The Set of Serine Peptidases of the Tenebrio molitor Beetle: Transcriptomic Analysis on Different Developmental Stages
Abstract
1. Introduction
2. Results
2.1. General Characteristics of T. molitor Predicted SPs/SPHs of the S1A Subfamily
2.1.1. Identified Set of Peptidase-like Sequences
2.1.2. Annotation of Predicted Protein Sequences of T. molitor SPs
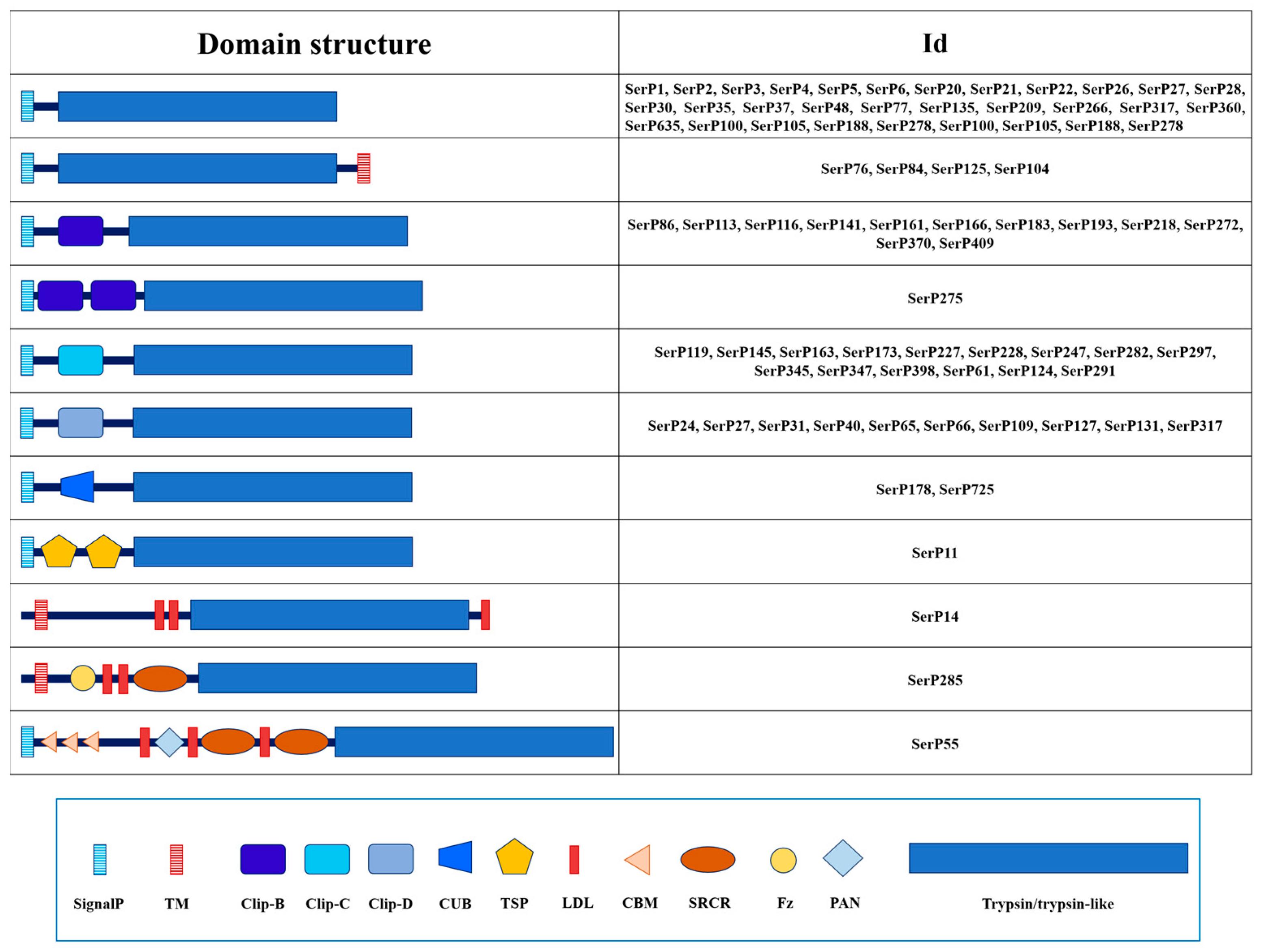
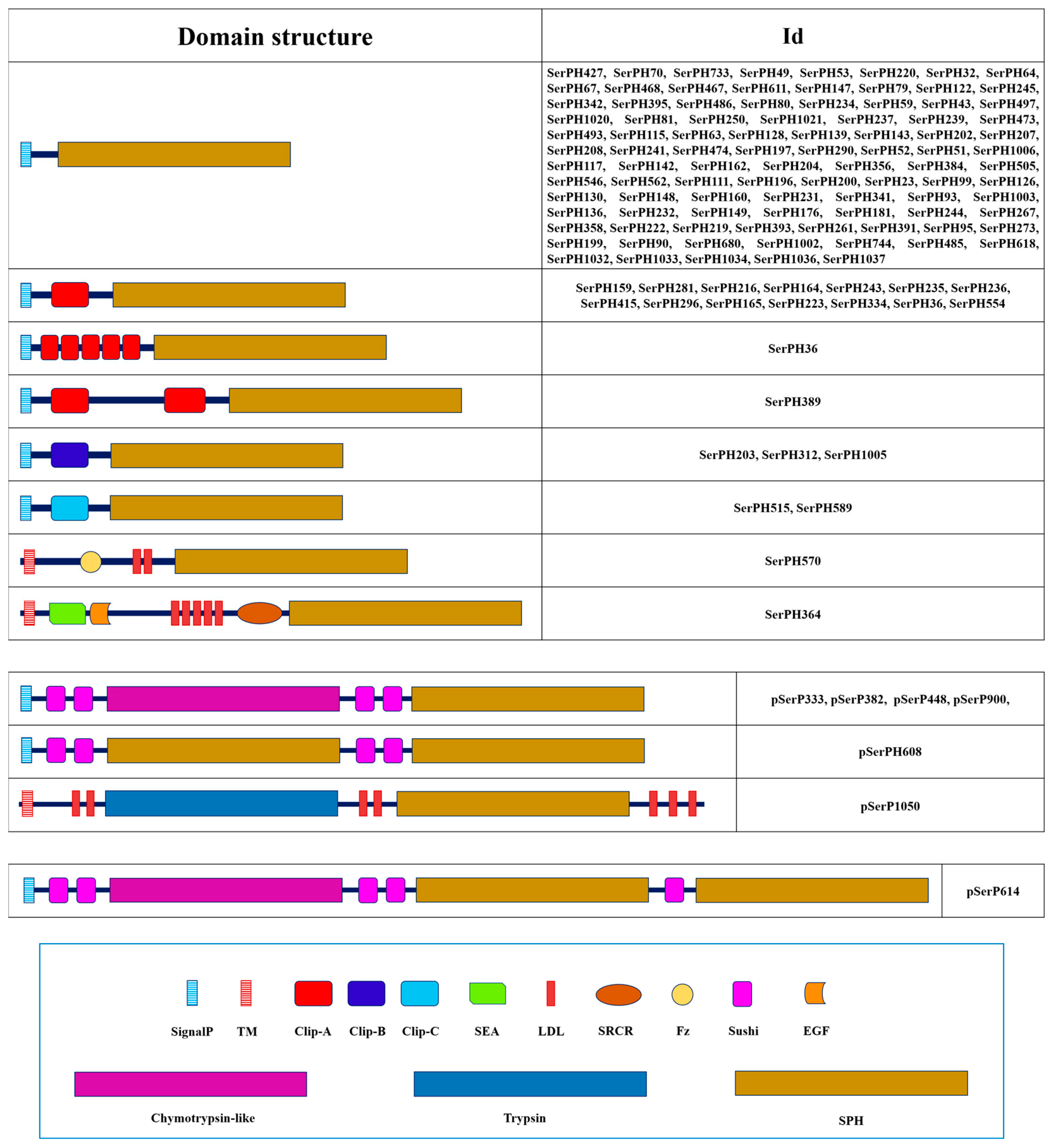
2.1.3. Domain Organization
2.2. Trypsins and Trypsin-like Peptidases
| № | Name | NCBI ID (Protein) | Preproenzyme/Mature Enzyme (aa) | SignalP (aa) | Regulatory Domain | Propeptide Cleavage Site | Active Site | S1 Subsite | Enzyme Specificity | Mm Mature, Da | pI | TM (Position) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SerP1 | ABC88729 | 258 | 227 | 16 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 22,742 | 6.9 | - |
| 2 | SerP2 | QWS65012 | 252 | 227 | 16 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 23,618 | 4.3 | - |
| 3 | SerP3 | QWS65044 | 259 | 228 | 16 | - | K|IVGG | H | D | S | D | G | G | Trypsin | 24,386 | 5.0 | - |
| 4 | SerP4 | QWS65013 | 250 | 225 | 15 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 24,140 | 5.2 | - |
| 5 | SerP5 | QWS65045 | 333 | 236 | 24 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 26,035 | 9.2 | - |
| 6 | SerP6 | QWS65014 | 258 | 226 | 17 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 23,414 | 3.8 | - |
| 7 | SerP20 | QWS65048 | 361 | 238 | 17 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 26,395 | 9.0 | - |
| 8 | SerP21 | QWS65049 | 276 | 228 | 22 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 24,732 | 4.5 | - |
| 9 | SerP22 | QWS65050 | 290 | 242 | 17 | - | R|VVGG | H | D | S | D | G | G | Trypsin | 25,975 | 6.2 | - |
| 10 | SerP26 | QWS65055 | 254 | 227 | 23 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 24,214 | 5.8 | - |
| 11 | SerP28 | QWS65056 | 310 | 241 | 26 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 27,033 | 7.6 | - |
| 12 | SerP30 | QWS65015 | 249 | 226 | 16 | - | K|IIGG | H | D | S | D | G | G | Trypsin | 24,862 | 8.9 | - |
| 13 | SerP35 | QWS65057 | 260 | 231 | 21 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 24,884 | 5.6 | - |
| 14 | SerP37 | QWS65058 | 298 | 251 | 19 | - | R|VVGG | H | D | S | D | G | G | Trypsin | 27,327 | 6.2 | - |
| 15 | SerP48 | QWS65017 | 321 | 295 | 22 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 32,018 | 6.7 | - |
| 16 | SerP76 | QWS65019 | 387 | 362 | 18 | - | K|IIGG | H | D | S | D | G | G | Trypsin | 39,417 | 5.7 | 367–386 |
| 17 | SerP77 | QWS65060 | 288 | 252 | 17 | - | K|IVGG | H | D | S | D | G | G | Trypsin | 27,164 | 8.3 | - |
| 18 | SerP84 | QWS65020 | 332 | 308 | 20 | - | K|VVGG | H | D | S | D | G | G | Trypsin | 33,286 | 5.0 | 313–330 |
| 19 | SerP104 | QWS65061 | 323 | 300 | 18 | - | K|IVGG | H | D | S | D | G | G | Trypsin | 32,646 | 4.2 | 300–323 |
| 20 | SerP125 | QWS65024 | 278 | 254 | 19 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 27,535 | 4.8 | 257–275 |
| 21 | SerP135 | QWS65027 | 292 | 246 | 22 | - | G|IIGG | H | D | S | D | G | G | Trypsin | 26,850 | 9.5 | - |
| 22 | SerP209 | QWS65033 | 258 | 227 | 16 | - | R|IIGG | H | D | S | D | G | G | Trypsin | 22,943 | 4.8 | - |
| 23 | SerP266 | QWS65037 | 281 | 256 | 18 | - | K|IVGG | H | D | S | D | G | G | Trypsin | 27,895 | 8.8 | - |
| 24 | SerP360 | CAH1374004 | 286 | 249 | 19 | - | K|IVGG | H | D | S | D | G | G | Trypsin | 27,480 | 4.7 | - |
| 25 | SerP635 | WJL97986 | 249 | 224 | 19 | - | R|IVGG | H | D | S | D | G | G | Trypsin | 24,044 | 4.1 | - |
| 26 | SerP100 | WJL97987 | 293 | 263 | 23 | - | R|IIGG | H | D | S | D | G | A | Trypsin-like | 28,605 | 8.8 | - |
| 27 | SerP105 | CAH1374591 | 305 | 243 | 23 | - | L|IIGG | H | D | S | D | G | A | Trypsin-like | 26,155 | 5.9 | - |
| 28 | SerP188 | KAJ3637256 | 303 | 271 | 20 | - | R|IVGG | H | D | S | D | G | A | Trypsin-like | 29,751 | 8.3 | - |
| 29 | SerP278 | CAH1363947 | 298 | 256 | 18 | - | R|IIGG | H | D | S | D | G | A | Trypsin-like | 27,716 | 6.8 | - |
| 30 | SerP86 | QWS65021 | 458 | 258 | 22 | Clip-B | R|ILDG | H | D | S | D | G | G | Trypsin | 28,226 | 8.4 | - |
| 31 | SerP113 | QWS65022 | 386 | 255 | 23 | Clip-B | R|IING | H | D | S | D | G | G | Trypsin | 28,255 | 7.7 | - |
| 32 | SerP116 | QWS65063 | 381 | 257 | 16 | Clip-B | K|IVNG | H | D | S | D | G | G | Trypsin | 28,382 | 6.4 | - |
| 33 | SerP141 | QWS65028 | 435 | 259 | 21 | Clip-B | R|IFGG | H | D | S | D | G | G | Trypsin | 28,844 | 9.2 | - |
| 34 | SerP161 | WJL97988 | 278 | 254 | 20 | Clip-B | R|ITSG | H | D | S | D | G | G | Trypsin | 27,807 | 7.7 | - |
| 35 | SerP166 | QWS65064 | 376 | 259 | 15 | Clip-B | K|LVND | H | D | S | D | G | G | Trypsin | 28,449 | 4.8 | - |
| 36 | SerP183 SPE | BAG14262 | 383 | 265 | 18 | Clip-B | R|IYGG | H | D | S | D | G | G | Trypsin | 29,203 | 7.6 | - |
| 37 | SerP193 | QWS65032 | 375 | 247 | 22 | Clip-B | R|ILGG | H | D | S | D | G | G | Trypsin | 27,564 | 6.2 | - |
| 38 | SerP272 | QWS65038 | 404 | 297 | 17 | Clip-B | K|IYGG | H | D | S | D | G | G | Trypsin | 32,710 | 8.0 | - |
| 39 | SerP275 | QWS65065 | 430 | 257 | 23 | Clip-B (2) | K|IVGG | H | D | S | D | G | G | Trypsin | 28,969 | 8.5 | - |
| 40 | SerP370 | QWS65041 | 407 | 257 | 21 | Clip-B | K|ISNG | H | D | S | D | G | G | Trypsin | 28,048 | 6.4 | - |
| 41 | SerP409 | QWS65042 | 447 | 234 | 22 | Clip-B | K|IGKG | H | D | S | D | G | G | Trypsin | 26,142 | 8.8 | - |
| 42 | SerP218 | CAH1363991 | 356 | 263 | 22 | Clip-B | K|VSGG | H | D | S | D | A | T | Trypsin-like | 29,129 | 6.3 | - |
| 43 | SerP119 | QWS65023 | 387 | 253 | 19 | Clip-C | L|IVGG | H | D | S | D | G | G | Trypsin | 28,333 | 8.1 | - |
