Abstract
The association between vitamin D deficiency and cardiovascular disease remains a controversial issue. This study aimed to further elucidate the role of vitamin D signaling in the development of left ventricular (LV) hypertrophy and dysfunction. To ablate the vitamin D receptor (VDR) specifically in cardiomyocytes, VDRfl/fl mice were crossed with Mlcv2-Cre mice. To induce LV hypertrophy experimentally by increasing cardiac afterload, transverse aortic constriction (TAC) was employed. Sham or TAC surgery was performed in 4-month-old, male, wild-type, VDRfl/fl, Mlcv2-Cre, and cardiomyocyte-specific VDR knockout (VDRCM-KO) mice. As expected, TAC induced profound LV hypertrophy and dysfunction, evidenced by echocardiography, aortic and cardiac catheterization, cardiac histology, and LV expression profiling 4 weeks post-surgery. Sham-operated mice showed no differences between genotypes. However, TAC VDRCM-KO mice, while having comparable cardiomyocyte size and LV fibrosis to TAC VDRfl/fl controls, exhibited reduced fractional shortening and ejection fraction as measured by echocardiography. Spatial transcriptomics of heart cryosections revealed more pronounced pro-inflammatory and pro-fibrotic gene regulatory networks in the stressed cardiac tissue niches of TAC VDRCM-KO compared to VDRfl/fl mice. Hence, our study supports the notion that vitamin D signaling in cardiomyocytes plays a protective role in the stressed heart.
1. Introduction
The increasing burden of cardiovascular diseases, including left ventricular hypertrophy (LVH), hypertension, coronary artery disease, and diabetes, places a significant strain on clinical resources and socioeconomic systems [1]. Despite remarkable progress in cardiovascular research and improved therapeutic strategies against major risk factors, morbidity and mortality due to heart failure complications are still a major concern.
Over the past three decades, a large number of clinical and preclinical studies have supported a role of vitamin D signaling in cardiovascular health. In this context, large epidemiological studies have presented strong evidence linking vitamin D deficiency with cardiovascular disease, even after adjusting for traditional cardiovascular risk factors [2,3,4]. However, it is still controversial whether vitamin D supplementation has a role in the therapy of cardiovascular diseases, as large intervention trials failed to show a beneficial effect on cardiovascular endpoints [5,6,7].
Apart from its well-established role in bone health and mineral metabolism [8,9], vitamin D signaling has been observed in many tissues expressing the vitamin D receptor (VDR), and it is thought that all direct regulatory actions of the vitamin D hormone, 1α,25-dihydroxyvitamin D3, are mediated via the VDR [10]. The expression of the VDR in the heart and in blood vessels suggests possible direct effects of vitamin D signaling in these tissues [10,11]. In the heart, the presence of the VDR has been demonstrated in cardiomyocytes close to T tubuli as well as in fibroblasts, and cardiac hypertrophy leads to an upregulation of VDR expression at the mRNA and protein levels [12,13]. Indeed, a plethora of preclinical studies employing different animal models has unveiled a multitude of mechanisms underlying the beneficial cardiovascular effects of vitamin D [14,15,16,17,18].
Nonetheless, inconsistencies in data, especially regarding the role of VDR signaling in cardiomyocytes, persist. While Li et al. [19] reported cardiomyocyte hypertrophy, hypertension, and upregulated renin secretion in global VDR knockout mice on a normal diet, we failed to find hypertension, increased renin secretion, or LVH in 3-month-old global VDR knockout mice on a rescue diet, which normalizes mineral homeostasis [20]. Chen et al. [21] reported that deletion of the VDR specifically in cardiomyocytes results in LVH under baseline conditions and upon induction with isoproterenol via the pro-hypertrophic calcineurin/NFAT/MCIP 1 signaling pathway. In contrast, we observed no difference in the hypertrophic response of wild-type (WT) mice and those lacking VDR globally subjected to transverse aortic constriction (TAC), a standard model of experimental LVH induced by chronically increased afterload [22]. In addition, we found no difference in heart function between WT mice and global VDR knockout mice after myocardial infarction in an earlier study [23].
A potential pitfall in studies addressing the cardiovascular role of vitamin D signaling is the pleiotropic effects of vitamin D in many different tissues, making it difficult to compare studies using global and conditional, tissue-specific VDR ablation models. The VDR is expressed ubiquitously, and approximately 3% of the human genome is directly regulated by vitamin D [10]. Hence, ablation of vitamin D signaling results in complex changes in global VDR knockout models.
