neurosci. 2009, 10, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Eichenbaum, H.; Dudchenko, P.; Wood, E.; Shapiro, M.; Tanila, H. The hippocampus, memory, and place cells: Is it spatial memory or a memory space? Neuron 1999, 23, 209–226. [Google Scholar] [CrossRef]
- Schroyen, G.; Blommaert, J.; Van Weehaeghe, D.; Sleurs, C.; Vandenbulcke, M.; Dedoncker, N.; Hatse, S.; Goris, A.; Koole, M.; Smeets, A.; et al. Neuroinflammation and Its Association with Cognition, Neuronal Markers and Peripheral Inflammation after Chemotherapy for Breast Cancer. Cancers 2021, 13, 4198. [Google Scholar] [CrossRef]
- Zhang, S.; Xiao, M.; Liu, Z.; Mo, Y.; Liu, H.; Xu, B. The functional significance of ATM phosphorylation of Bub3 on Serine 135 in sensitivity to DNA damaging agents. J. Clin. Oncol. 2021, 39, e15048. [Google Scholar] [CrossRef]
- Silva, P.M.A.; Bousbaa, H. BUB3, beyond the Simple Role of Partner. Pharmaceutics 2022, 14, 1084. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Müller, K.; Meineke, V. Radiation-induced alterations in cytokine production by skin cells. Exp. Hematol. 2007, 35, 96–104. [Google Scholar] [CrossRef] [PubMed]
- de Andrade, C.B.V.; Ramos, I.P.R.; de Moraes, A.C.N.; do Nascimento, A.L.R.; Salata, C.; Goldenberg, R.C.D.S.; de Carvalho, J.J.; de Almeida, C.E.V. Radiotherapy-Induced Skin Reactions Induce Fibrosis Mediated by TGF-β1 Cytokine. Dose Response 2017, 15, 1559325817705019. [Google Scholar] [CrossRef]
- Todd, T.P.; Bucci, D.J. Retrosplenial Cortex and Long-Term Memory: Molecules to Behavior. Neural Plast. 2015, 2015, 414173. [Google Scholar] [CrossRef] [PubMed]
- Alexander, A.S.; Robinson, J.C.; Dannenberg, H.; Kinsky, N.R.; Levy, S.J.; Mau, W.; Chapman, G.W.; Sullivan, D.W.; Hasselmo, M.E. Neurophysiological coding of space and time in the hippocampus, entorhinal cortex, and retrosplenial cortex. Brain Neurosci. Adv. 2020, 4, 2398212820972871. [Google Scholar] [CrossRef]
- Saade, M.; Araujo de Souza, G.; Scavone, C.; Kinoshita, P.F. The Role of GPNMB in Inflammation. Front. Immunol. 2021, 12, 674739. [Google Scholar] [CrossRef] [PubMed]
- Hüttenrauch, M.; Ogorek, I.; Klafki, H.; Otto, M.; Stadelmann, C.; Weggen, S.; Wiltfang, J.; Wirths, O. Glycoprotein NMB: A novel Alzheimer’s disease associated marker expressed in a subset of activated microglia. Acta Neuropathol. Commun. 2018, 6, 108. [Google Scholar] [CrossRef]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef]
- Song, W.M.; Colonna, M. The identity and function of microglia in neurodegeneration. Nat. Immunol. 2018, 19, 1048–1058. [Google Scholar] [CrossRef]
- Butovsky, O.; Weiner, H.L. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 2018, 19, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Preston, A.N.; Cervasio, D.A.; Laughlin, S.T. Visualizing the brain’s astrocytes. Methods Enzymol. 2019, 622, 129–151. [Google Scholar] [CrossRef]
- Guzman-Martinez, L.; Maccioni, R.B.; Andrade, V.; Navarrete, L.P.; Pastor, M.G.; Ramos-Escobar, N. Neuroinflammation as a Common Feature of Neurodegenerative Disorders. Front. Pharmacol. 2019, 10, 1008. [Google Scholar] [CrossRef] [PubMed]
- Maurya, S.K.; Bhattacharya, N.; Mishra, S.; Bhattacharya, A.; Banerjee, P.; Senapati, S.; Mishra, R. Microglia Specific Drug Targeting Using Natural Products for the Regulation of Redox Imbalance in Neurodegeneration. Front. Pharmacol. 2021, 12, 654489. [Google Scholar] [CrossRef]