| 44 | SerP145 | QWS65029 | 370 | 241 | 22 | Clip-C | H|IVGG | H | D | S | D | G | G | Trypsin | 26,781 | 7.7 | - |
| 45 | SerP163 | QWS65030 | 354 | 254 | 21 | Clip-C | V|IAFG | H | D | S | D | G | G | Trypsin | 28,041 | 5.7 | - |
| 46 | SerP173 | QWS65031 | 362 | 249 | 21 | Clip-C | F|VFGG | H | D | S | D | G | G | Trypsin | 27,495 | 4.9 | - |
| 47 | SerP227 | QWS65034 | 376 | 251 | 23 | Clip-C | L|IVGG | H | D | S | D | G | G | Trypsin | 27,969 | 5.8 | - |
| 48 | SerP228 SAE | QWS65035 | 374 | 250 | 20 | Clip-C | L|IVGG | H | D | S | D | G | G | Trypsin | 27,849 | 6.2 | - |
| 49 | SerP247 | QWS65036 | 379 | 257 | 18 | Clip-C | T|IISM | H | D | S | D | G | G | Trypsin | 28,343 | 6.1 | - |
| 50 | SerP282 | QWS65039 | 349 | 270 | 17 | Clip-C | G|ITGG | H | D | S | D | G | G | Trypsin | 29,212 | 6.0 | - |
| 51 | SerP297 | QWS65066 | 350 | 255 | 18 | Clip-C | V|EYEE | H | D | S | D | G | G | Trypsin | 28,238 | 5.7 | - |
| 52 | SerP345 | QWS65040 | 359 | 234 | 22 | Clip-C | L|IVGG | H | D | S | D | G | G | Trypsin | 26,360 | 6.5 | - |
| 53 | SerP347 | QWS65067 | 367 | 256 | 25 | Clip-C | G|IAIG | H | D | S | D | G | G | Trypsin | 28,001 | 5.8 | - |
| 54 | SerP398 | CAH1365893 | 385 | 253 | 19 | Clip-C | L|IIGG | H | D | S | D | G | G | Trypsin | 28,360 | 8.9 | - |
| 55 | SerP61 | CAH1377522 | 422 | 246 | 26 | Clip-C | L|IVGG | H | D | S | D | G | A | Trypsin-like | 27,368 | 8.7 | - |
| 56 | SerP124 | CAH1383174 | 371 | 250 | 20 | Clip-C | L|IVGG | H | D | S | D | S | G | Trypsin-like | 27,690 | 6.0 | - |
| 57 | SerP291 | WJL97989 | 357 | 251 | 20 | Clip-C | Q|IWGG | H | D | S | D | G | T | Trypsin-like | 28,108 | 7.1 | - |
| 58 | SerP15 | QWS65047 | 516 | 235 | 23 | Clip-D | R|IVGG | H | D | S | D | G | G | Trypsin | 25,699 | 9.2 | - |
| 59 | SerP24 | QWS65051 | 810 | 243 | 19 | Clip-D | R|IVGG | H | D | S | D | G | G | Trypsin | 27,555 | 5.4 | - |
| 60 | SerP27 | QWS65052 | 369 | 242 | 19 | Clip-D | R|IVGG | H | D | S | D | G | G | Trypsin | 26,704 | 9.0 | - |
| 61 | SerP31 | CAH1379474 | 557 | 244 | 15 | Clip-D | K|IVGG | H | D | S | D | G | G | Trypsin | 26,888 | 6.5 | - |
| 62 | SerP40 | QWS65016 | 392 | 241 | 21 | Clip-D | G|NPGG | H | D | S | D | G | G | Trypsin | 26,535 | 5.5 | - |
| 63 | SerP65 | QWS65053 | 619 | 240 | 20 | Clip-D | R|IVGG | H | D | S | D | G | G | Trypsin | 26,001 | 9.2 | - |
| 64 | SerP66 | QWS65059 | 523 | 245 | 29 | Clip-D | R|VVGG | H | D | S | D | G | G | Trypsin | 27,560 | 9.1 | - |
| 65 | SerP109 | QWS65062 | 964 | 247 | 17 | Clip-D | R|IVGG | H | D | S | D | G | G | Trypsin | 26,987 | 7.8 | - |
| 66 | SerP127 | QWS65025 | 376 | 247 | 22 | Clip-D | R|IVNG | H | D | S | D | G | G | Trypsin | 27,075 | 7.0 | - |
| 67 | SerP131 | QWS65026 | 375 | 247 | 22 | Clip-D | R|VVNG | H | D | S | D | G | G | Trypsin | 26,799 | 8.4 | - |
| 68 | SerP317 | QWS65054 | 389 | 246 | 16 | Clip-D | R|IIGG | H | D | S | D | G | G | Trypsin | 27,195 | 6.2 | - |
| 69 | SerP178 | KAJ3638924 | 409 | 242 | 27 | CUB | R|IVGG | H | D | S | D | G | A | Trypsin-like | 26,019 | 5.0 | - |
| 70 | SerP725 | KAJ3638922 | 405 | 246 | 23 | CUB | K|IVGG | H | D | S | D | G | A | Trypsin-like | 26,660 | 4.9 | - |
| 71 | SerP285 Corin | CAH1378270 | 965 | 247 | - | Fz, LDL (2), SRCR | R|IVGG | H | D | S | D | G | G | Trypsin | 27,268 | 5.9 | 338–359 |
| 72 | SerP14 | QWS65046 | 1289 | 286 | - | LDL (3) | R|IVGG | H | D | S | D | G | G | Trypsin | 31,448 | 5.9 | 68–94 |
| 73 | SerP11 TSP | QWS65043 | 447 | 231 | 19 | TSP (2) | K|IIGG | H | D | S | D | G | G | Trypsin | 26,306 | 9.5 | - |
| 74 | SerP55 Tequila | QWS65018 | 1672 | 245 | 23 | CBM (3), LDL (3), SRCR (2) PAN | R|VVRG | H | D | S | D | G | G | Trypsin | 26,947 | 5.9 | - |
2.3. Chymotrypsin-like Peptidases
2.4. Elastase-like Peptidases
2.5. Non-Annotated Serine Peptidases
2.6. Serine Peptidase Homologs
2.7. Polypeptidases
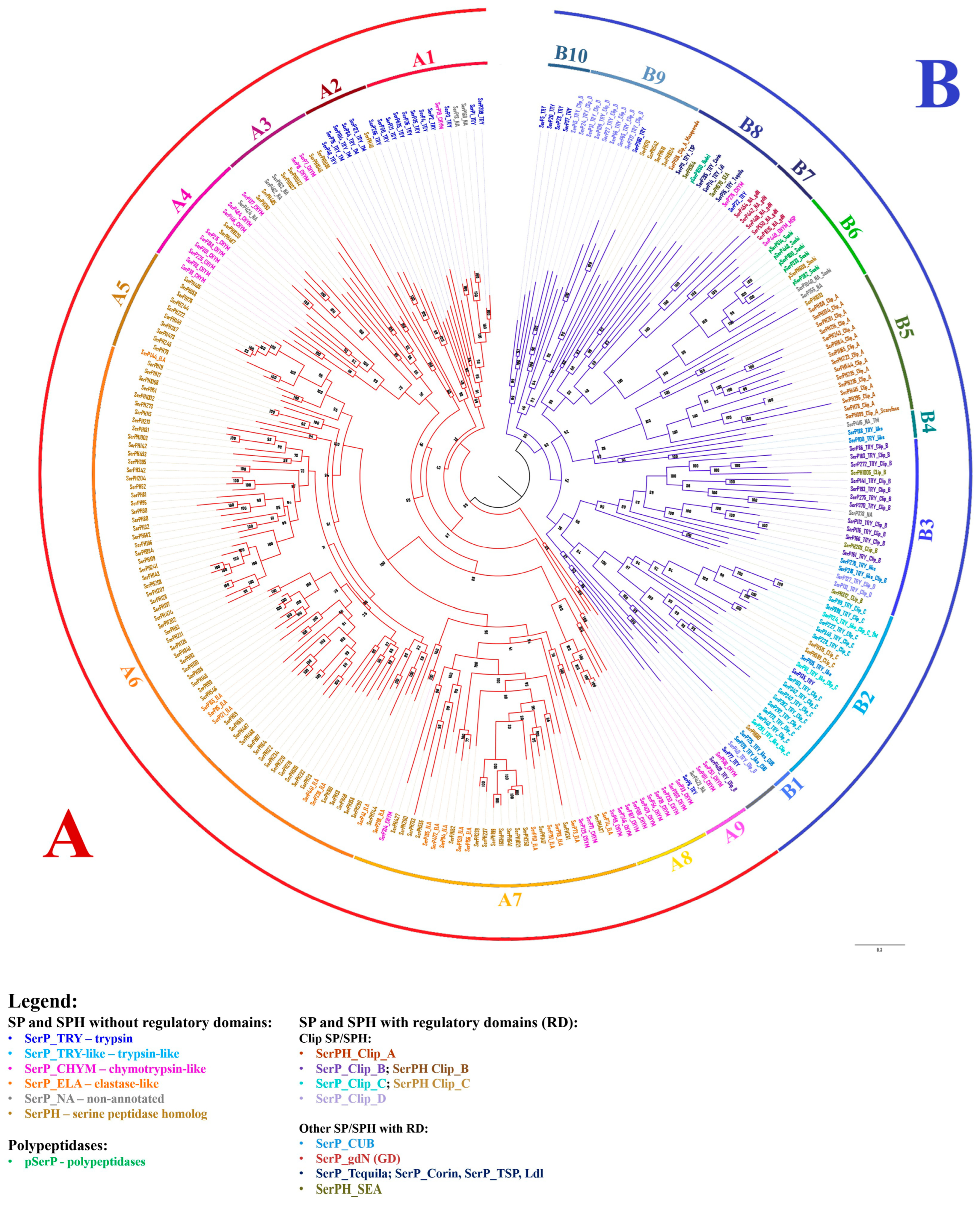
2.8. Phylogenetic Analysis of SPs and SPHs in T. molitor
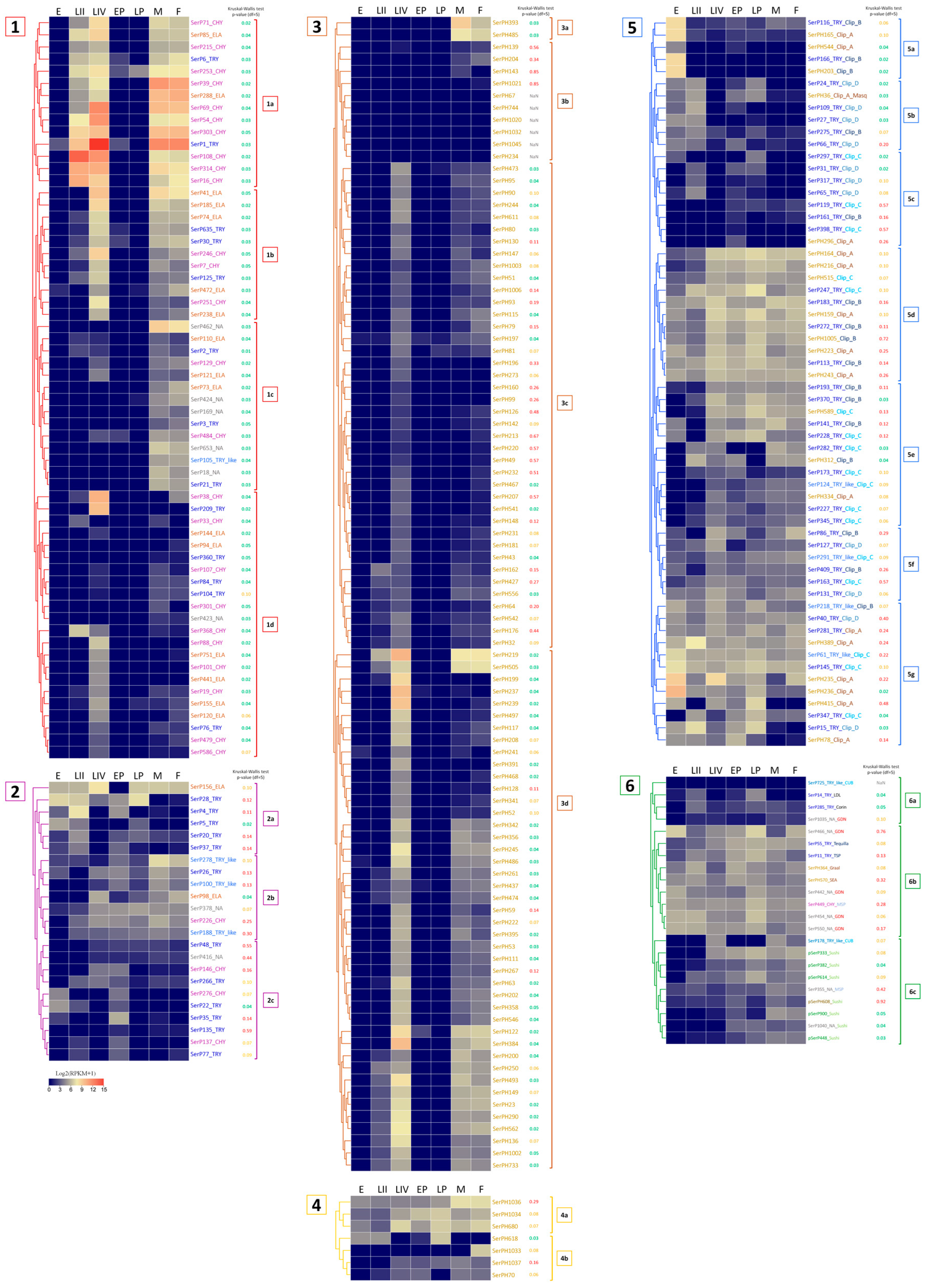
2.9. Expression Profiling of SP and SPH Genes in Different Life Stages of T. molitor
2.9.1. Embryonic Stage: Eggs
2.9.2. Metamorphosis: Early and Late Pupae
2.9.3. Feeding Stages: Larvae and Imago (Adults)
2.9.4. Constitutively Expressed SP-Related Proteins of T. molitor
3. Discussion
4. Materials and Methods
4.1. Preparation of Biological Material, RNA Isolation and cDNA Sequencing
4.2. Transcriptomes Assembly
4.2.1. Assembly of Larval Gut Sequences
4.2.2. Assemblies of Different Developmental Stages
4.2.3. The Total T. molitor Transcriptome Assembly
4.3. SP/SPH Identification in the Transcriptomes
4.4. Analysis of Protein Sequences
4.5. Phylogenetic Analysis
4.6. Expression Profiling of SP and SPH at Different Developmental Stages
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ikeda, M.; Yaginuma, T.; Kobayashi, M.; Yamashita, O. cDNA cloning, sequencing and temporal expression of the protease responsible for vitellin degradation in the silkworm, Bombyx mori. Comp. Biochem. Physiol. B 1991, 99, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Choo, Y.M.; Lee, K.S.; Yoon, H.J.; Lee, S.B.; Kim, J.H.; Sohn, H.D.; Jin, B.R. A serine protease from the midgut of the bumblebee, Bombus ignites (Hymenoptera: Apidae): cDNA cloning, gene structure, expression and enzyme activity. Eur. J. Entomol. 2007, 104, 1–7. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Kriventseva, E.V.; Meister, S.; Xi, Z.; Alvarez, K.S.; Bartholomay, L.C.; Barillas-Mury, C.; Bian, G.; Blandin, S.; Christensen, B.M.; et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 2007, 316, 1738–1743. [Google Scholar] [CrossRef] [PubMed]
