Development of a New Binary Matrix for the Comprehensive Analysis of Lipids and Pigments in Micro- and Macroalgae Using MALDI-ToF/ToF Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Suitability of Selected Matrices for Lipid Analysis
2.2. Analysis of Wakame Lipid Extract
2.3. Analysis of Micro and Macro Algae Extracts
3. Materials and Methods
3.1. Chemicals
3.2. Instrumentation
3.3. Sample Preparation
3.3.1. Extraction of Lipids
3.3.2. MALDI MS Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chacón-Lee, T.L.; González-Mariño, G.E. Microalgae for “Healthy” Foods-Possibilities and Challenges. Compr. Rev. Food Sci. Food Saf. 2010, 9, 655–675. [Google Scholar] [CrossRef] [PubMed]
- Becker, E.W. Micro-algae as a source of protein. Biotechnol. Adv. 2007, 25, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Morais, E.G.; Schüler, L.; Pereira, H.; Maia, I.; Gangadhar, K.N.; Costa, J.A.V.; Morais, M.G.; Varela, J.; Barreira, L. Microalgae as source of edible lipids. In Cultured Microalgae for the Food Industry: Current and Potential Applications; Academic Press: Cambridge, MA, USA, 2021; pp. 147–175. [Google Scholar] [CrossRef]
- Herminia, D.; Leonel Pereira, S.K. Functional Ingredients from Algae for Foods and Nutraceuticals; Elsevier B.V: Amsterdam, The Netherlands, 2023; ISBN 9780323988193. [Google Scholar]
- Yamada, H.; Yamazaki, Y.; Koike, S.; Hakozaki, M.; Nagahora, N.; Yuki, S.; Yano, A.; Tsurumi, K.; Okumura, T. Lipids, fatty acids and hydroxy-fatty acids of Euphausia pacifica. Sci. Rep. 2017, 7, 9944. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.K.; Prasad, P.; Shang, X.; Keum, Y.-S. Advances in Lipid Extraction Methods—A Review. Int. J. Mol. Sci. 2021, 22, 13643. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Kliman, M.; Claude, E.; Geromanos, S.; Astarita, G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal. Bioanal. Chem. 2015, 407, 4995–5007. [Google Scholar] [CrossRef]
- Criscuolo, A.; Zeller, M.; Fedorova, M. Evaluation of Lipid In-Source Fragmentation on Different Orbitrap-based Mass Spectrometers. J. Am. Soc. Mass Spectrom. 2020, 31, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. TrAC Trends Anal. Chem. 2014, 61, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Kendel, M.; Wielgosz-Collin, G.; Bertrand, S.; Roussakis, C.; Bourgougnon, N.B.; Bedoux, G. Lipid composition, fatty acids and sterols in the seaweeds ulva armoricana, and solieria chordalis from brittany (France): An analysis from nutritional, chemotaxonomic, and antiproliferative activity perspectives. Mar. Drugs 2015, 13, 5606–5628. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Guihéneuf, F.; Stengel, D.B. Fatty acid contents and profiles of 16 macroalgae collected from the Irish Coast at two seasons. J. Appl. Phycol. 2014, 26, 451–463. [Google Scholar] [CrossRef]