- Liu, C.Y.; Wang, X.; Liu, C.; Zhang, H.L. Pharmacological Targeting of Microglial Activation: New Therapeutic Approach. Front. Cell. Neurosci. 2019, 13, 514. [Google Scholar] [CrossRef] [PubMed]
- Popa-Wagner, A.; Schröder, E.; Schmoll, H.; Walker, L.C.; Kessler, C. Upregulation of MAP1B and MAP2 in the rat brain after middle cerebral artery occlusion: Effect of age. J. Cereb. Blood Flow Metab. 1999, 19, 425–434. [Google Scholar] [CrossRef]
- Geisert, E.E.; Johnson, H.G.; Binder, L.I. Expression of microtubule-associated protein 2 by reactive astrocytes. Proc. Natl. Acad. Sci. USA 1990, 87, 3967–3971. [Google Scholar] [CrossRef]
- Lin, R.C.; Matesic, D.F. Immunohistochemical demonstration of neuron-specific enolase and microtubule-associated protein 2 in reactive astrocytes after injury in the adult forebrain. Neuroscience 1994, 60, 11–16. [Google Scholar] [CrossRef]
- Nakano, T.; Oka, K. Transition of Ki-67 index of uterine cervical tumors during radiation therapy. Immunohistochemical study. Cancer 1991, 68, 517–523. [Google Scholar] [CrossRef]
- Kang, J.O.; Hong, S.E.; Kim, S.K.; Kim, C.J.; Lee, T.H.; Chang, H.K.; Shin, M.S.; Kim, H. Adaptive responses induced by low dose radiation in dentate gyrus of rats. J. Korean Med. Sci. 2006, 21, 1103–1107. [Google Scholar] [CrossRef]
- Nagy, Z.; Esiri, M.M.; Smith, A.D. Expression of cell division markers in the hippocampus in Alzheimer’s disease and other neurodegenerative conditions. Acta Neuropathol. 1997, 93, 294–300. [Google Scholar] [CrossRef]
- Budelier, M.M.; He, Y.; Barthelemy, N.R.; Jiang, H.; Li, Y.; Park, E.; Henson, R.L.; Schindler, S.E.; Holtzman, D.M.; Bateman, R.J. A map of neurofilament light chain species in brain and cerebrospinal fluid and alterations in Alzheimer’s disease. Brain Commun. 2022, 4, fcac045. [Google Scholar] [CrossRef]
- Dietmann, A.S.; Kruse, N.; Stork, L.; Gloth, M.; Brück, W.; Metz, I. Neurofilament light chains in serum as biomarkers of axonal damage in early MS lesions: A histological-serological correlative study. J. Neurol. 2023, 270, 1416–1429. [Google Scholar] [CrossRef] [PubMed]
- Unal-Cevik, I.; Kilinç, M.; Gürsoy-Ozdemir, Y.; Gurer, G.; Dalkara, T. Loss of NeuN immunoreactivity after cerebral ischemia does not indicate neuronal cell loss: A cautionary note. Brain Res. 2004, 1015, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Dan, Q.-Q.; Zhao, N.; Li, J.-Y.; Li, J.; Chang, Q.; Su, P.; Cen, J.-C. Functional implication of synaptophysin upregulation with traumatic brain injury adult rats. Ibrain 2016, 2, 1–6. [Google Scholar] [CrossRef]
- Gudi, V.; Gai, L.; Herder, V.; Tejedor, L.S.; Kipp, M.; Amor, S.; Sühs, K.W.; Hansmann, F.; Beineke, A.; Baumgärtner, W.; et al. Synaptophysin Is a Reliable Marker for Axonal Damage. J. Neuropathol. Exp. Neurol. 2017, 76, 109–125. [Google Scholar] [CrossRef]
- Brown, G.C.; Neher, J.J. Microglial phagocytosis of live neurons. Nat. Rev. Neurosci. 2014, 15, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Adrian, M.; Weber, M.; Tsai, M.C.; Glock, C.; Kahn, O.I.; Phu, L.; Cheung, T.K.; Meilandt, W.J.; Rose, C.M.; Hoogenraad, C.C. Polarized microtubule remodeling transforms the morphology of reactive microglia and drives cytokine release. Nat. Commun. 2023, 14, 6322. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef]