The objective of this study was to gain further insight into the role of the VDR in cardiac hypertrophy and in LVH pathogenesis by specifically deleting the VDR in cardiomyocytes of mice and employing TAC as an experimental model of LVH induction. To address the complex effects of VDR signaling on gene regulatory networks in an unbiased, holistic manner, we performed spatial transcriptomics on cryosections of TAC hearts from mice with cardiomyocyte-specific deletion of the VDR and those of controls. We found that while LVH developed independently of cardiomyocyte VDR presence, cardiomyocyte-specific VDR deficiency was associated with LV functional impairment and more pronounced pro-inflammatory and pro-fibrotic gene regulatory networks in stressed cardiac tissue niches.
2. Results
2.1. Generation and Characterization of Cardiomyocyte-Specific VDR Knockout Mice
To decipher the effects of vitamin D deficiency on cardiac function, we generated a cardiomyocyte-specific VDR knockout mouse model using Cre-LoxP technology. Mice with a floxed VDR gene carrying loxP sequences surrounding exon 3 were previously created [24]. For specific deletion of the VDR gene in ventricular cardiomyocytes, we mated VDRfl/fl mice with mice expressing Cre recombinase under the ventricle-specific Mlc2v promoter [25] (Figure 1A). The resulting genotypes included wild-type (WT), VDRfl/fl, Mlc2vCre/wt, and VDRfl/fl/Mlc2vCre/wt mice, henceforth referred to as cardiomyocyte-specific VDR knockout mice (VDRCM-KO). We observed germline VDR deletion when the Cre allele was maternally inherited and therefore solely used male Cre-expressing mice for breeding in this study. To rule out effects of Mlc2v haploinsufficiency, we initially performed TAC experiments in wt/wt, wt/Cre+, and VDRfl/fl mice. Wt/wt, wt/Cre+, and VDRfl/fl mice showed comparable increases in systolic (SP) and pulse (PP) pressures as well as similar values for fractional shortening (FS) and ejection fraction (EF) after TAC (Supplementary Figure S1), corroborating earlier reports demonstrating that Mlc2v-Cre mice do not show an altered cardiac response to TAC-induced pressure overload compared with wt/wt mice [26].