- Coniglio, D.; Bianco, M.; Ventura, G.; Calvano, C.D.; Losito, I.; Cataldi, T.R.I. Lipidomics of the edible brown alga wakame (Undaria pinnatifida) by liquid chromatography coupled to electrospray ionization and tandem mass spectrometry. Molecules 2021, 26, 4480. [Google Scholar] [CrossRef]
- Bolik, S.; Albrieux, C.; Schneck, E.; Demé, B.; Jouhet, J. Sulfoquinovosyldiacylglycerol and phosphatidylglycerol bilayers share biophysical properties and are good mutual substitutes in photosynthetic membranes. Biochim. Biophys. Acta-Biomembr. 2022, 1864, 184037. [Google Scholar] [CrossRef]
- Coniglio, D.; Calvano, C.D.; Ventura, G.; Losito, I.; Cataldi, T.R.I. Arsenosugar Phospholipids (As-PL) in Edible Marine Algae: An Interplay between Liquid Chromatography with Electrospray Ionization Multistage Mass Spectrometry and Phospholipases A1 and A2 for Regiochemical Assignment. J. Am. Soc. Mass Spectrom. 2020, 31, 1260–1270. [Google Scholar] [CrossRef]
- Coniglio, D.; Ventura, G.; Calvano, C.D.; Losito, I.; Cataldi, T.R.I. Positional Assignment of C-C Double Bonds in Fatty Acyl Chains of Intact Arsenosugar Phospholipids Occurring in Seaweed Extracts by Epoxidation Reactions. J. Am. Soc. Mass Spectrom. 2022, 33, 823–831. [Google Scholar] [CrossRef]
- Francesconi, K.A.; Edmonds, J.S.; Stick, R.V. Arsenic Compounds from the Kidney of the Giant Clam Tridacna maxima: Isolation and Identification of an Arsenic-Containing Nucleoside. J. Chem. Soc. Perkin Trans. 1 1992, 1, 1349–1357. [Google Scholar] [CrossRef]
- Francesconi, K.A. Arsenic species in seafood: Origin and human health implications. Pure Appl. Chem. 2010, 82, 373–381. [Google Scholar] [CrossRef]
- Zhao, Y.F.; Wu, J.F.; Shang, D.R.; Ning, J.S.; Ding, H.Y.; Zhai, Y.X. Arsenic species in edible seaweeds using in vitro biomimetic digestion determined by high-performance liquid chromatography inductively coupled plasma mass spectrometry. Int. J. Food Sci. 2014, 2014, 436347. [Google Scholar] [CrossRef] [PubMed]
- Calvano, C.D.; Italiano, F.; Catucci, L.; Agostiano, A.; Cataldi, T.R.I.; Palmisano, F.; Trotta, M. The lipidome of the photosynthetic bacterium Rhodobacter sphaeroides R26 is affected by cobalt and chromate ions stress. Biometals 2014, 27, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Hotimchenko, S.V. Fatty acid composition of algae from habitats with varying amounts of illumination. Russ. J. Mar. Biol. 2002, 28, 218–220. [Google Scholar] [CrossRef]
- Peralta-García, E.; Caamal-Fuentes, E.; Robledo, D.; Hernández-Núñez, E.; Freile-Pelegrín, Y. Lipid characterization of red alga Rhodymenia pseudopalmata (Rhodymeniales, Rhodophyta). Phycol. Res. 2017, 65, 58–68. [Google Scholar] [CrossRef]