- Schöneberg, T.; Meister, J.; Knierim, A.B.; Schulz, A. The G protein-coupled receptor GPR34—The past 20 years of a grownup. Pharmacol. Ther. 2018, 189, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Santpere, G.; Garcia-Esparcia, P.; Andres-Benito, P.; Lorente-Galdos, B.; Navarro, A.; Ferrer, I. Transcriptional network analysis in frontal cortex in Lewy body diseases with focus on dementia with Lewy bodies. Brain Pathol. 2018, 28, 315–333. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wei, M.; Ye, T.; Liu, Y.; Qi, D.; Cheng, X. Identification of the molecular subgroups in Alzheimer’s disease by transcriptomic data. Front. Neurol. 2022, 13, 901179. [Google Scholar] [CrossRef] [PubMed]
- Kempf, S.J.; Moertl, S.; Sepe, S.; von Toerne, C.; Hauck, S.M.; Atkinson, M.J.; Mastroberardino, P.G.; Tapio, S. Low-dose ionizing radiation rapidly affects mitochondrial and synaptic signaling pathways in murine hippocampus and cortex. J. Proteome Res. 2015, 14, 2055–2064. [Google Scholar] [CrossRef] [PubMed]
- Vavilova, J.D.; Boyko, A.A.; Troyanova, N.I.; Ponomareva, N.V.; Fokin, V.F.; Fedotova, E.Y.; Streltsova, M.A.; Kust, S.A.; Grechikhina, M.V.; Shustova, O.A.; et al. Alterations in Proteostasis System Components in Peripheral Blood Mononuclear Cells in Parkinson Disease: Focusing on the HSP70 and p62 Levels. Biomolecules 2022, 12, 493. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, T.; Ma, Z.; Zhou, C.; Wang, X.; Li, J. HSPA1A, HSPA2, and HSPA8 Are Potential Molecular Biomarkers for Prognosis among HSP70 Family in Alzheimer’s Disease. Dis. Markers 2022, 2022, 9480398. [Google Scholar] [CrossRef] [PubMed]
- Gerrits, E.; Brouwer, N.; Kooistra, S.M.; Woodbury, M.E.; Vermeiren, Y.; Lambourne, M.; Mulder, J.; Kummer, M.; Möller, T.; Biber, K.; et al. Distinct amyloid-β and tau-associated microglia profiles in Alzheimer’s disease. Acta Neuropathol. 2021, 141, 681–696. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, J.; Xu, H.; Chang, M.; Lv, C.; Xue, W.; Song, Z.; Zhang, L.; Zhang, X.; Tian, X. Retigabine ameliorates acute stress-induced impairment of spatial memory retrieval through regulating USP2 signaling pathways in hippocampal CA1 area. Neuropharmacology 2018, 135, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Markarian, M.; Krattli, R.P.; Baddour, J.D.; Alikhani, L.; Giedzinski, E.; Usmani, M.T.; Agrawal, A.; Baulch, J.E.; Tenner, A.J.; Acharya, M.M. Glia-Selective Deletion of Complement. Cancer Res. 2021, 81, 1732–1744. [Google Scholar] [CrossRef]
- Park, H.; Lee, S.; Shrestha, P.; Kim, J.; Park, J.A.; Ko, Y.; Ban, Y.H.; Park, D.Y.; Ha, S.J.; Koh, G.Y.; et al. AMIGO2, a novel membrane anchor of PDK1, controls cell survival and angiogenesis via Akt activation. J. Cell. Biol. 2015, 211, 619–637. [Google Scholar] [CrossRef]
- Reid, M.M.; Kautzmann, M.I.; Andrew, G.; Obenaus, A.; Mukherjee, P.K.; Khoutorova, L.; Ji, J.X.; Roque, C.R.; Oria, R.B.; Habeb, B.F.; et al. NPD1 Plus RvD1 Mediated Ischemic Stroke Penumbra Protection Increases Expression of Pro-homeostatic Microglial and Astrocyte Genes. Cell. Mol. Neurobiol. 2023, 43, 3555–3573. [Google Scholar] [CrossRef]
- Ma, H.; Zhou, Y.; Li, Z.; Zhu, L.; Li, H.; Zhang, G.; Wang, J.; Gong, H.; Xu, D.; Hua, W.; et al. Single-Cell RNA-Sequencing Analyses Revealed Heterogeneity and Dynamic Changes of Metabolic Pathways in Astrocytes at the Acute Phase of Ischemic Stroke. Oxidative Med. Cell. Longev. 2022, 2022, 1817721. [Google Scholar] [CrossRef]
- McMorran, B.J.; McCarthy, F.E.; Gibbs, E.M.; Pang, M.; Marshall, J.L.; Nairn, A.V.; Moremen, K.W.; Crosbie-Watson, R.H.; Baum, L.G. Differentiation-related glycan epitopes identify discrete domains of the muscle glycocalyx. Glycobiology 2016, 26, 1120–1132. [Google Scholar] [CrossRef] [PubMed]