- Kan, H.; Kim, C.H.; Kwon, H.M.; Park, J.W.; Roh, K.B.; Lee, H.; Park, B.J.; Zhang, R.; Zhang, J.; Söderhäll, K.; et al. Molecular control of phenoloxidase-induced melanin synthesis in an insect. J. Biol. Chem. 2008, 283, 25316–25323. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Vilcinskas, A.; Kanost, M.R. Immunity in lepidopteran insects. Adv. Exp. Med. Biol. 2010, 708, 181–204. [Google Scholar] [CrossRef] [PubMed]
- Veillard, F.; Troxler, L.; Reichhart, J.M. Drosophila melanogaster clip-domain serine proteases: Structure, function and regulation. Biochimie 2016, 22, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.D. Insect Hemolymph Immune Complexes. Subcell Biochem. 2020, 94, 123–161. [Google Scholar] [CrossRef] [PubMed]
- Contreras, E.G.; Glavic, Á.; Brand, A.H.; Sierralta, J.A. The serine protease homolog, scarface, is sensitive to nutrient availability and modulates the development of the Drosophila blood-brain barrier. J. Neurosci. 2021, 41, 6430–6448. [Google Scholar] [CrossRef] [PubMed]
- Perona, J.J.; Craik, C.S. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995, 4, 337–360. [Google Scholar] [CrossRef]
- Bao, Y.Y.; Qin, X.; Yu, B.; Chen, L.B.; Wang, Z.C.; Zhang, C.X. Genomic insights into the serine protease gene family and expression profile analysis in the planthopper, Nilaparvata lugens. BMC Genom. 2014, 15, 507. [Google Scholar] [CrossRef]
- Ross, J.; Jiang, H.; Kanost, M.; Wanga, Y. Serine proteases and their homologs in the Drosophila melanogaster genome: An initial analysis of sequence conservation and phylogenetic relationships. Gene 2003, 304, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Jiang, H. Building a platform for predicting functions of serine protease-related proteins in Drosophila melanogaster and other insects. Insect Biochem. Mol. Biol. 2018, 103, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Gulati, M.; Jiang, H. Serine protease-related proteins in the malaria mosquito, Anopheles gambiae. Insect Biochem. Mol. Biol. 2017, 88, 48–62. [Google Scholar] [CrossRef] [PubMed]
- Brackney, D.E.; Isoe, J.; Black, W.C.; Zamora, J.; Foy, B.D.; Miesfeld, R.L.; Olson, K.E. Expression profiling and comparative analyses of seven midgut serine proteases from the yellow fever mosquito, Aedes aegypti. J. Insect Physiol. 2010, 56, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Soares, T.S.; Watanabe, R.M.O.; Lemos, F.J.A.; Tanaka, A.S. Molecular characterization of genes encoding trypsinlike enzymes from Aedes aegypti larvae and identification of digestive enzymes. Gene 2011, 489, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; He, Y.; Hu, Y.; Zhang, X.; Wang, Y.; Zou, Z.; Chen, Y.; Blissard, G.W.; Kanost, M.R.; Jiang, H. Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem. Mol. Biol. 2015, 62, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Cao, X.; Jiang, H. Digestion-related proteins in the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 2020, 126, 103457. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, G.H.; Dong, Z.M.; Duan, J.; Xu, P.Z.; Cheng, T.C.; Xiang, Z.H.; Xia, Q.Y. Genome-wide identification and expression analysis of serine proteases and homologs in the silkworm Bombyx mori. BMC Genom. 2010, 11, 405. [Google Scholar] [CrossRef]
- Liu, H.; Heng, J.; Wang, L.; Tang, X.; Guo, P.; Li, Y.; Xia, Q.; Zhao, P. Identification, characterization, and expression analysis of clip-domain serine protease genes in the silkworm, Bombyx mori. Dev. Comp. Immunol. 2020, 105, 103584. [Google Scholar] [CrossRef]
- Lin, H.; Xia, X.; Yu, L.; Vasseur, L.; Gurr, G.M.; Yao, F.; Yang, G.; You, M. Genome-wide identification and expression profiling of serine proteases and homologs in the diamondback moth, Plutella xylostella (L.). BMC Genom. 2015, 16, 1054. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xing, B.Q.; Wang, L.K.; Yuan, L.L.; Manzoor, M.; Li, F.; Wu, S. Identification of serine protease, serine protease homolog and prophenoloxidase genes in Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 2021, 24, 1144–1152. [Google Scholar] [CrossRef]
- Zou, Z.; Lopez, D.L.; Kanost, M.R.; Evans, J.D.; Jiang, H. Comparative analysis of serine protease-related genes in the honey bee genome: Possible involvement in embryonic development and innate immunity. Insect Mol. Biol. 2006, 15, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lin, Z.; Fang, Q.; Wang, J.; Yan, Z.; Zou, Z.; Song, Q.; Ye, G. The genomic and transcriptomic analyses of serine proteases and their homologs in an endoparasitoid, Pteromalus puparum. Dev. Comp. Immunol. 2017, 77, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Oppert, B.; Muszewska, A.; Steczkiewicz, K.; Šatović-Vukšić, E.; Plohl, M.; Fabrick, J.A.; Vinokurov, K.S.; Koloniuk, I.; Johnston, J.S.; Smith, T.P.L.; et al. The Genome of Rhyzopertha dominica (Fab.) (Coleoptera: Bostrichidae): Adaptation for Success. Genes 2022, 13, 446. [Google Scholar] [CrossRef] [PubMed]
- Tribolium Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, S.; Chen, M.-S.; Elpidina, E.N.; Vinokurov, K.S.; Smith, C.M.; Marshall, J.; Oppert, B. Sequence analysis and molecular characterization of larval midgut cDNA transcripts encoding peptidases from the yellow mealworm, Tenebrio molitor L. Insect Mol. Biol. 2007, 16, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Tsybina, T.A.; Dunaevsky, Y.E.; Belozersky, M.A.; Zhuzhikov, D.P.; Oppert, B.; Elpidina, E.N. Digestive proteinases of yellow mealworm (Tenebrio molitor) larvae: Purification and characterization of a trypsin-like proteinase. Biochemistry 2005, 70, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Elpidina, E.N.; Tsybina, T.A.; Dunaevsky, Y.E.; Belozersky, M.A.; Zhuzhikov, D.P.; Oppert, B. A chymotrypsin-like proteinase from the midgut of Tenebrio molitor larvae. Biochimie 2005, 87, 771–779. [Google Scholar] [CrossRef]