- Van Ginneken, V.J.T.; Helsper, J.P.F.G.; De Visser, W.; Van Keulen, H.; Brandenburg, W.A. Polyunsaturated fatty acids in various macroalgal species from north Atlantic and tropical seas. Lipids Health Dis. 2011, 10, 104. [Google Scholar] [CrossRef]
- Monopoli, A.; Ventura, G.; Aloia, A.; Ciriaco, F.; Nacci, A.; Cataldi, T.R.I.; Calvano, C.D. Synthesis and Investigation of Novel CHCA-Derived Matrices for Matrix-Assisted Laser Desorption/Ionization Mass Spectrometric Analysis of Lipids. Molecules 2022, 27, 2565. [Google Scholar] [CrossRef] [PubMed]
- Monopoli, A.; Calvano, C.D.; Nacci, A.; Palmisano, F. Boronic acid chemistry in MALDI MS: A step forward in designing a reactive matrix with molecular recognition capabilities. Chem. Commun 2014, 50, 4322–4324. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zheng, H.; Wang, J.; Hou, J.; He, Q.; Liu, H.; Xiong, C.; Kong, X.; Nie, Z. Carbon nanodots as a matrix for the analysis of low-molecular-weight molecules in both positive- and negative-ion matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and quantification of glucose and uric acid in real samples. Anal. Chem. 2013, 85, 6646–6652. [Google Scholar] [CrossRef] [PubMed]
- Calvano, C.D.; Cataldi, T.R.I.; Kögel, J.F.; Monopoli, A.; Palmisano, F.; Sundermeyer, J. Superbasic alkyl-substituted bisphosphazene proton sponges: A new class of deprotonating matrices for negative ion matrix-assisted ionization/laser desorption mass spectrometry of low molecular weight hardly ionizable analytes. Rapid Commun. Mass Spectrom. 2016, 30, 1680–1686. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.R.; Lee, Y.J. MALDI-MS analysis and imaging of small molecule metabolites with 1,5-diaminonaphthalene (DAN). J. Mass Spectrom. 2014, 49, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Vermillion-Salsbury, R.L.; Hercules, D.M. 9-Aminoacridine as a matrix for negative mode matrix-assisted laser desorption/ionization. Rapid Commun. Mass Spectrom. 2002, 16, 1575–1581. [Google Scholar] [CrossRef]
- Monopoli, A.; Cotugno, P.; Cortese, M.; Calvano, C.D.; Ciminale, F.; Nacci, A. Selective N-Alkylation of Arylamines with Alkyl Chloride in Ionic Liquids: Scope and Applications. European J. Org. Chem. 2012, 2012, 3105–3111. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, D.; Bai, H.; Liu, H.; Lin, S.; Jiang, Y. Carbon Dots and 9AA as a Binary Matrix for the Detection of Small Molecules by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2016, 27, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; He, L. A binary matrix for background suppression in MALDI-MS of small molecules. Proc. Anal. Bioanal. Chem. 2007, 387, 1939–1944. [Google Scholar] [CrossRef]