- Dukay, B.; Walter, F.R.; Vigh, J.P.; Barabási, B.; Hajdu, P.; Balassa, T.; Migh, E.; Kincses, A.; Hoyk, Z.; Szögi, T.; et al. Neuroinflammatory processes are augmented in mice overexpressing human heat-shock protein B1 following ethanol-induced brain injury. J. Neuroinflamm. 2021, 18, 22. [Google Scholar] [CrossRef] [PubMed]
- Demyanenko, G.P.; Mohan, V.; Zhang, X.; Brennaman, L.H.; Dharbal, K.E.; Tran, T.S.; Manis, P.B.; Maness, P.F. Neural cell adhesion molecule NrCAM regulates Semaphorin 3F-induced dendritic spine remodeling. J. Neurosci. 2014, 34, 11274–11287. [Google Scholar] [CrossRef]
- Takano, T.; Wallace, J.T.; Baldwin, K.T.; Purkey, A.M.; Uezu, A.; Courtland, J.L.; Soderblom, E.J.; Shimogori, T.; Maness, P.F.; Eroglu, C.; et al. Chemico-genetic discovery of astrocytic control of inhibition in vivo. Nature 2020, 588, 296–302. [Google Scholar] [CrossRef]
- Kedrov, A.V.; Durymanov, M.; Anokhin, K.V. The Arc gene: Retroviral heritage in cognitive functions. Neurosci. Biobehav. Rev. 2019, 99, 275–281. [Google Scholar] [CrossRef]
- Rosi, S.; Andres-Mach, M.; Fishman, K.M.; Levy, W.; Ferguson, R.A.; Fike, J.R. Cranial irradiation alters the behaviorally induced immediate-early gene arc (activity-regulated cytoskeleton-associated protein). Cancer Res. 2008, 68, 9763–9770. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Yang, G.Y.; Liu, N.; Xu, P.; Chen, Y.L.; Zhou, Z.; Luo, Z.G.; Ding, X. P4-ATPase ATP8A2 acts in synergy with CDC50A to enhance neurite outgrowth. FEBS Lett. 2012, 586, 1803–1812. [Google Scholar] [CrossRef]
- Deng, J.; Song, L.; Yang, Z.; Zheng, S.; Du, Z.; Luo, L.; Liu, J.; Jin, X.; Yang, J. Neonatal LPS exposure reduces ATP8A2 level in the prefrontal cortex in mice via increasing IFN-γ level. Brain Res. Bull. 2021, 171, 103–112. [Google Scholar] [CrossRef]
- Bernstein, H.-G.; Kirches, E.; Bogerts, B.; Lendeckel, U.; Keilhoff, G.; Zempeltzi, M.; Steiner, J.; Tenbrock, K.; Dobrowolny, H.; Kyttaris, V.C.; et al. Wide distribution of CREM immunoreactivity in adult and fetal human brain, with an increased expression in dentate gyrus neurons of Alzheimer’s as compared to normal aging brains. Amino Acids 2013, 45, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Vyas, Y.; Montgomery, J.M. The role of postsynaptic density proteins in neural degeneration and regeneration. Neural Regen. Res. 2016, 11, 906–907. [Google Scholar] [CrossRef] [PubMed]
- Vogel, F.D.; Krenn, M.; Westphal, D.S.; Graf, E.; Wagner, M.; Leiz, S.; Koniuszewski, F.; Augé-Stock, M.; Kramer, G.; Scholze, P.; et al. A de novo missense variant in GABRA4 alters receptor function in an epileptic and neurodevelopmental phenotype. Epilepsia 2022, 63, e35–e41. [Google Scholar] [CrossRef]
- Janeczek, P.; Colson, N.; Dodd, P.R.; Lewohl, J.M. Sex Differences in the Expression of the α5 Subunit of the GABA. Alcohol. Clin. Exp. Res. 2020, 44, 423–434. [Google Scholar] [CrossRef]