- Oppert, B.; Dowd, S.E.; Bouffard, P.; Li, L.; Conesa, A.; Lorenzen, M.D.; Toutges, M.; Marshall, J.; Huestis, D.L.; Fabrick, J.; et al. Transcriptome profiling of the intoxication response of Tenebrio molitor larvae to Bacillus thuringiensis Cry3Aa protoxin. PLoS ONE 2012, 7, e34624. [Google Scholar] [CrossRef]
- Zhiganov, N.I.; Tereshchenkova, V.F.; Oppert, B.; Filippova, I.Y.; Belyaeva, N.V.; Dunaevsky, Y.E.; Belozersky, M.A.; Elpidina, E.N. The dataset of predicted trypsin serine peptidases and their inactive homologs in Tenebrio molitor transcriptomes. Data Brief 2021, 38, 107301. [Google Scholar] [CrossRef] [PubMed]
- Gorbunov, A.A.; Akentyev, F.I.; Gubaidullin, I.I.; Zhiganov, N.I.; Tereshchenkova, V.F.; Elpidina, E.N.; Kozlov, D.G. Biosynthesis and Secretion of Serine Peptidase SerP38 from Tenebrio molitor in the Yeast Komagataella kurtzmanii. Appl. Biochem. Microbiol. 2021, 57, 917–924. [Google Scholar] [CrossRef]
- Tereshchenkova, V.F.; Zhiganov, N.I.; Akentyev, P.I.; Gubaidullin, I.I.; Kozlov, D.G.; Belyaeva, N.V.; Filippova, I.Y.; Elpidina, E.N. Preparation and properties of the recombinant Tenebrio molitor SerPH122—Proteolytically active homolog of serine peptidase. Appl. Biochem. Microbiol. 2021, 57, 579–585. [Google Scholar] [CrossRef]
- Wu, C.Y.; Xiao, K.R.; Wang, L.Z.; Wang, J.; Song, Q.S.; Stanley, D.; Wei, S.J.; Zhu, J.Y. Identification and expression profiling of serine protease-related genes in Tenebrio molitor. Arch. Insect Biochem. Physiol. 2022, 111, e21963. [Google Scholar] [CrossRef] [PubMed]
- Errico, S.; Spagnoletta, A.; Verardi, A.; Moliterni, S.; Dimatteo, S.; Sangiorgio, P. Tenebrio molitor as a source of interesting natural compounds, their recovery processes, biological effects, and safety aspects. Compr. Rev. Food Sci. Food Saf. 2022, 21, 148–197. [Google Scholar] [CrossRef] [PubMed]
- Moncada-Pazos, A.; Cal, S.; Lopez-Otín, C. Polyserases. In Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: London, UK, 2013; pp. 2990–2994. [Google Scholar]
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Botos, I.; Meyer, E.; Nguyen, M.; Swanson, S.M.; Koomen, J.M.; Russell, D.H.; Meyer, E.F. The structure of an insect chymotrypsin. J. Mol. Biol. 2000, 298, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J. Introduction: Serine peptidases and their clans. In Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: London, UK, 2013; pp. 2491–2523. [Google Scholar]
- Baird, T.T., Jr.; Craik, C.S. Trypsin. In Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: London, UK, 2013; pp. 2594–2600. [Google Scholar]
- Kanost, M.R.; Jiang, H. Clip-domain serine proteases as immune factors in insect hemolymph. Curr. Opin. Insect Sci. 2015, 11, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.R.; Sato, P.M.; Terra, W.R. Insect chymotrypsins: Chloromethyl ketone inactivation and substrate specificity relative to possible coevolutional adaptation of insects and plants. Arch. Insect Biochem. Physiol. 2009, 70, 188–203. [Google Scholar] [CrossRef]
- Whitworth, S.T.; Blum, M.S.; Travis, J. Proteolytic enzymes from larvae of the fire ant, Solenopsis invicta. Isolation and characterization of four serine endopeptidases. J. Biol. Chem. 1998, 273, 14430–14434. [Google Scholar] [CrossRef]
- Tereshchenkova, V.F.; Zhiganov, N.I.; Gubaeva, A.S.; Akentyev, F.I.; Dunaevsky, Y.E.; Kozlov, D.G.; Belozersky, M.A.; Elpidina, E.N. Characteristics of recombinant chymotrypsin-like peptidase from the midgut of Tenebrio molitor larvae. Appl. Biochem. Microbiol. 2024, 60, 420–430. [Google Scholar] [CrossRef]
- Tsu, C.A.; Perona, J.J.; Schellenberger, V.; Turck, C.W.; Craik, C.S. The substrate specificity of Uca pugilator collagenolytic serine protease 1 correlates with the bovine type I collagen cleavage sites. J. Biol. Chem. 1994, 269, 19565–19572. [Google Scholar] [CrossRef] [PubMed]
- Tsu, C.A.; Craik, C.S. Substrate recognition by recombinant serine collagenase 1 from Uca pugilator. J. Biol. Chem. 1996, 271, 11563–11570. [Google Scholar] [CrossRef] [PubMed]
- Bode, W.; Meyer, E., Jr.; Powers, J.C. Human leukocyte and porcine pancreatic elastase: X-ray crystal structures, mechanism, substrate specificity, and mechanism-based inhibitors. Biochemistry 1989, 28, 1951–1963. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, E.B.; Salgado, M.C.O. Pancreatic elastases. In Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: London, UK, 2013; pp. 2639–2645. [Google Scholar] [CrossRef]
- DeLotto, R. Gastrulation defective, a complement factor C2/B-like protease, interprets a ventral prepattern in Drosophila. EMBO Rep. 2001, 2, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, S.L.; Fischer, K. Pseudoproteases: Mechanisms and function. Biochem. J. 2015, 468, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Cal, S.; Moncada-Pazos, A.; Lopez-Otin, C. Expanding the complexity of the human degradome: Polyserases and their tandem serine protease domains. Front. Biosci. 2007, 12, 4661–4669. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Skinner, M.L.; Kauffman, S.W.; Chao, J.; Chao, L.; Thaler, C.D.; Chai, K.X. Prostasin is a glycosylphosphatidylinositol-anchored active serine protease. J. Biol. Chem. 2001, 276, 21434–21442. [Google Scholar] [CrossRef] [PubMed]
- Scarman, A.L.; Hooper, J.D.; Boucaut, K.J.; Sit, M.; Webb, G.C.; Normyle, J.F.; Antalis, T.M. Organization and chromosomal localization of the murine Testisin gene encoding a serine protease temporally expressed during spermatogenesis. Eur. J. Biochem. 2001, 268, 1250–1258. [Google Scholar] [CrossRef]