- Calvano, C.D.; Monopoli, A.; Ditaranto, N.; Palmisano, F. 1,8-Bis(dimethylamino)naphthalene/9-aminoacridine: A new binary matrix for lipid fingerprinting of intact bacteria by matrix assisted laser desorption ionization mass spectrometry. Anal. Chim. Acta 2013, 798, 56–63. [Google Scholar] [CrossRef]
- Shanta, S.R.; Zhou, L.H.; Park, Y.S.; Kim, Y.H.; Kim, Y.; Kim, K.P. Binary matrix for MALDI imaging mass spectrometry of phospholipids in both ion modes. Anal. Chem. 2011, 15, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Lee, H.K.; Yue, G.G.L.; Chung, A.C.K.; Lau, C.B.S.; Cai, Z. A novel binary matrix consisting of graphene oxide and caffeic acid for the analysis of scutellarin and its metabolites in mouse kidney by MALDI imaging. Analyst 2021, 146, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.N.; Wang, H.Y.J.; Woods, A.S. In Situ Structural Characterization of Glycerophospholipids and Sulfatides in Brain Tissue Using MALDI-MS/MS. J. Am. Soc. Mass Spectrom. 2007, 18, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Calvano, C.D.; Ventura, G.; Trotta, M.; Bianco, G.; Cataldi, T.R.I.; Palmisano, F. Electron-Transfer Secondary Reaction Matrices for MALDI MS Analysis of Bacteriochlorophyll a in Rhodobacter sphaeroides and Its Zinc and Copper Analogue Pigments. J. Am. Soc. Mass Spectrom. 2017, 28, 125–135. [Google Scholar] [CrossRef]
- Calvano, C.D.; Ventura, G.; Cataldi, T.R.I.; Palmisano, F. Improvement of chlorophyll identification in foodstuffs by MALDI ToF/ToF mass spectrometry using 1,5-diaminonaphthalene electron transfer secondary reaction matrix. Anal. Bioanal. Chem. 2015, 407, 6369–6379. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.J.; Hsu, F.-F. Structural characterization of phospholipids and sphingolipids by in-source fragmentation MALDI/TOF mass spectrometry. Anal. Bioanal. Chem. 2022, 414, 2089–2102. [Google Scholar] [CrossRef] [PubMed]
- Baquer, G.; Sementé, L.; Ràfols, P.; Martín-Saiz, L.; Bookmeyer, C.; Fernández, J.A.; Correig, X.; García-Altares, M. rMSIfragment: Improving MALDI-MSI lipidomics through automated in-source fragment annotation. J. Cheminform. 2023, 15, 80. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Martinez-Chapa, S.O. Principles and Mechanism of MALDI-ToF-MS Analysis; Springer: Singapore, 2017; pp. 1–19. [Google Scholar]
- Köfeler, H.C.; Ahrends, R.; Baker, E.S.; Ekroos, K.; Han, X.; Hoffmann, N.; Holcapek, M.; Wenk, M.R.; Liebisch, G. Recommendations for good practice in ms-based lipidomics. J. Lipid Res. 2021, 62, 100138. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, D.; Calligaris, D.; Smargiasso, N.; De Pauw, E. Ultraviolet Laser Induced Hydrogen Transfer Reaction: Study of the First Step of MALDI In-Source Decay Mass Spectrometry. J. Phys. Chem. B 2013, 117, 2321–2327. [Google Scholar] [CrossRef]