- Márquez, J.; Martín-Rufián, M.; Segura, J.A.; Matés, J.M.; Campos-Sandoval, J.A.; Alonso, F.J. Brain glutaminases. Biomol. Concepts 2010, 1, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.W.D.; Liu, Y.; Bradshaw, P.C.; Wang, K. In Silico Preliminary Association of Ammonia Metabolism Genes GLS, CPS1, and GLUL with Risk of Alzheimer’s Disease, Major Depressive Disorder, and Type 2 Diabetes. J. Mol. Neurosci. 2018, 64, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Jia, E.; Pan, M.; Liu, Z.; Zhou, Y.; Zhao, X.; Dong, J.; Bai, Y.; Ge, Q. Transcriptomic profiling of differentially expressed genes and related pathways in different brain regions in Parkinson’s disease. Neurosci. Lett. 2020, 732, 135074. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Pang, X.; Guo, W.; Zhu, C.; Yu, L.; Song, X.; Wang, K.; Pang, C. An Exploration of the Coherent Effects between METTL3 and NDUFA10 on Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 10111. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Sun, Y.; Wang, G.Y.; Yin, J.; Li, C.J.; Luo, Y.Y.; Luan, Z.L. Positive Association of Human SHC3 Gene with Schizophrenia in a Northeast Chinese Han Population. Psychiatry Investig. 2020, 17, 934–940. [Google Scholar] [CrossRef]
- Valsecchi, V.; Laudati, G.; Cuomo, O.; Sirabella, R.; Del Prete, A.; Annunziato, L.; Pignataro, G. The hypoxia sensitive metal transcription factor MTF-1 activates NCX1 brain promoter and participates in remote postconditioning neuroprotection in stroke. Cell Death Dis. 2021, 12, 423. [Google Scholar] [CrossRef]
- Sirabella, R.; Sisalli, M.J.; Costa, G.; Omura, K.; Ianniello, G.; Pinna, A.; Morelli, M.; Di Renzo, G.M.; Annunziato, L.; Scorziello, A. NCX1 and NCX3 as potential factors contributing to neurodegeneration and neuroinflammation in the A53T transgenic mouse model of Parkinson’s Disease. Cell Death Dis. 2018, 9, 725. [Google Scholar] [CrossRef]
- Lee, U.; Choi, C.; Ryu, S.H.; Park, D.; Lee, S.E.; Kim, K.; Kim, Y.; Chang, S. SCAMP5 plays a critical role in axonal trafficking and synaptic localization of NHE6 to adjust quantal size at glutamatergic synapses. Proc. Natl. Acad. Sci. USA 2021, 118, e2011371118. [Google Scholar] [CrossRef] [PubMed]
- Prasad, H.; Rao, R. The Na+/H+ exchanger NHE6 modulates endosomal pH to control processing of amyloid precursor protein in a cell culture model of Alzheimer disease. J. Biol. Chem. 2015, 290, 5311–5327. [Google Scholar] [CrossRef] [PubMed]
- Kivisäkk, P.; Carlyle, B.C.; Sweeney, T.; Quinn, J.P.; Ramirez, C.E.; Trombetta, B.A.; Mendes, M.; Brock, M.; Rubel, C.; Czerkowicz, J.; et al. Increased levels of the synaptic proteins PSD-95, SNAP-25, and neurogranin in the cerebrospinal fluid of patients with Alzheimer’s disease. Alzheimer’s Res. Ther. 2022, 14, 58. [Google Scholar] [CrossRef]
- Hoerder-Suabedissen, A.; Korrell, K.V.; Hayashi, S.; Jeans, A.; Ramirez, D.M.O.; Grant, E.; Christian, H.C.; Kavalali, E.T.; Wilson, M.C.; Molnár, Z. Cell-Specific Loss of SNAP25 from Cortical Projection Neurons Allows Normal Development but Causes Subsequent Neurodegeneration. Cereb. Cortex 2019, 29, 2148–2159. [Google Scholar] [CrossRef] [PubMed]
- Giovagnoni, C.; Ali, M.; Eijssen, L.M.T.; Maes, R.; Choe, K.; Mulder, M.; Kleinjans, J.; Del Sol, A.; Glaab, E.; Mastroeni, D.; et al. Altered sphingolipid function in Alzheimer’s disease; a gene regulatory network approach. Neurobiol. Aging 2021, 102, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Jia, L.; Li, F.; Jia, J. Identification of KIAA0513 and Other Hub Genes Associated With Alzheimer Disease Using Weighted Gene Coexpression Network Analysis. Front. Genet. 2020, 11, 981. [Google Scholar] [CrossRef] [PubMed]
- Grønborg, M.; Pavlos, N.J.; Brunk, I.; Chua, J.J.; Münster-Wandowski, A.; Riedel, D.; Ahnert-Hilger, G.; Urlaub, H.; Jahn, R. Quantitative comparison of glutamatergic and GABAergic synaptic vesicles unveils selectivity for few proteins including MAL2, a novel synaptic vesicle protein. J. Neurosci. 2010, 30, 2–12. [Google Scholar] [CrossRef]