- Rickert, K.W.; Kelley, P.; Byrne, N.J.; Diehl, R.E.; Hall, D.L.; Montalvo, A.M.; Reid, J.C.; Shipman, J.M.; Thomas, B.W.; Munshi, S.K.; et al. Structure of human prostasin, a target for the regulation of hypertension. J. Biol. Chem. 2008, 283, 34864–34872. [Google Scholar] [CrossRef]
- Lee, K.Y.; Zhang, R.; Kim, M.S.; Park, J.W.; Park, H.Y.; Kawabata, S.; Lee, B.L. A zymogen form of masquerade-like serine proteinase homologue is cleaved during pro-phenoloxidase activation by Ca2+ in coleopteran and Tenebrio molitor larvae. Eur. J. Biochem. 2002, 269, 4375–4383. [Google Scholar] [CrossRef]
- Piao, S.; Song, Y.-L.; Kim, J.H.; Park, S.Y.; Park, J.W.; Lee, B.L.; Oh, B.-H.; Ha, N.-C. Crystal structure of a clip-domain serine protease and functional roles of the clip domains. EMBO J. 2005, 24, 4404–4414. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Lu, Z.; Dai, H.; Velde, D.V.; Prakash, O.; Jiang, H. The solution structure of clip domains from Manduca sexta prophenoloxidase activating proteinase-2. Biochemistry 2007, 46, 11431–11439. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Kim, S.J.; Kan, H.; Kwon, H.M.; Roh, K.B.; Jiang, R.; Yang, Y.; Park, J.W.; Lee, H.H.; Ha, N.C.; et al. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J. Biol. Chem. 2008, 283, 7599–7607. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, Y.; Yang, F.; Jiang, H. Manduca sexta hemolymph protease-1, activated by an unconventional non-proteolytic mechanism, mediates immune responses. Insect Biochem. Mol. Biol. 2017, 84, 23–31. [Google Scholar] [CrossRef]
- Bork, P.; Beckmann, G. The CUB domain: A widespread module in developmentally regulated proteins. J. Mol. Biol. 1993, 231, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Font, B.; Eichenberger, D.; Moreau, C.; Ricard-Blum, S.; Hulmes, D.J.; Moali, C. Insights into how CUB domains can exert specific functions while sharing a common fold: Conserved and specific features of the CUB1 domain contribute to the molecular basis of procollagen C-proteinase enhancer-1 activity. J. Biol. Chem. 2007, 282, 16924–16933. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Kim, C.H.; Kim, J.H.; Je, B.R.; Roh, K.B.; Kim, S.J.; Lee, H.H.; Ryu, J.H.; Lim, J.H.; Oh, B.H.; et al. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc. Natl. Acad. Sci. USA 2007, 104, 6602–6607. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.S.; Stevens, L.M.; Sieverman, K.J.; Nguyen, J.; Stein, D. A ventrally localized protease in the Drosophila egg controls embryo dorsoventral polarity. Curr. Biol. 2012, 22, 1013–1018. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false Discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Keller, M.; Sneh, B.; Strizhov, N.; Prudovsky, E.; Regev, A.; Koncz, C.; Schell, J.; Zilberstein, A. Digestion of δ-endotoxin by gut proteases may explain reduced sensitivity of advanced instar larvae of Spodoptera littoralis to CryIC. Insect Biochem. Mol. Biol. 1996, 26, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Zalunin, I.A.; Elpidina, E.N.; Oppert, B. The role of proteolysis in the biological activity of Bt insecticidal crystal proteins. In Bt Resistance—Characterization and Strategies for GM Crops Producing Bacillus thuringiensis Toxins; Soberón, M., Gao, Y., Bravo, A., Eds.; CAB International Publishers: Wallingford, UK, 2015; pp. 107–118. [Google Scholar]
- Oppert, B.; Elpidina, E.N.; Toutges, M.; Mazumdar-Leighton, S. Microarray analysis reveals strategies of Tribolium castaneum larvae to compensate for cysteine and serine protease inhibitors. Comp. Biochem. Physiol. D Genom. Proteom. 2010, 5, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Martynov, A.G.; Elpidina, E.N.; Perkin, L.; Oppert, B. Functional analysis of C1 family cysteine peptidases in the larval gut of Tenebrio molitor and Tribolium castaneum. BMC Genom. 2015, 16, 75. [Google Scholar] [CrossRef] [PubMed]
- Broehan, G.; Arakane, Y.; Beeman, R.W.; Kramer, K.J.; Muthukrishnan, S.; Merzendorfer, H. Chymotrypsin-like peptidases from Tribolium castaneum: A role in molting revealed by RNA interference. Insect Biochem. Mol. Biol. 2010, 40, 274–283. [Google Scholar] [CrossRef] [PubMed]
- LeMosy, E.K.; Tan, Y.Q.; Hashimoto, C. Activation of a protease cascade involved in patterning the Drosophila embryo. Proc. Natl. Acad. Sci. USA 2001, 98, 5055–5060. [Google Scholar] [CrossRef] [PubMed]
- Muta, T.; Hashimoto, R.; Miyata, T.; Nishimura, H.; Toh, Y.; Iwanaga, S. Proclotting enzyme from horseshoe crab hemocytes. cDNA cloning, disulfide locations, and subcellular localization. J. Biol. Chem. 1990, 265, 22426–22433. [Google Scholar] [CrossRef]
- Munier, A.I.; Medzhitov, R.; Janeway, C.A.; Doucet, D.; Capovilla, M.; Lagueux, M. Graal: A Drosophila gene coding for several mosaic serine proteases. Insect Biochem. Mol. Biol. 2004, 34, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. 1999, 41, 95–98. [Google Scholar]
- Kaur, S.; Stinson, S.A.; di Cenzo, G.C. Whole genome assemblies of Zophobas morio and Tenebrio molitor. G3 2023, 13, jkad079. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, T.; Andere, A.A.; Kelstrup, H.; Emery, V.J.; Picard, C.J. The yellow mealworm (Tenebrio molitor) genome: A resource for the emerging insects as food and feed industry. J. Insects Food Feed 2020, 6, 445–455. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Lomize, A.L.; Hage, J.M.; Pogozheva, I.D. Membranome 2.0: Database for proteome-wide profiling of bitopic proteins and their dimers. Bioinformatics 2018, 34, 1061–1062. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Tool kit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Statistics Kingdom. Available online: https://www.statskingdom.com/index.html (accessed on 1 April 2024).