- Cheng, H.; Sun, G.; Yang, K.; Gross, R.W.; Han, X. Selective desorption/ionization of sulfatides by MALDI-MS facilitated using 9-aminoacridine as matrix. J. Lipid Res. 2010, 51, 1599–1609. [Google Scholar] [CrossRef]
- Thomas, A.; Charbonneau, J.L.; Fournaise, E.; Chaurand, P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: Enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal. Chem. 2012, 84, 2048–2054. [Google Scholar] [CrossRef] [PubMed]
- Molin, L.; Seraglia, R.; Dani, F.R.; Moneti, G.; Traldi, P. The double nature of 1,5-diaminonaphthalene as matrix-assisted laser desorption/ionization matrix: Some experimental evidence of the protonation and reduction mechanisms. Rapid Commun. Mass Spectrom. 2011, 25, 3091–3096. [Google Scholar] [CrossRef] [PubMed]
- Karas, M.; Krüger, R. Ion Formation in MALDI: The Cluster Ionization Mechanism. Chem. Rev. 2003, 103, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Miller, Z.M.; Zhang, J.D.; Donald, W.A.; Prell, J.S. Gas-Phase Protonation Thermodynamics of Biological Lipids: Experiment, Theory, and Implications. Anal. Chem. 2020, 92, 10365–10374. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.H.; Shimizu, T.; Spener, F.; Van Meer, G.; Wakelam, M.J.O.; Dennis, E.A. Update of the LIPID MAPS Comprehensive Classification System for Lipids. J. Lipid Res. 2009, 50, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Ellis, S.R.; Paine, M.R.L.; Eijkel, G.B.; Pauling, J.K.; Husen, P.; Jervelund, M.W.; Hermansson, M.; Ejsing, C.S.; Heeren, R.M.A. Automated, Parallel Mass Spectrometry Imaging and Structural Identification of Lipids. Nat. Methods 2018, 15, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Schlotterbeck, J.; Kolb, A.; Lämmerhofer, M. Free fatty acid profiling in marine algae extract by LC-MS/MS and isolation as well as quantification of the ω-3 fatty acid hexadeca-4,7,10,13-tetraenoic acid. J. Sep. Sci. 2018, 41, 4286–4295. [Google Scholar] [CrossRef] [PubMed]
- Schwarts, S.J.; Von Elbe, J.H. Kinetics of chlorophyll degradation to pyropheophytin in vegetables. J. Food Sci. 1983, 48, 1303–1306. [Google Scholar] [CrossRef]
- Lopes, D.; Melo, T.; Meneses, J.; Abreu, M.H.; Pereira, R.; Domingues, P.; Lillebø, A.I.; Calado, R.; Rosário Domingues, M. A New Look for the Red Macroalga Palmaria palmata: A seafood with polar lipids rich in EPA and with antioxidant properties. Mar. Drugs 2019, 17, 533. [Google Scholar] [CrossRef]
- Yao, L.; Gerde, J.A.; Lee, S.L.; Wang, T.; Harrata, K.A. Microalgae lipid characterization. J. Agric. Food Chem. 2015, 63, 1773–1787. [Google Scholar] [CrossRef]