- Konar, A.; Thakur, M.K. Neuropsin Expression Correlates with Dendritic Marker MAP2c Level in Different Brain Regions of Aging Mice. Mol. Neurobiol. 2015, 51, 1130–1138. [Google Scholar] [CrossRef]
- Benice, T.S.; Rizk, A.; Kohama, S.; Pfankuch, T.; Raber, J. Sex-differences in age-related cognitive decline in C57BL/6J mice associated with increased brain microtubule-associated protein 2 and synaptophysin immunoreactivity. Neuroscience 2006, 137, 413–423. [Google Scholar] [CrossRef]
- VanGuilder, H.D.; Farley, J.A.; Yan, H.; Van Kirk, C.A.; Mitschelen, M.; Sonntag, W.E.; Freeman, W.M. Hippocampal dysregulation of synaptic plasticity-associated proteins with age-related cognitive decline. Neurobiol. Dis. 2011, 43, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Tfilin, M.; Turgeman, G. Interleukine-17 Administration Modulates Adult Hippocampal Neurogenesis and Improves Spatial Learning in Mice. J. Mol. Neurosci. 2019, 69, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Alam, R.; Mrad, Y.; Hammoud, H.; Saker, Z.; Fares, Y.; Estephan, E.; Bahmad, H.F.; Harati, H.; Nabha, S. New insights into the role of fibroblast growth factors in Alzheimer’s disease. Mol. Biol. Rep. 2022, 49, 1413–1427. [Google Scholar] [CrossRef]
- Wefel, J.S.; Kesler, S.R.; Noll, K.R.; Schagen, S.B. Clinical characteristics, pathophysiology, and management of noncentral nervous system cancer-related cognitive impairment in adults. CA Cancer J. Clin. 2015, 65, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.H.; Shim, S.; Wang, T.; Yoon, Y.; Jang, W.S.; Myung, J.K.; Park, S.; Kim, K.H. In vivo characterization of early-stage radiation skin injury in a mouse model by two-photon microscopy. Sci. Rep. 2016, 6, 19216. [Google Scholar] [CrossRef]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef]
- Vider, J.; Croaker, A.; Cox, A.J.; Raymond, E.; Rogers, R.; Adamson, S.; Doyle, M.; O’Brien, B.; Cripps, A.W.; West, N.P. Comparison of skin biopsy sample processing and storage methods on high dimensional immune gene expression using the Nanostring nCounter system. Diagn. Pathol. 2020, 15, 57. [Google Scholar] [CrossRef]
- Wu, X.; Nelson, M.; Basu, M.; Srinivasan, P.; Lazarski, C.; Zhang, P.; Zheng, P.; Sandler, A.D. MYC oncogene is associated with suppression of tumor immunity and targeting Myc induces tumor cell immunogenicity for therapeutic whole cell vaccination. J. Immunother. Cancer 2021, 9, e001388. [Google Scholar] [CrossRef]
- Ma, C.; Hunt, J.B.; Kovalenko, A.; Liang, H.; Selenica, M.B.; Orr, M.B.; Zhang, B.; Gensel, J.C.; Feola, D.J.; Gordon, M.N.; et al. Myeloid Arginase 1 Insufficiency Exacerbates Amyloid-β Associated Neurodegenerative Pathways and Glial Signatures in a Mouse Model of Alzheimer’s Disease: A Targeted Transcriptome Analysis. Front. Immunol. 2021, 12, 628156. [Google Scholar] [CrossRef]


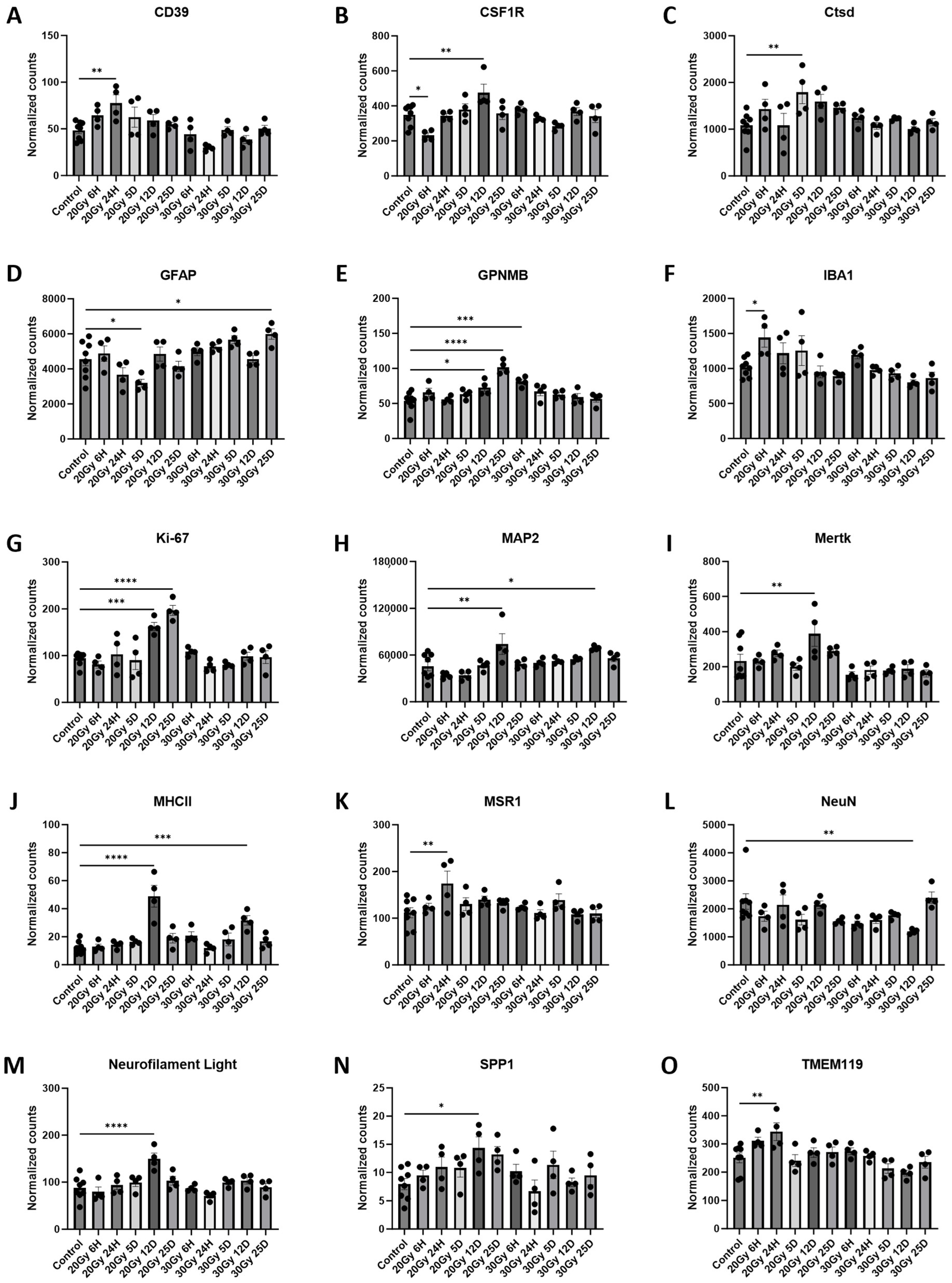
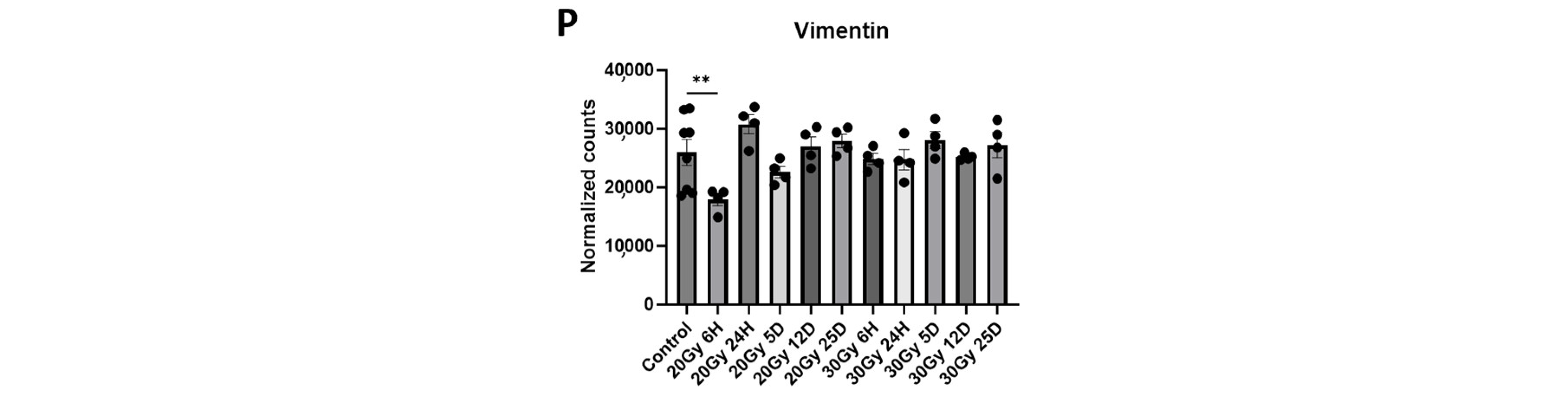
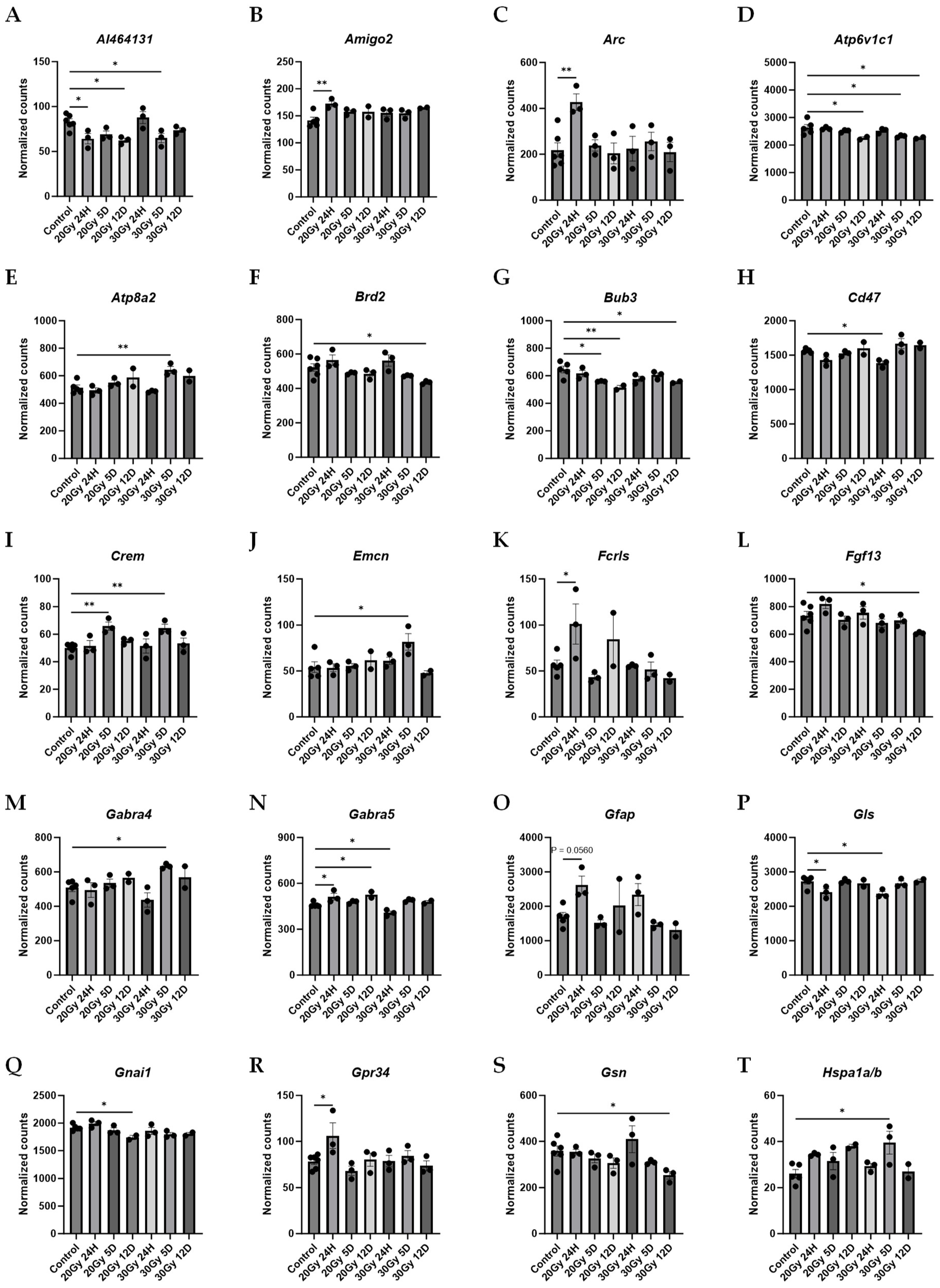
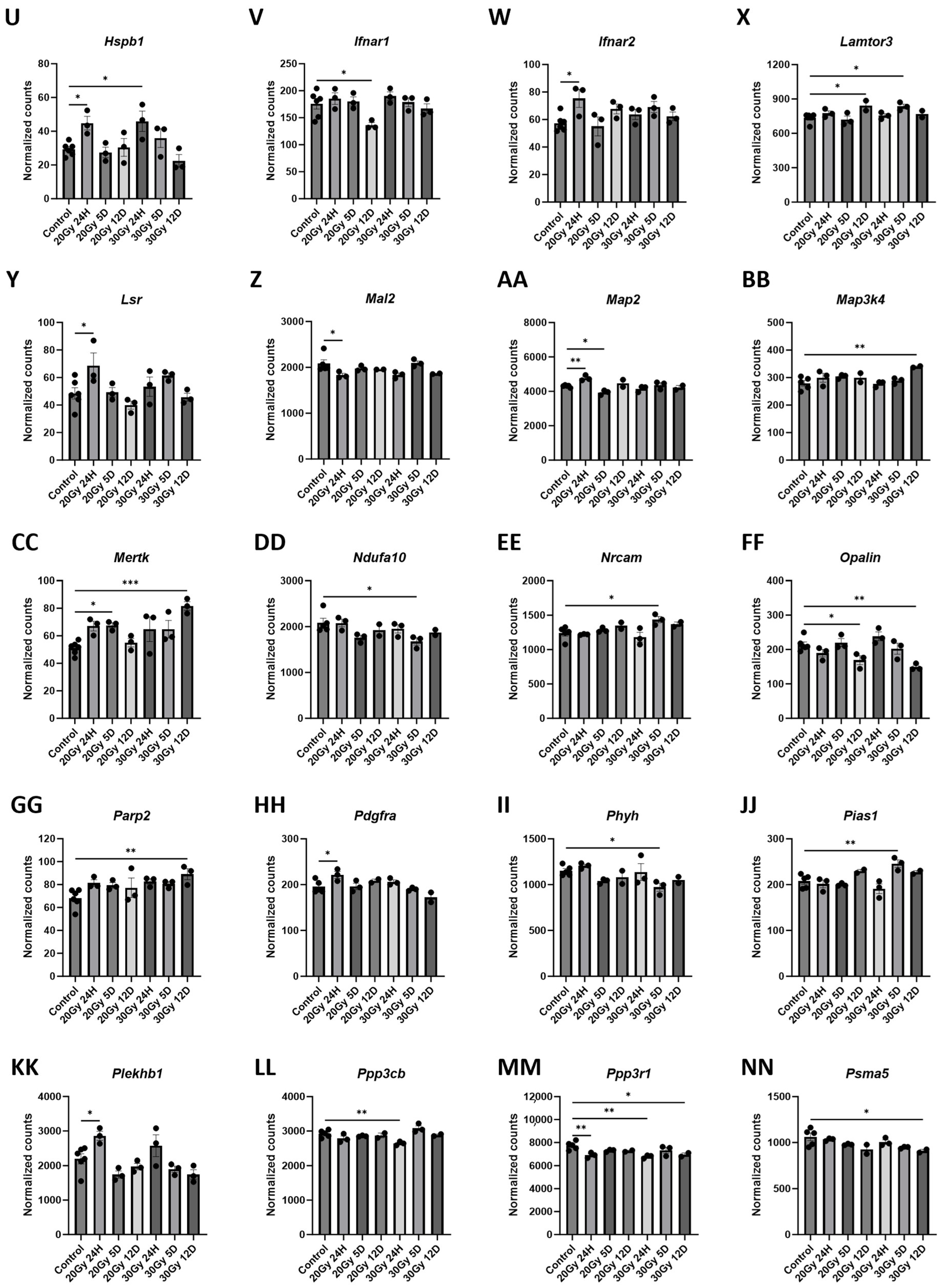
| Pathway Source | Pathway Name | p-Value | Adjusted p-Value | Odds Ratio | Combined Score | Genes Included in Pathway |
|---|---|---|---|---|---|---|
| BioPlanet 2019 | MAPK signaling pathway | 1.478 × 10−7 | 0.00005026 | 12.88 | 202.56 | PDGFRA, PPP3CB, PPP3R1, MAP2, STMN1, HSPB1, FGF13, LAMTOR3, MAP3K4 |
| WikiPathway 2023 Human | MAPK signaling pathway WP382 | 0.000004232 | 0.0007153 | 12.28 | 151.95 | PPP3CB, PPP3R1, STMN1, HSPB1, FGF13, LAMTOR3, MAP3K4 |