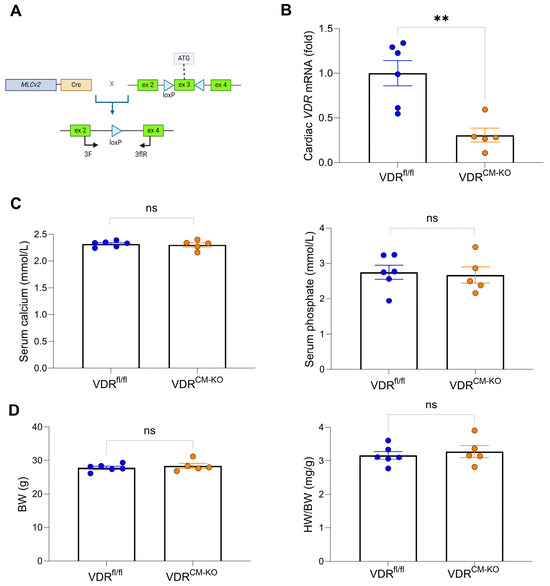
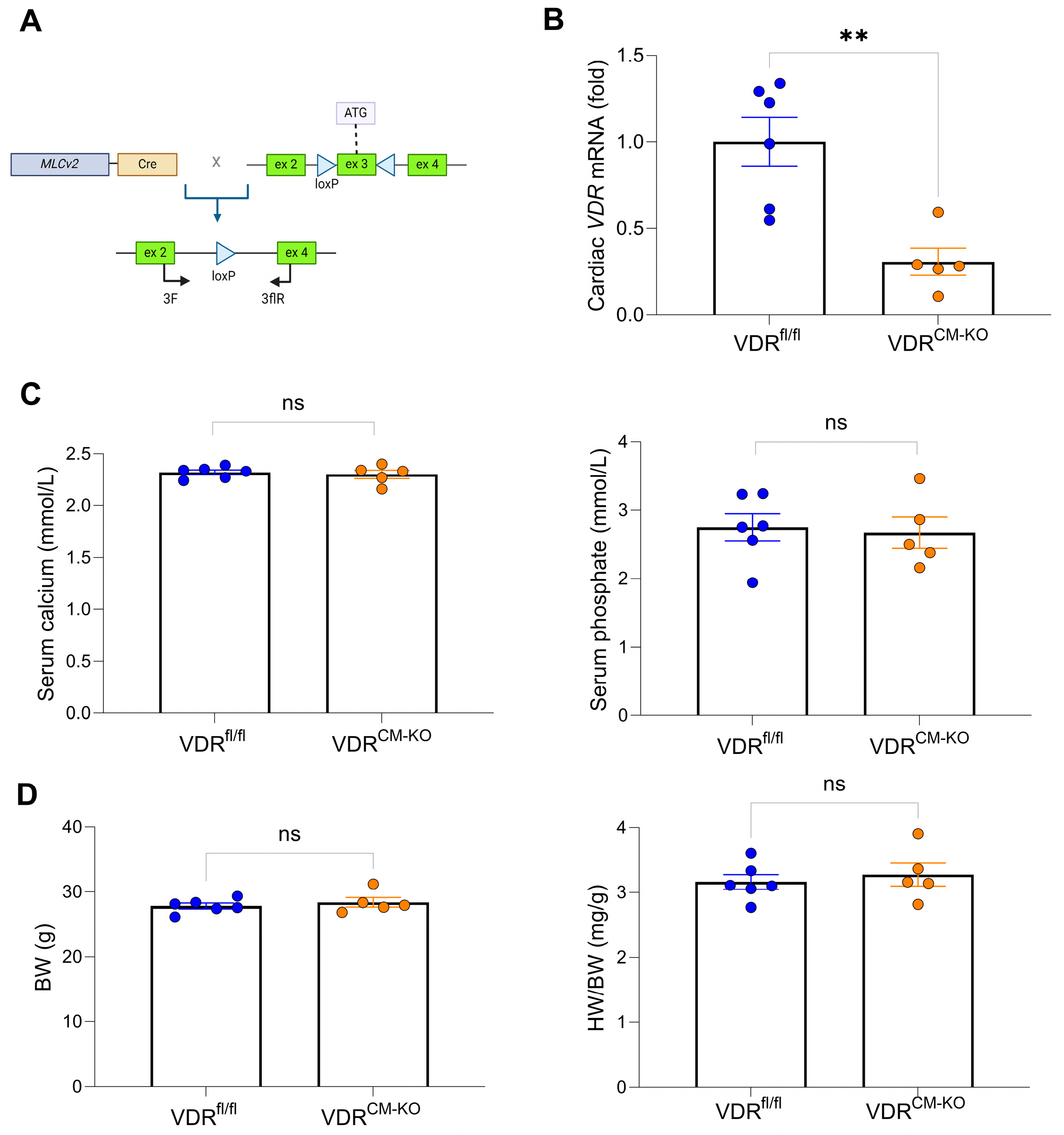
Figure 1.
Gene targeting strategy, cardiac VDR mRNA expression, as well as unaltered mineral metabolism and cardiac phenotype in VDRCM-KO mice. (A) Schematic illustration of Cre-LoxP strategy to generate cardiomyocyte-specific VDR knockout mice, showing the murine VDR allele with exons 1–3 and location of LoxP sites (triangles) surrounding exon 3. (B) Quantitative real-time PCR analysis reveals ~70% reduction of VDR mRNA levels at the whole heart level in VDRCM-KO relative to VDRfl/fl mice (n = 6 VDRfl/fl; n = 5 VDRCM-KO). (C) Serum concentration of calcium and phosphate (n = 6 VDRfl/fl; n = 5 VDRCM-KO) as well as (D) body weight and heart weight-to-body weight ratio (HW/BW) in VDRCM-KO mice are comparable to those of VDRfl/fl controls (n = 6 VDRfl/fl; n = 5 VDRCM-KO). Data are given as bar dot plots with SEM. ** p < 0.01 by unpaired t-test; ns, not significant. (A) was created with BioRender.com.
The VDRCM-KO mice were born at the anticipated Mendelian frequency without any gross phenotypic abnormalities. Recombination efficacy was confirmed by a ~70% reduction of cardiac VDR mRNA expression in VDRCM-KO mice relative to controls (Figure 1B and Supplementary Table S1). This level of reduction in VDR mRNA expression is expected in VDRCM-KO mice at the whole heart level because VDR expression is ubiquitous and other cell types, such as endothelial cells or fibroblasts, contribute to VDR expression in the heart. Serum levels of calcium and phosphate remained unaltered in VDRCM-KO mice (Figure 1C). Moreover, VDRCM-KO mice showed normal body weight and heart weight-to-body weight (HW/BW) ratio, relative to age-matched VDRfl/fl controls (Figure 1D), suggesting that cardiomyocyte-specific VDR inactivation does not lead to a pathologic cardiac phenotype under standard resting conditions in 5-month-old male mice.