- Osório, C.; Machado, S.; Peixoto, J.; Bessada, S.; Pimentel, F.B.; Alves, R.C.; Oliveira, M.B.P.P. Pigments content (Chlorophylls, fucoxanthin and phycobiliproteins) of different commercial dried algae. Separations 2020, 7, 1–14. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dier, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
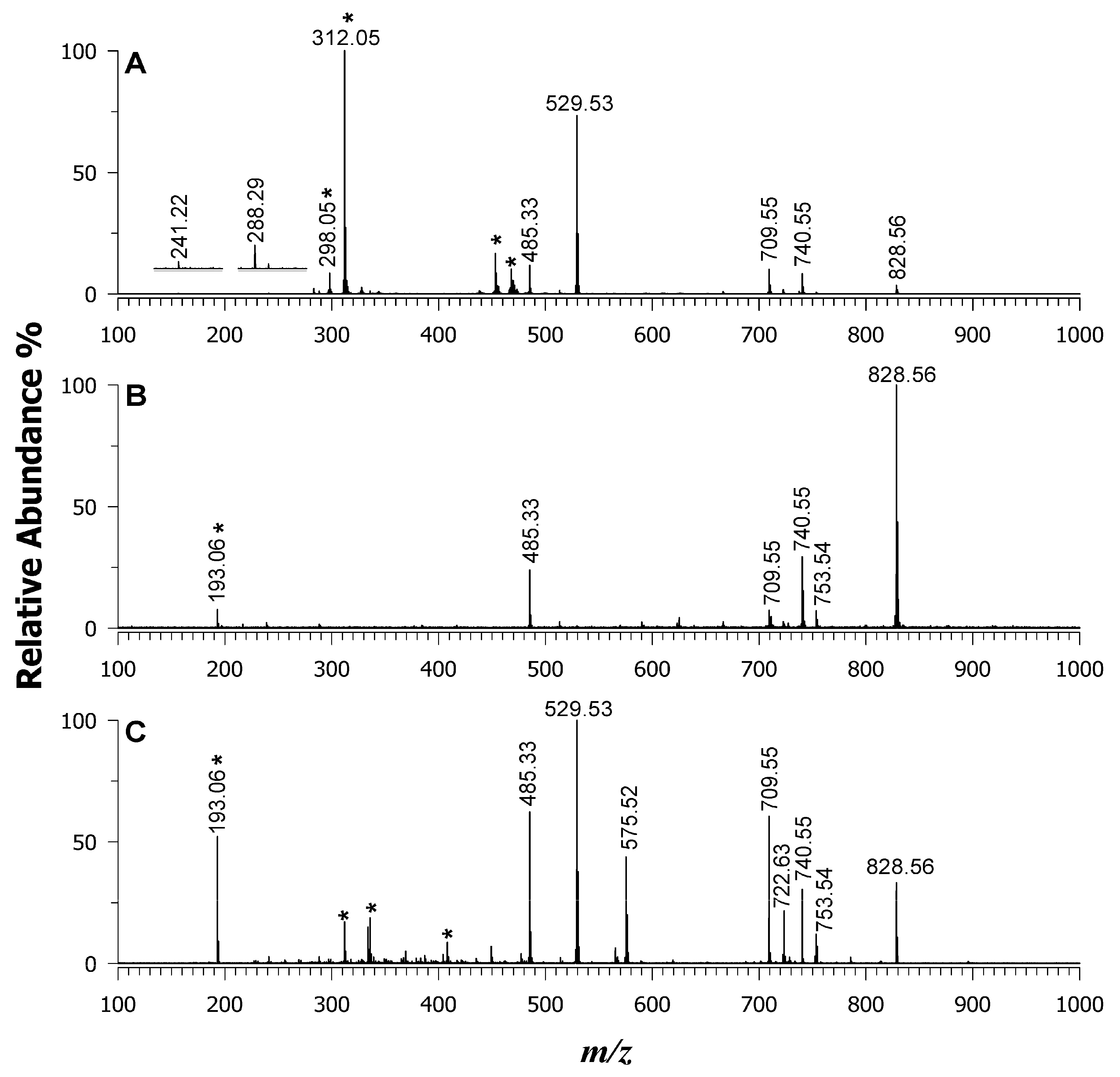


| Lipid Species | [M-H]– m/z | DAN | 9AA | DAN/9AA (Equimolar) |
|---|---|---|---|---|
| FA (15:0) | 241.22 | 0.9 | ||
| FA (18:1(d7)) | 288.29 | 1.8 | ||
| PC 15:0_18:1(d7) | 752.60 [M]–• | 3.3 | ||
| LPC 18:1(d7) | 528.38 [M]–• | 5.6 | ||
| PE 15:0_18:1(d7) | 709.55 | 11 | 6.2 | 61 |
| LPE 18:1(d7) | 485.33 | 12 | 24 | 63 |
| PG 15:0_18:1(d7) | 740.55 | 8.5 | 30 | 31 |
| PI 15:0_18:1(d7) | 828.56 | 3.9 | 100 | 33 |
| PS 15:0_18:1(d7) | 753.54 | 4.4 | 12 | |
| SM 18:1_18:1(d9) | 722.63 [M-CH3]–• | 2.7 | 22 | |
| C15 ceramide-d7 | 529.53/ | 74 | 100 | |
| 575.52 [M-H+HCOOH]— | 44 |
| Observed m/z | Theoretical m/z | Error (ppm) | Suggested Identification | Matrix | ||
|---|---|---|---|---|---|---|
| DAN | 9AA | Binary | ||||
| 199.16 | 199.170 | −50 | [FA 11:0–H]— | X | ||
| 213.18 | 213.186 | −28 | [FA 12:0–H]— | X | X | |
| 225.18 | 225.186 | −27 | [FA 14:1–H]— | X | X | |
| 227.20 | 227.202 | −9 | [FA 14:0–H]— | X | X | |
| 251.21 | 251.202 | 32 | [FA 16:2–H]— | X | X | |
| 253.22 | 253.217 | 12 | [FA 16:1–H]— | X | X | X |
| 255.23 | 255.233 | −12 | [FA 16:0–H]— | X | X | X |
| 271.23 | 271.228 | 7 | [FA 16:0;1–H]— | X | X | X |
| 275.21 | 275.202 | 29 | [FA 18:4–H]— | X | X | X |
| 277.22 | 277.217 | 11 | [FA 18:3–H]— | X | X | X |
| 279.24 | 279.233 | 25 | [FA 18:2–H]— | X | X | X |
| 281.26 | 281.249 | 39 | [FA 18:1–H]— | X | X | X |
| 283.26 | 283.248 | 42 | [FA 18:0–H]— | X | X | X |
| 291.22 | 291.233 | −45 | [FA 18:4;1–H]— | X | X | X |
| 293.22 | 293.212 | 27 | [FA 18:3;1–H]— | X | X | X |
| 295.27 | 295.263 | 24 | [FA 18:2;1–H]— | X | X | X |
| 297.25 | 297.244 | 20 | [FA 18:1;1–H]— | X | X | X |
| 299.25 | 299.259 | −30 | [FA 18:0;1–H]— | X | ||
| 301.22 | 301.217 | 10 | [FA 20:5–H]— | X | ||
| 303.24 | 303.233 | 23 | [FA 20:4–H]— | X | X | X |
| 307.26 | 307.264 | −13 | [FA 20:2–H]— | X | X | X |
| 309.27 | 309.280 | −32 | [FA 20:1–H]— | X | X | X |
| 311.28 | 311.295 | −48 | [FA 20:0–H]— | X | ||
| 317.22 | 317.212 | 25 | [FA 20:5;1–H]— | X | X | X |
| 319.23 | 319.228 | 6 | [FA 20:4;1–H]— | X | X | X |
| 325.26 | 325.275 | −46 | [FA 20:1;1–H]— | X | X | X |
| 327.23 | 327.233 | −9 | [FA 22:6–H]— | X | ||
| 333.27 | 333.280 | −30 | [FA 22:3–H]— | X | X | X |
| 335.29 | 335.296 | −18 | [FA 22:2–H]— | X | X | X |
| 339.33 | 339.326 | 12 | [FA 22:0–H]— | X | ||
| 343.21 | 343.228 | −52 | [FA 22:6;1–H]— | X | ||
| 351.27 | 351.290 | −57 | [FA 22:2;1–H]— | X | X | X |