| KEGG 2021 Human | MAPK signaling pathway | 0.00001354 | 0.001032 | 10.20 | 114.36 | PDGFRA, PPP3CB, PPP3R1, STMN1, HSPB1, LAMTOR3, MAP3K4 |
| KEGG 2021 Human | Glutamatergic synapse | 0.00001394 | 0.001032 | 18.57 | 207.63 | PPP3CB, PPP3R1, SHANK3, GNAI1, GLS |
| KEGG 2021 Human | Natural killer cell mediated cytotoxicity | 0.00002730 | 0.001347 | 16.05 | 168.68 | IFNAR2, PPP3CB, PPP3R1, SHC3, IFNAR1 |
| WikiPathway 2023 Human | Immune response to tuberculosis WP4197 | 0.00002763 | 0.002335 | 61.69 | 647.58 | IFNAR2, PIAS1, IFNAR1 |
| BioPlanet 2019 | Natural killer cell-mediated cytotoxicity | 0.00003385 | 0.003487 | 15.32 | 157.67 | IFNAR2, PPP3CB, PPP3R1, SHC3, IFNAR1 |
| BioPlanet 2019 | CD8/T cell receptor downstream pathway | 0.00003400 | 0.003487 | 24.85 | 255.71 | IFNAR2, PPP3CB, PPP3R1, IFNAR1 |
| BioPlanet 2019 | Interferon alpha signaling regulation | 0.00004103 | 0.003487 | 53.27 | 538.12 | IFNAR2, PTPN1, IFNAR1 |
| WikiPathway 2023 Human | Regulatory circuits of the STAT3 signaling pathway WP4538 | 0.00005838 | 0.002459 | 21.48 | 209.43 | IFNAR2, PDGFRA, STMN1, IFNAR1 |
| WikiPathway 2023 Human | SARS coronavirus and innate immunity WP4912 | 0.00007173 | 0.002459 | 43.40 | 414.11 | IFNAR2, TRAF3, IFNAR1 |
| KEGG 2021 Human | JAK-STAT signaling pathway | 0.00007523 | 0.002784 | 12.86 | 122.12 | IFNAR2, PDGFRA, PIAS1, IFNAR1, GFAP |
| WikiPathway 2023 Human | Type I interferon induction and signaling during SARS-CoV-2 infection WP4868 | 0.00007927 | 0.002459 | 41.84 | 395.12 | IFNAR2, TRAF3, IFNAR1 |
| WikiPathway 2023 Human | Host pathogen interaction of human coronaviruses interferon induction WP4880 | 0.00008730 | 0.002459 | 40.40 | 377.58 | IFNAR2, TRAF3, IFNAR1 |
| KEGG 2021 Human | GABAergic synapse | 0.00009775 | 0.002893 | 18.69 | 172.59 | GABRA5, GABRA4, GNAI1, GLS |
| BioPlanet 2019 | EGF receptor (ErbB1) signaling pathway | 0.0001145 | 0.007789 | 36.61 | 332.19 | PTPN1, GSN, GNAI1 |
| WikiPathway 2023 Human | mBDNF and proBDNF regulation of GABA neurotransmission WP4829 | 0.0001468 | 0.003544 | 33.46 | 295.37 | GABRA5, GABRA4, CREM |
| BioPlanet 2019 | Transmission across chemical synapses | 0.0001592 | 0.009019 | 10.90 | 95.32 | SNAP25, GABRA5, GABRA4, GNAI1, GLS |
| KEGG 2021 Human | Kaposi sarcoma-associated herpesvirus infection | 0.0001712 | 0.004223 | 10.72 | 93.00 | IFNAR2, PPP3CB, PPP3R1, TRAF3, IFNAR1 |
| Elsevier Pathway Collection | Cochlear hair cell synapse proteins mutations (age-related/congenital) | 0.0002545 | 0.01874 | 109.55 | 906.71 | SNAP25, GNAI1 |
| Elsevier Pathway Collection | HRH1/3 -> synaptic transmission | 0.0002545 | 0.01874 | 109.55 | 906.71 | SNAP25, GNAI1 |
| Elsevier Pathway Collection | AXL receptor tyrosine kinase signaling | 0.0002774 | 0.01874 | 26.61 | 217.92 | IFNAR2, HSPB1, IFNAR1 |
| BioPlanet 2019 | Interferon-alpha signaling pathway | 0.0003175 | 0.01375 | 95.86 | 772.11 | IFNAR2, IFNAR1 |
| KEGG 2021 Human | Osteoclast differentiation | 0.0003836 | 0.008110 | 12.89 | 101.42 | IFNAR2, PPP3CB, PPP3R1, IFNAR1 |
| BioPlanet 2019 | Cardiac protection against reactive oxygen species | 0.0003874 | 0.01375 | 85.20 | 669.33 | GABRA5, GABRA4 |
| Elsevier Pathway Collection | Histamine in arousal regulation | 0.0003874 | 0.01874 | 85.20 | 669.33 | SNAP25, GNAI1 |
| BioPlanet 2019 | GABA A and B receptor activation | 0.0003961 | 0.01375 | 23.41 | 183.36 | GABRA5, GABRA4, GNAI1 |