2.2. Development of Afterload-Induced Cardiac Hypertrophy Is Not Modulated by Loss of VDR in Cardiomyocytes
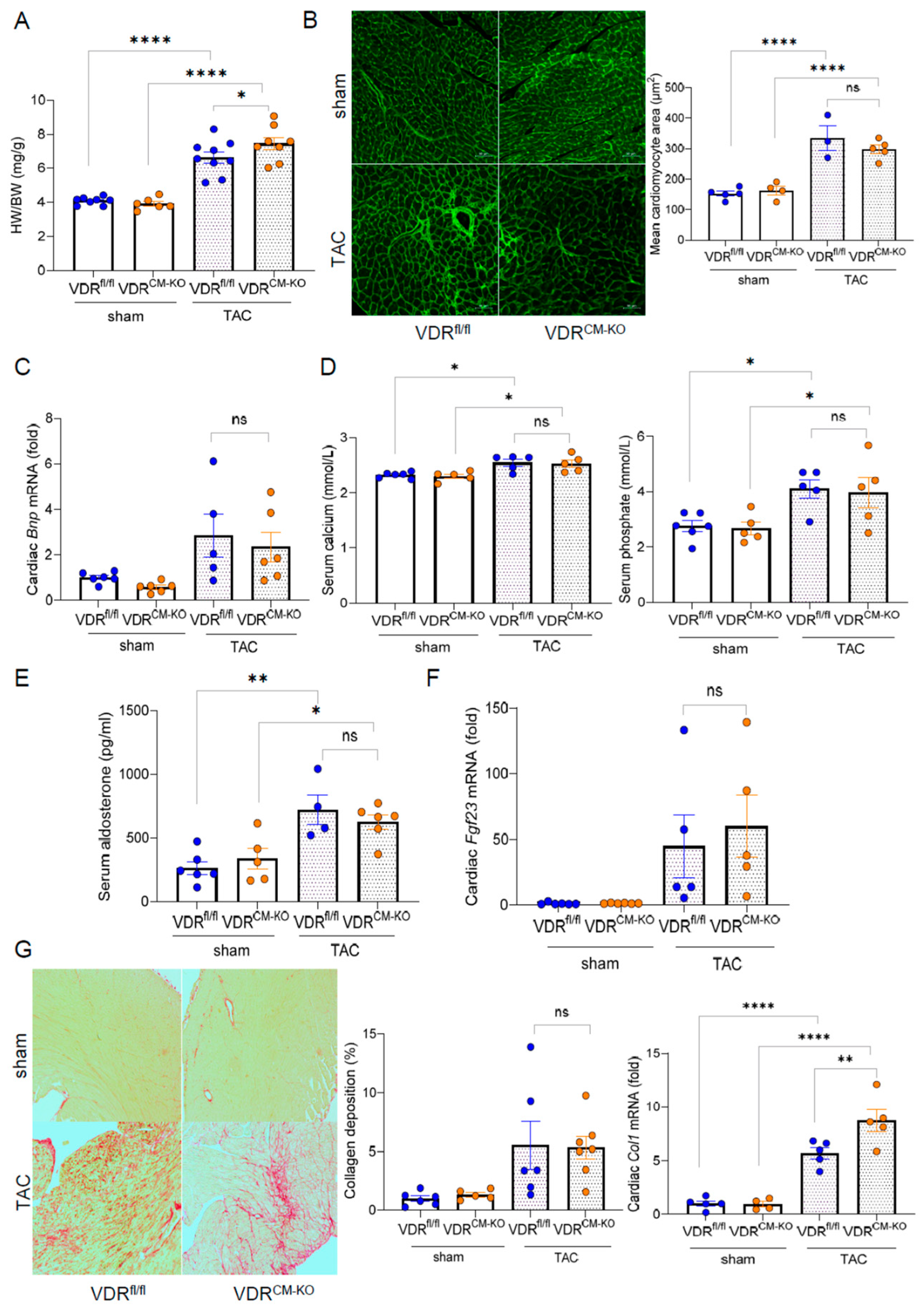
To examine the tissue-specific effects of vitamin D signaling in LVH, we employed TAC, a pressure overload–induced cardiac hypertrophy model. In line with previous studies, which show heart failure with reduced ejection fraction (HFrEF) development after 4 weeks of TAC [27], we terminated our experiments at this timepoint. TAC significantly increased the HW/BW ratio compared to sham-operated animals in all genotypes (Figure 2A). Based on previous findings [21], we hypothesized that loss of VDR in cardiomyocytes could accelerate cardiac pathology when subjected to pressure overload. Despite a slight increase in HW/BW ratio observed in TAC VDRCM-KO compared to TAC VDRfl/fl mice (Figure 2A), histological examination by FITC-labeled wheat germ agglutinin (WGA) staining revealed a similar increase in mean cardiomyocyte cross-sectional area post-TAC (Figure 2B). This indicates that cardiomyocytes respond to the afterload-induced hypertrophic stimulus independent of VDR. In addition, cardiac mRNA expression of the hypertrophy marker brain natriuretic peptide (BNP) was comparably increased after TAC in VDRfl/fl and VDRCM-KO mice (Figure 2C). Moreover, TAC mice were characterized by hyperphosphatemia, hypercalcemia, and increased serum aldosterone concentrations, independent of genotype (Figure 2D,E). It is well known that TAC is associated with an upregulation of cardiac mRNA expression of fibroblast growth factor-23 (FGF23) [22,28]. In line with these earlier reports, we observed approximately 50-fold upregulation of cardiac Fgf23 transcription in response