| 353.29 | 353.306 | −45 | [FA 22:1;1–H]— | X | X | X |
| 369.32 | 369.337 | −46 | [FA 23:0;1–H]— | X | ||
| 381.32 | 381.337 | −45 | [FA 24:1;1–H]— | X | ||
| 424.27 | 424.247 | 54 | [LPE 14:0–H]— | X | ||
| 450.27 | 450.263 | 16 | [LPE 16:1–H]— | X | X | |
| 453.24 | 453.225 | 33 | [LPG 14:1–H]— | X | ||
| 479.27 | 479.242 | 58 | [LPG 16:2–H]— | X | ||
| 481.28 | 481.257 | 48 | [LPG 16:1–H]— | X | X | X |
| 483.27 | 483.273 | −6 | [LPG 16:0–H]— | X | X | |
| 505.28 | 505.257 | 46 | [LPG 18:3–H]— | X | ||
| 507.29 | 507.273 | 34 | [LPG 18:2–H]— | X | X | X |
| 509.31 | 509.288 | 43 | [LPG 18:1–H]— | X | X | X |
| 515.23 | 515.226 | 8 | [LPI 12:0–H]— | X | X | X |
| 528.33 | 528.310 | 38 | [LPC 20:4–CH3]— | X | ||
| 536.43 | 536.438 | −15 | [Lycopene]–• | X | ||
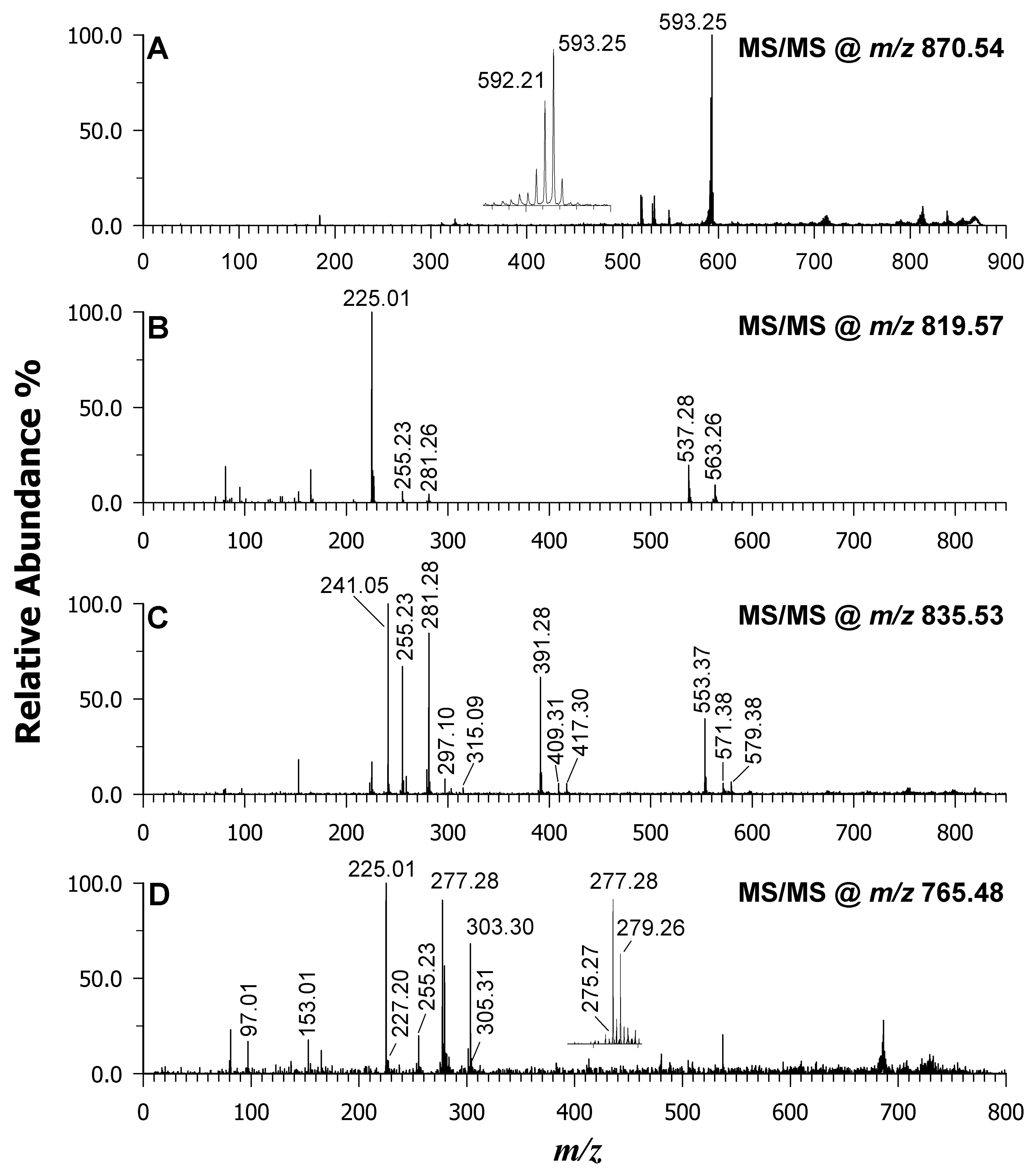
| 555.28 | 555.284 | −7 | [SQMG 16:0–H]— | X | X | |
| 579.26 | 579.284 | −41 | [SQMG 18:2–H]— | X | X | |
| 581.31 | 581.300 | 17 | [SQMG 18:1–H]— | X | X | |
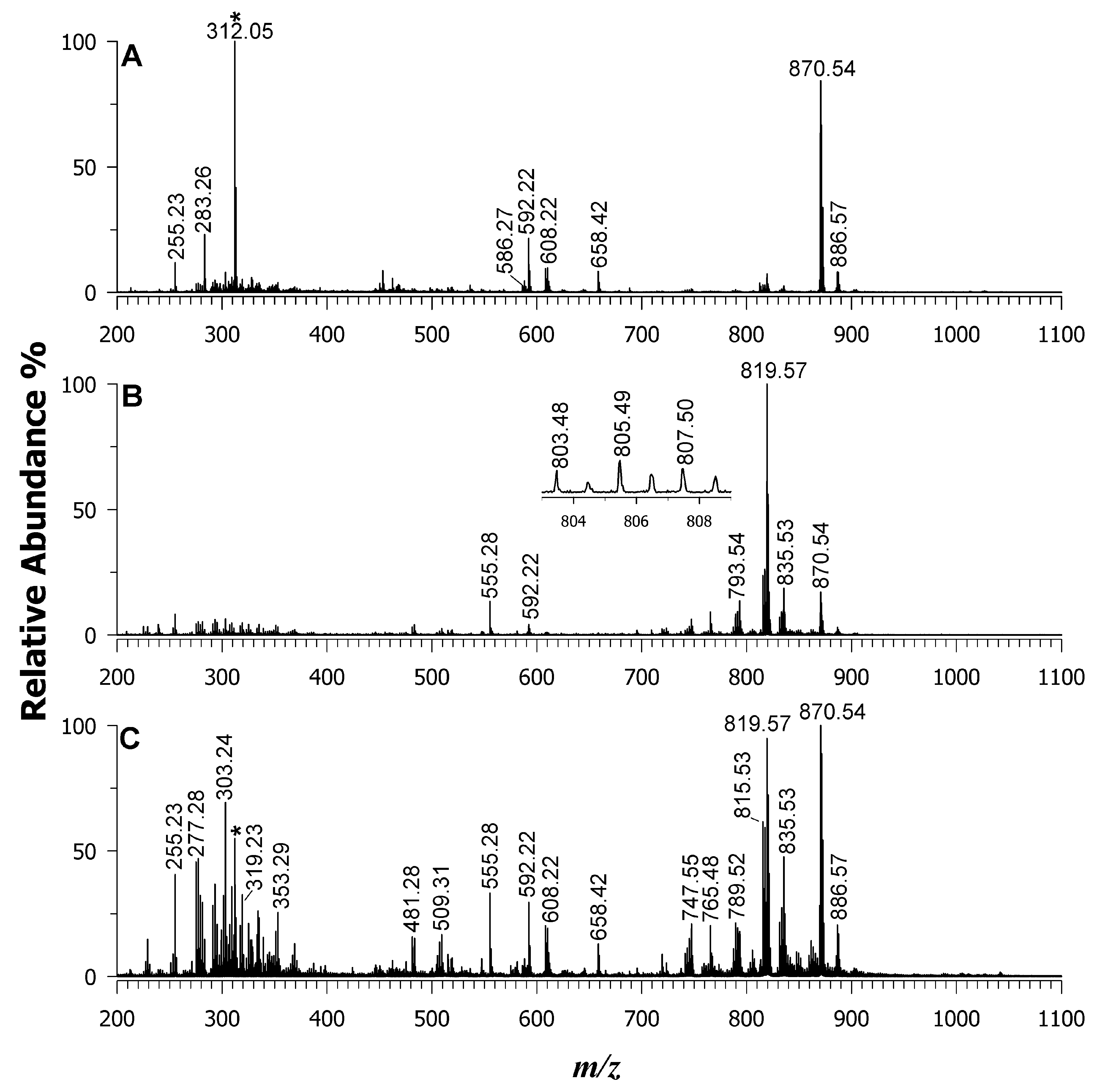
| 586.27 | 586.280 | −17 | [Phycocyanobilin diacid]–• [Phycoerythrobilin]–• | X | X | |
| 590.28 | 590.310 | −51 | [Phycourobilin]–• | X | X | |
| 591.29 | 591.261 | 49 | [Pheo a-phytil–H]–• | X | X | |
| 592.22 | 592.269 | -83 | [Pheo a-phytil]–• | X | X | |
| 608.22 | 608.263 | −71 | [Pheo b-phytil]–• | X | X | |
| 658.42 | 658.424 | −6 | Fucoxanthin | X | X | |
| 688.50 | 688.492 | 12 | [PE 32:1–H]— | X | ||
| 695.50 | 695.466 | 49 | [PA 36:4–H]— | X | ||
| 709.39 | 709.420 | −42 | [SQDG 26:0–H]— | X | X | |
| 717.51 | 717.471 | 54 | [PG 32:2–H]— | X | X | |
| 719.49 | 719.487 | 4 | [PG 32:1–H]— | X | X | |
| 719.466 | 33 | [PA 38:6–H]— | ||||
| 723.53 | 723.497 | 46 | [PA 38:4–H]— | X | X | |
| 737.48 | 737.439 | 56 | [PG 36:4–H]— | X | X | |
| 741.48 | 741.450 | 40 | [PA 40:9–H]— | X | ||
| 743.50 | 743.487 | 17 | [PG 34:3–H]— | X | X | |
| 743.466 | 46 | [PA 40:8–H]— | ||||
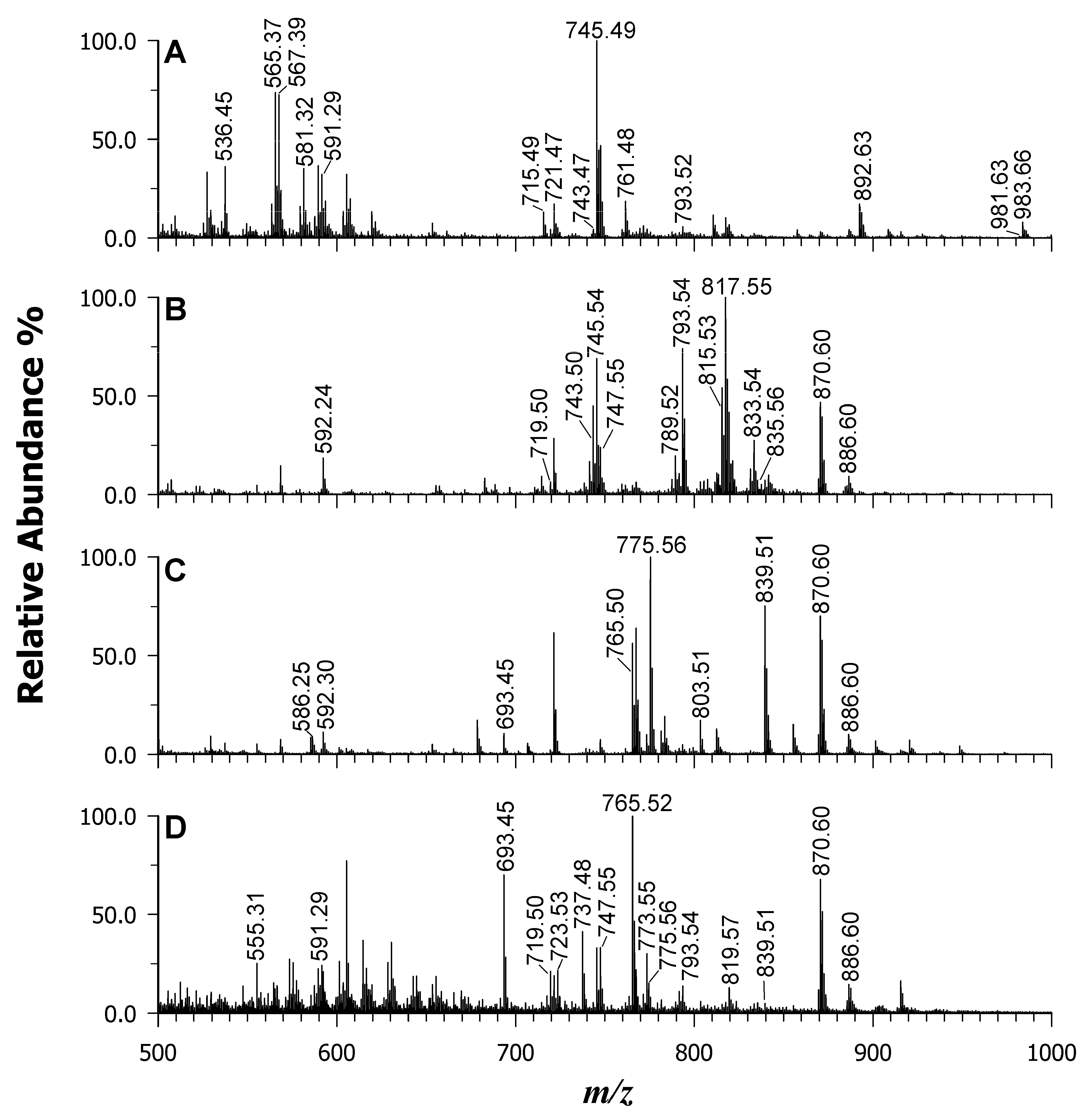
| 745.54 | 745.503 | 50 | [PG 34:2–H]— | X | X | X |
| 747.55 | 747.518 | 43 | [PG 34:1–H]— | X | X | X |
| 765.48 | 765.483 | −4 | [SQDG 30:0–H]— | X | X | |
| 765.471 | 12 | [PG 36:6–H]— | ||||
| 773.55 | 773.534 | 21 | [PG 36:2–H]— | X | ||
| 775.56 | 775.549 | 14 | [PG 36:1–H]— | X | ||
| 787.50 | 787.467 | 42 | [SQDG 32:3–H]— | X | X | |
| 789.52 | 789.483 | 47 | [SQDG 32:2–H]— | X | X | |
| 791.53 | 791.498 | 40 | [SQDG 32:1–H]— | X | X | |
| 793.54 | 793.503 | 47 | [PG 38:6–H]— | X | X | |
| 793.514 | 33 | [SQDG 32:0–H]— | ||||
| 801.52 | 801.456 | 80 | [PI 32:4–H]— | X | X | |
| 801.565 | −56 | [PG 38:2–H]— | ||||
| 803.51 | 803.472 | 47 | [PI 32:3–H]— | X | X | |
| 803.581 | −88 | [PG 38:1–H]— | ||||
| 805.49 | 805.487 | 4 | [PI 32:2–H]— | X | X | |
| 807.50 | 807.503 | −4 | [PI 32:1–H]— | X | X | |