| № | Name | NCBI ID (Protein) | Preproenzyme/Mature Enzyme (aa) | SignalP (aa) | Regulatory Domain | Propeptide Cleavage Site | Active Site | S1 Subsite | Enzyme Specificity | Mm Mature, Da | pI | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SerP16 | CAH1383061 | 275 | 246 | 16 | - | H|ITNG | H | D | S | S | G | S | Chymotrypsin-like | 25,749 | 3.9 |
| 2 | SerP69 | ABC88746 | 275 | 230 | 16 | - | R|IISG | H | D | S | S | G | S | Chymotrypsin-like | 22,899 | 8.8 |
| 3 | SerP71 | CAG9035017 | 271 | 235 | 21 | - | R|IING | H | D | S | S | G | A | Chymotrypsin-like | 24,308 | 4.1 |
| 4 | SerP303 | CAG9018553 | 281 | 237 | 18 | - | R|ITGG | H | D | S | S | G | A | Chymotrypsin-like | 25,047 | 4.2 |
| 5 | SerP129 | CAH1365737 | 265 | 230 | 18 | - | R|IISG | H | D | S | G | A | S | Chymotrypsin-like | 24,439 | 4.0 |
| 6 | SerP314 | ABC88747 | 266 | 232 | 16 | - | R|IVGG | H | D | S | G | A | S | Chymotrypsin-like | 24,475 | 4.2 |
| 7 | SerP7 | CAG9037665 | 279 | 246 | 16 | - | R|IING | H | D | S | G | G | S | Chymotrypsin-like | 25,707 | 3.9 |
| 8 | SerP39 | CAG9029806 | 267 | 225 | 16 | - | R|IIGG | H | D | S | G | G | S | Chymotrypsin-like | 23,838 | 4.3 |
| 9 | SerP54 | CAH1375188 | 276 | 233 | 16 | - | R|IIGG | H | D | S | G | G | S | Chymotrypsin-like | 24,736 | 4.0 |
| 10 | SerP107 | WJL97990 | 277 | 234 | 16 | - | R|IIGG | H | D | S | G | G | S | Chymotrypsin-like | 25,428 | 4.3 |
| 11 | SerP108 | CAH1375189 | 276 | 233 | 16 | - | R|IIGG | H | D | S | G | G | S | Chymotrypsin-like | 24,987 | 3.8 |
| 12 | SerP246 | CAH1375190 | 275 | 233 | 16 | - | R|IIGG | H | D | S | G | G | S | Chymotrypsin-like | 24,822 | 3.9 |
| 13 | SerP253 | CAH1367742 | 277 | 241 | 21 | - | R|IIGG | H | D | S | G | G | S | Chymotrypsin-like | 25,998 | 4.1 |
| 14 | SerP479 | WJL97991 | 276 | 233 | 16 | - | R|IIGG | H | D | S | G | G | S | Chymotrypsin-like | 25,012 | 4.2 |
| 15 | SerP33 | WJL97992 | 256 | 217 | 24 | - | R|IVGG | H | D | S | G | S | G | Chymotrypsin-like | 22,618 | 4.2 |
| 16 | SerP251 | CAH1372320 | 255 | 232 | 17 | - | R|IIVG | H | D | S | G | S | G | Chymotrypsin-like | 24,576 | 5.2 |
| 17 | SerP101 | ABC88734 | 258 | 235 | 17 | - | R|IVNG | H | D | S | G | S | G | Chymotrypsin-like | 25,014 | 6.6 |
| 18 | SerP19 | WJL97993 | 252 | 227 | 16 | - | R|IVGG | H | D | S | S | S | G | Chymotrypsin-like | 23,900 | 4.5 |
| 19 | SerP38 | QRE01764 | 258 | 229 | 16 | - | R|VVGG | H | D | S | G | G | D | Chymotrypsin-like | 24,410 | 5.3 |
| 20 | SerP88 | ABC88737 | 258 | 229 | 18 | - | R|VVGG | H | D | S | G | G | D | Chymotrypsin-like | 24,896 | 5.3 |
| 21 | SerP226 | WJL97994 | 258 | 221 | 22 | - | R|LIGG | H | D | S | G | G | D | Chymotrypsin-like | 23,606 | 4.2 |
| 22 | SerP146 | CAH1383003 | 262 | 222 | 18 | - | R|IVGG | H | D | S | G | G | D | Chymotrypsin-like | 23,993 | 4.5 |
| 23 | SerP276 | KAJ3628034 | 284 | 247 | 15 | - | R|IIHG | H | D | S | G | G | D | Chymotrypsin-like | 27,432 | 6.9 |
| 24 | SerP301 | CAH1380401 | 244 | 221 | 17 | - | R|IFGG | H | D | S | G | S | D | Chymotrypsin-like | 23,620 | 4.1 |
| 25 | SerP368 | WJL97995 | 247 | 233 | - | - | R|IFGG | H | D | S | A | G | D | Chymotrypsin-like | 24,560 | 4.2 |
| 26 | SerP137 | CAH1379909 | 248 | 218 | 19 | - | K|IVGG | H | D | S | A | G | D | Chymotrypsin-like | 23,683 | 5.4 |
| 27 | SerP484 | CAH1368908 | 247 | 226 | 16 | - | R|IVGG | H | D | S | A | G | D | Chymotrypsin-like | 24,734 | 5.0 |
| 28 | SerP215 | CAH1380399 | 254 | 231 | 17 | - | R|IFGG | H | D | S | G | A | D | Chymotrypsin-like | 24,822 | 4.4 |
| 29 | SerP586 | KAJ3636193 | 270 | 224 | - | - | L|KDNG | H | D | S | T | G | S | Chymotrypsin-like | 24,961 | 5.0 |
| 30 | SerP449 MSP | BAG14264 | 632 | 258 | 23 | LDL (4), Sushi | L|IVNG | H | D | S | S | S | G | Chymotrypsin-like | 28,757 | 6.4 |
| № | Name | NCBI ID (Protein) | Preproenzyme/Mature Enzyme (aa) | SignalP (aa) | Regulatory Domain | Propeptide Cleavage Site | Active Site | S1 Subsite | Enzyme Specificity | Mm Mature, Da | pI | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SerP41 | ABC88760 | 266 | 233 | 19 | - | R|IVGG | H | D | S | G | I | S | Elastase-like | 25,006 | 4.4 |
| 2 | SerP121 | CAH1368236 | 274 | 236 | 16 | - | R|IIGG | H | D | S | G | I | S | Elastase-like | 26,285 | 4.5 |
| 3 | SerP144 | WJL97996 | 268 | 234 | 19 | - | R|IIGG | H | D | S | G | I | S | Elastase-like | 25,448 | 4.4 |
| 4 | SerP238 | KAJ3632560 | 264 | 234 | 21 | - | R|IVGG | H | D | S | G | I | S | Elastase-like | 25,330 | 4.2 |
| 5 | SerP441 | KAJ3632561 | 267 | 234 | 22 | - | R|IIGG | H | D | S | G | I | S | Elastase-like | 25,072 | 4.3 |
| 6 | SerP156 | CAH1380384 | 267 | 236 | 19 | - | R|IING | H | D | S | A | V | S | Elastase-like | 25,326 | 4.6 |
| 7 | SerP94 | WJL97997 | 266 | 232 | 21 | - | H|IVAG | H | D | S | G | V | N | Elastase-like | 24,874 | 4.8 |
| 8 | SerP120 | WJL97998 | 268 | 232 | 19 | - | H|IILG | H | D | S | G | V | S | Elastase-like | 24,988 | 4.7 |
| 9 | SerP288 | CAH1375483 | 266 | 232 | 16 | - | R|IVGG | H | D | S | G | V | S | Elastase-like | 24,259 | 4.0 |
| 10 | SerP472 | CAH1380701 | 272 | 235 | 17 | - | R|IVNG | H | D | S | S | V | A | Elastase-like | 25,265 | 4.4 |
| 11 | SerP73 | KAJ3638657 | 267 | 232 | 16 | - | R|IING | H | D | S | S | V | S | Elastase-like | 24,485 | 4.1 |
| 12 | SerP74 | KAH0820461 | 261 | 229 | 16 | - | R|IING | H | D | S | S | V | S | Elastase-like | 23,423 | 8.6 |
| 13 | SerP110 | KAH0813654 | 266 | 231 | 16 | - | R|IING | H | D | S | S | V | S | Elastase-like | 24,831 | 4.2 |
| 14 | SerP98 | CAH1365740 | 267 | 232 | 16 | - | R|IING | H | D | S | S | V | S | Elastase-like | 24,969 | 4.2 |
| 15 | SerP751 | CAH1365741 | 267 | 232 | 16 | - | R|IING | H | D | S | S | V | S | Elastase-like | 24,360 | 4.0 |
| 16 | SerP185 | KAJ3620429 | 266 | 233 | 17 | - | R|IING | H | D | S | S | T | S | Elastase-like | 24,779 | 4.9 |
| 17 | SerP155 | KAJ3632649 | 265 | 235 | 16 | - | R|IIGG | H | D | S | G | F | S | Elastase-like | 24,905 | 4.4 |
| 18 | SerP85 | ABC88761 | 267 | 237 | 16 | - | R|IIGG | H | D | S | G | Y | S | Elastase-like | 25,364 | 4.3 |