| WikiPathway 2023 Human | Interferon type I signaling pathways WP585 | 0.0004186 | 0.008844 | 22.95 | 178.49 | IFNAR2, PIAS1, IFNAR1 |
| Elsevier Pathway Collection | Dendritic cell activation in systemic lupus erythematosis | 0.0004420 | 0.01874 | 22.50 | 173.83 | IFNAR2, TRAF3, IFNAR1 |
| BioPlanet 2019 | GABA (A) receptor activation | 0.0004641 | 0.01375 | 76.68 | 588.52 | GABRA5, GABRA4 |
| Elsevier Pathway Collection | DRD2/4 -> membrane transport | 0.0004641 | 0.01874 | 76.68 | 588.52 | SNAP25, GNAI1 |
| Elsevier Pathway Collection | Endocannabinoids role in sleep regulation | 0.0004641 | 0.01874 | 76.68 | 588.52 | SNAP25, GNAI1 |
| Elsevier Pathway Collection | Microtubule cytoskeleton | 0.0004641 | 0.01874 | 76.68 | 588.52 | MAP2, STMN1 |
| KEGG 2021 Human | VEGF signaling pathway | 0.0005435 | 0.01005 | 20.89 | 157.06 | PPP3CB, PPP3R1, HSPB1 |
| Elsevier Pathway Collection | Dopamine mediated glutamate release/uptake circle in neuron in migraine | 0.0005476 | 0.01965 | 69.70 | 523.47 | SNAP25, GLS |
| Elsevier Pathway Collection | GRM2-4/6-8 (presynaptic) -> glutamate release attenuation | 0.0006377 | 0.02060 | 63.89 | 470.09 | SNAP25, GNAI1 |
| KEGG 2021 Human | Retrograde endocannabinoid signaling | 0.0006831 | 0.01060 | 11.00 | 80.19 | GABRA5, GABRA4, NDUFA10, GNAI1 |
| WikiPathway 2023 Human | SARS CoV 2 innate immunity evasion and cell specific immune response WP5039 | 0.0007549 | 0.01418 | 18.56 | 133.46 | IFNAR2, TRAF3, IFNAR1 |
| KEGG 2021 Human | Hepatitis C | 0.0008517 | 0.01060 | 10.35 | 73.15 | IFNAR2, TRAF3, PIAS1, IFNAR1 |
| WikiPathway 2023 Human | Insulin signaling WP481 | 0.0009139 | 0.01545 | 10.15 | 71.02 | SNAP25, PTPN1, SHC3, MAP3K4 |
| Group | Treatment | Timepoint |
|---|---|---|
| Control 6 H | None | 6 h |
| 20 Gy RT 6 H | 20 Gy RT | 6 h |
| 30 Gy RT 6 H | 30 Gy RT | 6 h |
| Control 24 H | None | 24 h |
| 20 Gy RT 24 H | 20 Gy RT | 24 h |
| 30 Gy RT 24 H | 30 Gy RT | 24 h |
| Control 5 D | None | 5 days |
| 20 Gy RT 5 D | 20 Gy RT | 5 days |
| 30 Gy RT 5 D | 30 Gy RT | 5 days |
| Control 12 D | None | 12 days |
| 20 Gy RT 12 D | 20 Gy RT | 12 days |
| 30 Gy RT 12 D | 30 Gy RT | 12 days |
| Control 25 D | None | 25 days |
| 20 Gy RT 25 D | 20 Gy RT | 25 days |
| 30 Gy RT 25 D | 30 Gy RT | 25 days |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demos-Davies, K.; Lawrence, J.; Coffey, J.; Morgan, A.; Ferreira, C.; Hoeppner, L.H.; Seelig, D. Longitudinal Neuropathological Consequences of Extracranial Radiation Therapy in Mice. Int. J. Mol. Sci. 2024, 25, 5731. https://doi.org/10.3390/ijms25115731
Demos-Davies K, Lawrence J, Coffey J, Morgan A, Ferreira C, Hoeppner LH, Seelig D. Longitudinal Neuropathological Consequences of Extracranial Radiation Therapy in Mice. International Journal of Molecular Sciences. 2024; 25(11):5731. https://doi.org/10.3390/ijms25115731
Chicago/Turabian StyleDemos-Davies, Kimberly, Jessica Lawrence, Jessica Coffey, Amy Morgan, Clara Ferreira, Luke H. Hoeppner, and Davis Seelig. 2024. "Longitudinal Neuropathological Consequences of Extracranial Radiation Therapy in Mice" International Journal of Molecular Sciences 25, no. 11: 5731. https://doi.org/10.3390/ijms25115731
APA StyleDemos-Davies, K., Lawrence, J., Coffey, J., Morgan, A., Ferreira, C., Hoeppner, L. H., & Seelig, D. (2024). Longitudinal Neuropathological Consequences of Extracranial Radiation Therapy in Mice. International Journal of Molecular Sciences, 25(11), 5731. https://doi.org/10.3390/ijms25115731