| 813.52 | 813.483 | 45 | [SQDG 34:4–H]— | X | X | X |
| 815.53 | 815.498 | 39 | [SQDG 34:3–H]— | X | X | X |
| 817.55 | 817.514 | 44 | [SQDG 34:2–H]— | X | X | X |
| 819.57 | 819.530 | 49 | [SQDG 34:1–H]— | X | X | X |
| 829.52 | 829.487 | 40 | [PI 34:4–H]— | X | X | |
| 829.514 | 7 | [SQDG 35:3–H]— | ||||
| 831.50 | 831.503 | −4 | [PI 34:3–H]— | X | X | X |
| 833.52 | 833.519 | 1 | [PI 34:2–H]— | X | X | X |
| 835.53 | 835.534 | −5 | [PI 34:1–H]— | X | X | X |
| 839.53 | 839.472 | 69 | [PI 35:6–H]— | X | X | |
| 839.498 | 38 | [SQDG 36:5–H]— | ||||
| 841.55 | 841.487 | 75 | [PI 35:5–H]— | X | X | |
| 841.514 | 43 | [SQDG 36:4–H]— | ||||
| 843.56 | 843.503 | 68 | [PI 35:4–H]— | X | X | |
| 843.530 | 36 | [SQDG 36:3–H]— | ||||
| 845.54 | 845.519 | 25 | [PI 35:3–H]— | X | ||
| 845.545 | −6 | [SQDG 36:2–H]— | ||||
| 847.53 | 847.534 | −5 | [PI 35:2–H]— | X | X | |
| 847.561 | −37 | [SQDG 36:1–H]— | ||||
| 849.54 | 849.550 | −12 | [PI 35:1–H]— | X | X | |
| 849.577 | −44 | [SQDG 36:0–H]— | ||||
| 851.55 | 851.566 | −19 | [PI 35:0–H]— | X | ||
| 861.52 | 861.483 | 43 | [SQDG 38:8–H]— | X | X | X |
| 863.53 | 863.566 | −42 | [PI 36:1–H]— | X | X | |
| 863.498 | 37 | [SQDG 38:7–H]— | ||||
| 865.54 | 865.581 | −47 | [PI 36:0–H]— | X | X | |
| 865.514 | 30 | [SQDG 38:6–H]— | ||||
| 867.55 | 867.530 | 23 | [SQDG 38:5–H]— | X | X | |
| 869.56 | 869.545 | 17 | [SQDG 38:4–H]— | X | X | |
| 870.54 | 870.566 | −30 | [Pheo a]–• | X | X | |
| 885.59 | 885.550 | 45 | [PI 38:4–H]— | X | X | X |
| 886.57 | 886.561 | 10 | [Pheo b]–• | X | X | |
| 957.50 | 957.506 | −6 | [As-PL 32:0–H]— | X | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianco, M.; Ventura, G.; Coniglio, D.; Monopoli, A.; Losito, I.; Cataldi, T.R.I.; Calvano, C.D. Development of a New Binary Matrix for the Comprehensive Analysis of Lipids and Pigments in Micro- and Macroalgae Using MALDI-ToF/ToF Mass Spectrometry. Int. J. Mol. Sci. 2024, 25, 5919. https://doi.org/10.3390/ijms25115919
Bianco M, Ventura G, Coniglio D, Monopoli A, Losito I, Cataldi TRI, Calvano CD. Development of a New Binary Matrix for the Comprehensive Analysis of Lipids and Pigments in Micro- and Macroalgae Using MALDI-ToF/ToF Mass Spectrometry. International Journal of Molecular Sciences. 2024; 25(11):5919. https://doi.org/10.3390/ijms25115919
Chicago/Turabian StyleBianco, Mariachiara, Giovanni Ventura, Davide Coniglio, Antonio Monopoli, Ilario Losito, Tommaso R. I. Cataldi, and Cosima D. Calvano. 2024. "Development of a New Binary Matrix for the Comprehensive Analysis of Lipids and Pigments in Micro- and Macroalgae Using MALDI-ToF/ToF Mass Spectrometry" International Journal of Molecular Sciences 25, no. 11: 5919. https://doi.org/10.3390/ijms25115919