| № | Name | NCBI ID (Protein) | Preproenzyme/Mature Enzyme (aa) | SignalP (aa) | Regulatory Domain | Propeptide Cleavage Site | Active Site | S1 Subsite | Enzyme Specificity | Mm Mature, Da | pI | TM (Position) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SerP18 | WJL97999 | 258 | 228 | 16 | - | K|IVWG | H | D | S | A | A | T | NA | 24,375 | 8.4 | - |
| 2 | SerP169 | CAH1378761 | 257 | 226 | 16 | - | K|IVGG | H | D | S | G | A | T | NA | 24,300 | 9.9 | - |
| 3 | SerP423 | CAH1372319 | 279 | 257 | 17 | - | R|IVNG | H | D | S | G | G | K | NA | 28,256 | 4.5 | - |
| 4 | SerP416 | KAH0820967 | 300 | - | 23 | - | - | H | D | S | Q | G | S | NA | - | - | 277–299 |
| 5 | SerP378 | WJL98000 | 357 | 253 | 22 | - | K|ISGG | H | D | S | R | G | I | NA | 28,551 | 8.1 | - |
| 6 | SerP424 | KAJ3633461 | 250 | 227 | 18 | - | R|IIGG | H | D | S | R | G | V | NA | 25,210 | 7.0 | - |
| 7 | SerP462 | KAH0817404 | 257 | - | 23 | - | - | H | D | S | T | S | F | NA | - | - | - |
| 8 | SerP653 | CAH1380361 | 252 | - | 16 | - | - | H | D | S | V | A | D | NA | - | - | - |
| 9 | SerP355 | WJL98001 | 551 | 267 | 19 | LDL (4), Sushi | L|IVNG | H | D | S | G | S | T | NA | 29,980 | 5.1 | - |
| 10 | SerP1040 | WJL98002 | 432 | 263 | 22 | Sushi | L|IING | H | D | S | S | S | S | NA | 27,132 | 7.7 | - |
| 11 | SerP454 | CAH1384889 | 476 | 257 | 15 | GD | L|ITHG | H | D | S | S | S | V | NA | 28,642 | 7.8 | - |
| 12 | SerP442 | CAH1384890 | 561 | 257 | 17 | GD | L|ISYG | H | D | S | T | G | I | NA | 28,750 | 7.7 | - |
| 13 | SerP466 | KAJ3628554 | 427 | 247 | 23 | GD | K|PANE | H | D | S | S | G | V | NA | 27,618 | 7.3 | - |
| 14 | SerP550 | CAH1380129 | 447 | 249 | 18 | GD | L|VLKG | H | D | S | G | A | I | NA | 27,949 | 8.9 | - |
| 15 | SerP1035 | CAH1380127 | 568 | 249 | 25 | GD | L|VVNG | H | D | S | G | S | V | NA | 27,582 | 9.7 | - |
| № | Name | NCBI ID (Protein) | Preproenzyme (aa) | SignalP (aa) | Regulatory Domain | Propeptide Cleavage Site | Active Site | S1 Subsite | Enzyme Specificity | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | pSerP448 | WKK29891 | 892 | 20 | Sushi (2) | L|IVGG | H | D | S | S | S | G | Chymotrypsin-like |
| Sushi (2) | L|IVKG | H | D | A | S | S | A | SPH | |||||
| 2 | pSerP900 | CAH1380589 | 891 | 22 | Sushi (2) | L|IVGG | H | D | S | S | S | G | Chymotrypsin-like |
| Sushi (2) | L|IVKG | H | D | A | S | S | A | SPH | |||||
| 3 | pSerP333 | CAH1382424 | 891 | 24 | Sushi (2) | L|IVSG | H | D | S | S | S | G | Chymotrypsin-like |
| Sushi (2) | L|IVNG | R | N | V | F | Q | V | SPH | |||||
| 4 | pSerP382 | WKK29892 | 837 | 23 | Sushi (2) | L|IVGG | H | D | S | S | A | G | Chymotrypsin-like |
| Sushi (2) | L|IIGG | Q | D | R | I | S | G | SPH | |||||
| 5 | pSerPH608 | WKK29893 | 895 | 23 | Sushi (2) | L|IVGG | H | D | G | S | S | G | SPH |
| Sushi (2) | L|IIGG | Y | D | G | S | F | T | SPH | |||||
| 6 | pSerP614 | WKK29894 | 1347 | 24 | Sushi (2) | L|IVNG | H | D | S | S | S | A | Chymotrypsin-like |
| Sushi (2) | L|IING | H | D | G | S | S | S | SPH | |||||
| Sushi | L|IVNG | Q | D | S | A | S | A | SPH | |||||
| 7 | pSerP1050 Nudel | CAH1374346 | 1830 | TM (58–80) | LDL (7) | R|VVGG | H | D | S | D | G | G | Trypsin |
| N|ITSQ | T | E | D | D | S | A | SPH | ||||||
| Expression, RPKM | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence Name | Regulatory Domains | Active Site | S1 Subsite | Annotation of Sequence | Eggs | Larvae II | Larvae IV | Early Pupae | Late Pupae | Males | Females | ||||
| SerPH236 | Clip_A | H | D | G | D | G | G | SPH | 1001 | 27 | 26 | 16 | 57 | 8 | 9 |
| SerPH235 | Clip_A | H | D | G | D | G | G | SPH | 518 | 16 | 360 | 33 | 44 | 4 | 71 |
| SerPH203 | Clip_B | H | D | G | D | G | A | SPH | 357 | 0 | 0 | 0 | 0 | 0 | 0 |
| SerP166 | Clip_B | H | D | S | D | G | G | Trypsin | 344 | 0 | 0 | 0 | 0 | 0 | 0 |
| SerPH165 | Clip_A | H | D | G | D | G | G | SPH | 331 | 4 | 1 | 3 | 4 | 2 | 2 |
| SerP116 | Clip_B | H | D | S | D | G | G | Trypsin | 329 | 2 | 3 | 6 | 3 | 13 | 9 |
| SerP145 | Clip_C | H | D | S | D | G | G | Trypsin | 150 | 53 | 37 | 61 | 89 | 53 | 57 |
| SerP28 | - | H | D | S | D | G | G | Trypsin | 147 | 114 | 13 | 32 | 186 | 0 | 1 |
| SerP466 | GD | H | D | S | S | G | V | N/A | 122 | 10 | 26 | 37 | 139 | 6 | 62 |
| SerP61 | Clip_C | H | D | S | D | G | A | Trypsin-like | 119 | 41 | 44 | 41 | 60 | 23 | 32 |
| SerP156 | - | H | D | S | A | V | S | Elastase-like | 65 | 68 | 321 | 0 | 106 | 113 | 91 |
| SerP454 | GD | H | D | S | S | S | V | N/A | 61 | 65 | 25 | 22 | 93 | 24 | 23 |
| SerP550 | GD | H | D | S | G | A | I | N/A | 61 | 56 | 32 | 34 | 54 | 13 | 25 |
| SerP442 | GD | H | D | S | T | G | I | N/A | 53 | 19 | 16 | 12 | 45 | 24 | 21 |
| SerPH389 Scarface | Clip_A_ | H | D | Y | D | D | G | SPH | 51 | 200 | 29 | 42 | 27 | 27 | 43 |
| SerP5 | - | H | D | S | D | G | G | Trypsin | 47 | 17 | 0 | 0 | 9 | 0 | 0 |
| SerP22 | - | H | D | S | D | G | G | Trypsin | 34 | 6 | 0 | 7 | 0 | 0 | 0 |
| Expression, RPKM | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence Name | Regulatory Domains | Active Site | S1 Subsite | Annotation of Sequence | Eggs | Larvae II | Larvae IV | Early Pupae | Late Pupae | Males | Females | ||||
| SerPH164 | Clip_A | H | D | G | D | G | G | SPH | 6 | 10 | 85 | 88 | 123 | 72 | 78 |
| SerPH1034 | - | Q | N | T | E | E | K | SPH | 9 | 7 | 35 | 70 | 92 | 31 | 53 |
| SerP247 | Clip_C | H | D | S | D | G | G | Trypsin | 9 | 73 | 48 | 65 | 161 | 19 | 34 |
| SerP145 | Clip_C | H | D | S | D | G | G | Trypsin | 150 | 53 | 37 | 61 | 89 | 53 | 57 |
| SerPH159 | Clip_A | H | D | G | D | G | A | SPH | 29 | 9 | 105 | 60 | 121 | 37 | 70 |
| SerPH78 | Clip_A | H | D | G | D | G | G | SPH | 27 | 24 | 11 | 58 | 7 | 0 | 1 |
| SerP35 | - | H | D | S | D | G | G | Trypsin | 4 | 0 | 0 | 58 | 0 | 1 | 0 |
| SerP228 | Clip_C | H | D | S | D | G | G | Trypsin | 17 | 0 | 35 | 52 | 80 | 15 | 14 |
| SerPH364 | SEA; EGF; LDL; SRCR | S | D | E | D | R | R | SPH | 15 | 52 | 7 | 52 | 27 | 16 | 29 |
| SerPH243 | Clip_A | H | D | G | D | G | A | SPH | 42 | 22 | 67 | 51 | 43 | 44 | 51 |
| SerP55 Tequila | CBM (3), LDL (3), SRCR (2) PAN | H | D | S | D | G | G | Trypsin | 5 | 10 | 25 | 51 | 58 | 46 | 40 |
| SerP113 | Clip_B | H | D | S | D | G | G | Trypsin | 27 | 35 | 82 | 49 | 75 | 45 | 34 |
| SerPH223 | Clip_A | H | D | G | D | G | G | SPH | 44 | 15 | 73 | 46 | 86 | 19 | 33 |
| SerPH216 | Clip_A | H | D | G | D | G | G | SPH | 2 | 5 | 50 | 46 | 73 | 64 | 31 |
| SerP11 TSP | TSP (2) | H | D | S | D | G | G | Trypsin | 5 | 23 | 14 | 44 | 72 | 25 | 10 |
| SerP28 | - | H | D | S | D | G | G | Trypsin | 147 | 114 | 13 | 32 | 186 | 0 | 1 |
| SerP247 | Clip_C | H | D | S | D | G | G | Trypsin | 9 | 73 | 48 | 65 | 161 | 19 | 34 |
| SerP466 | GD | H | D | S | S | G | V | N/A | 122 | 10 | 26 | 37 | 139 | 6 | 62 |
| SerPH164 | Clip_A | H | D | G | D | G | G | SPH | 6 | 10 | 85 | 88 | 123 | 72 | 78 |
| SerPH159 | Clip_A | H | D | G | D | G | A | SPH | 29 | 9 | 105 | 60 | 121 | 37 | 70 |
| SerPH415 | Clip_A | H | D | G | D | G | G | SPH | 47 | 0 | 34 | 14 | 118 | 0 | 0 |
| SerP15 | Clip_D | H | D | S | D | G | G | Trypsin | 35 | 142 | 9 | 35 | 113 | 1 | 0 |
| SerP156 | - | H | D | S | A | V | S | Elastase-like | 65 | 68 | 321 | 0 | 106 | 113 | 91 |
| SerPH680 | - | H | N | I | S | G | T | SPH | 17 | 7 | 115 | 30 | 98 | 61 | 105 |
| SerPH589 | Clip_C | R | D | S | D | G | A | SPH | 0 | 1 | 47 | 41 | 94 | 43 | 39 |
| SerP454 | GD | H | D | S | S | S | V | N/A | 61 | 65 | 25 | 22 | 93 | 24 | 23 |
| SerPH1034 | - | Q | N | T | E | E | K | SPH | 9 | 7 | 35 | 70 | 92 | 31 | 53 |
| SerP145 | Clip_C | H | D | S | D | G | G | Trypsin | 150 | 53 | 37 | 61 | 89 | 53 | 57 |
| SerPH223 | Clip_A | H | D | G | D | G | G | SPH | 44 | 15 | 73 | 46 | 86 | 19 | 33 |
| SerPH618 | - | S | D | G | V | Q | G | SPH | 26 | 27 | 0 | 2 | 85 | 0 | 0 |
| Expression, RPKM | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Active Site | Annotation of Sequence | S1 Subsite | Eggs | Larvae II | Larvae IV | Early Pupae | Late Pupae | Males | Females | Larval IV Gut | ||||
| SerP1 | H | D | S | Trypsin | D | G | G | 7 | 675 | 16,880 | 6 | 0 | 2563 | 2646 | 10,480 |
| SerP69 | H | D | S | Chymotrypsin-like | S | G | S | 0 | 37 | 2574 | 0 | 0 | 782 | 695 | 4107 |
| SerP108 | H | D | S | Chymotrypsin-like | G | G | S | 0 | 6151 | 2092 | 0 | 0 | 191 | 152 | 6195 |
| SerP54 | H | D | S | Chymotrypsin-like | G | G | S | 0 | 255 | 1934 | 0 | 0 | 207 | 299 | 2102 |
| SerP314 | H | D | S | Chymotrypsin-like | G | A | S | 0 | 1207 | 1087 | 0 | 0 | 413 | 312 | 5564 |
| SerP38 | H | D | S | Chymotrypsin-like | G | G | D | 0 | 4 | 976 | 0 | 0 | 0 | 26 | 1517 |
| SerP209 | H | D | S | Trypsin | D | G | G | 0 | 0 | 765 | 0 | 0 | 0 | 0 | 494 |
| SerP303 | H | D | S | Chymotrypsin-like | S | G | A | 0 | 450 | 736 | 0 | 0 | 718 | 770 | 419 |
| SerP41 | H | D | S | Elastase-like | G | I | S | 0 | 0 | 678 | 0 | 0 | 403 | 294 | 8062 |
| SerP16 | H | D | S | Chymotrypsin-like | S | G | S | 0 | 1193 | 512 | 0 | 0 | 114 | 270 | 151 |
| SerP246 | H | D | S | Chymotrypsin-like | G | G | S | 0 | 11 | 481 | 0 | 0 | 20 | 41 | 511 |
| SerP85 | H | D | S | Elastase-like | G | Y | S | 0 | 158 | 465 | 0 | 0 | 93 | 99 | 1394 |
| SerP185 | H | D | S | Elastase-like | S | T | S | 0 | 0 | 425 | 0 | 0 | 117 | 72 | 2417 |
| SerP71 | H | D | S | Chymotrypsin-like | S | G | A | 0 | 30 | 378 | 0 | 0 | 147 | 100 | 991 |
| SerP253 | H | D | S | Chymotrypsin-like | G | G | S | 0 | 160 | 331 | 6 | 27 | 256 | 271 | 517 |
| SerP156 | H | D | S | Elastase-like | A | V | S | 65 | 68 | 321 | 0 | 106 | 113 | 91 | 220 |
| SerP39 | H | D | S | Chymotrypsin-like | G | G | S | 0 | 52 | 220 | 0 | 0 | 1877 | 1324 | 718 |
| SerP251 | H | D | S | Chymotrypsin-like | G | S | G | 0 | 0 | 217 | 0 | 0 | 10 | 6 | 1164 |
| SerP74 | H | D | S | Elastase-like | S | V | S | 0 | 4 | 146 | 0 | 0 | 64 | 68 | 615 |
| SerP288 | H | D | S | Elastase-like | G | V | S | 0 | 37 | 140 | 0 | 0 | 788 | 1101 | 1681 |
| SerPH219 | S | D | V | SPH | G | I | S | 0 | 64 | 1243 | 0 | 0 | 282 | 295 | 3687 |
| SerPH384 | Q | D | S | SPH | G | I | S | 0 | 0 | 985 | 0 | 0 | 52 | 98 | 1813 |
| SerPH237 | Q | D | G | SPH | S | I | S | 0 | 0 | 965 | 1 | 0 | 1 | 0 | 355 |
| SerPH239 | Q | D | G | SPH | S | I | S | 0 | 0 | 800 | 0 | 0 | 0 | 2 | 733 |
| SerPH122 | H | D | T | SPH | G | L | S | 0 | 0 | 411 | 5 | 0 | 103 | 125 | 1223 |
| SerPH493 | Q | D | I | SPH | G | V | S | 0 | 9 | 347 | 0 | 0 | 89 | 35 | 991 |
| SerPH562 | Q | D | S | SPH | G | I | S | 0 | 8 | 264 | 0 | 0 | 51 | 67 | 576 |
| SerPH245 | H | D | T | SPH | G | M | T | 0 | 0 | 225 | 0 | 0 | 37 | 41 | 510 |
| SerPH290 | Q | D | M | SPH | G | R | S | 0 | 6 | 207 | 0 | 0 | 41 | 35 | 106 |
| SerPH136 | Q | D | T | SPH | G | L | S | 0 | 7 | 185 | 0 | 0 | 17 | 18 | 490 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhiganov, N.I.; Vinokurov, K.S.; Salimgareev, R.S.; Tereshchenkova, V.F.; Dunaevsky, Y.E.; Belozersky, M.A.; Elpidina, E.N. The Set of Serine Peptidases of the Tenebrio molitor Beetle: Transcriptomic Analysis on Different Developmental Stages. Int. J. Mol. Sci. 2024, 25, 5743. https://doi.org/10.3390/ijms25115743
Zhiganov NI, Vinokurov KS, Salimgareev RS, Tereshchenkova VF, Dunaevsky YE, Belozersky MA, Elpidina EN. The Set of Serine Peptidases of the Tenebrio molitor Beetle: Transcriptomic Analysis on Different Developmental Stages. International Journal of Molecular Sciences. 2024; 25(11):5743. https://doi.org/10.3390/ijms25115743
Chicago/Turabian StyleZhiganov, Nikita I., Konstantin S. Vinokurov, Ruslan S. Salimgareev, Valeriia F. Tereshchenkova, Yakov E. Dunaevsky, Mikhail A. Belozersky, and Elena N. Elpidina. 2024. "The Set of Serine Peptidases of the Tenebrio molitor Beetle: Transcriptomic Analysis on Different Developmental Stages" International Journal of Molecular Sciences 25, no. 11: 5743. https://doi.org/10.3390/ijms25115743
APA StyleZhiganov, N. I., Vinokurov, K. S., Salimgareev, R. S., Tereshchenkova, V. F., Dunaevsky, Y. E., Belozersky, M. A., & Elpidina, E. N. (2024). The Set of Serine Peptidases of the Tenebrio molitor Beetle: Transcriptomic Analysis on Different Developmental Stages. International Journal of Molecular Sciences, 25(11), 5743. https://doi.org/10.3390/ijms25115743