to TAC in both genotypes, with no differences between the genotypes (Figure 2F). We also investigated collagen remodeling and cardiac fibrosis, common LVH accompaniments, using picrosirius red (PSR) staining of paraffin sections. TAC-induced LVH was associated with a profound increase in interstitial fibrosis (Figure 2G). However, PSR staining did not demonstrate differences in cardiac fibrosis between TAC VDRCM-KO and VDRfl/fl mice (Figure 2G). In contrast, collagen type I (Col1a1) mRNA expression was upregulated in TAC VDRCM-KO mice, relative to TAC VDRfl/fl controls (Figure 2G). Nonetheless, as shown in the left and upper right panels of Figure 2G, this increase in Col1a1 transcription in TAC VDRCM-KO mice did not result in increased fibrosis as evidenced by PSR staining at 4 weeks post-TAC. Collectively, our data suggest that ablation of VDR expression in cardiomyocytes does not modulate afterload-induced cardiac hypertrophy, but results in higher collagen 1 levels at the transcriptional, but not at the histological level.
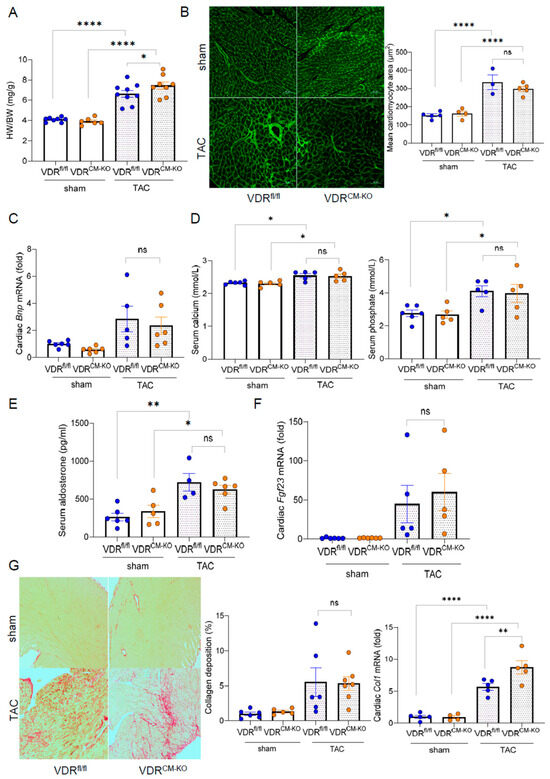
Figure 2.
Cardiomyocyte-specific VDR deletion does not aggravate left ventricular hypertrophy. (A) Heart weight-to-body weight (HW/BW) ratio in sham-operated and TAC VDRfl/fl and VDRCM-KO mice (n = 8 VDRfl/fl sham; n = 6 VDRCM-KO sham; n = 9 VDRfl/fl TAC; n = 8 VDRCM-KO TAC). (B) Representative histological images of FITC-WGA staining and quantification of cardiomyocyte size reveal similar cardiomyocyte hypertrophy between the genotypes post-TAC (n = 5 VDRfl/fl sham; n = 4 VDRCM-KOsham; n = 3 VDRfl/fl TAC; n = 5 VDRCM-KO TAC). Scale bar = 50 µm. (C) Relative mRNA expression of brain natriuretic peptide (BNP) is unchanged between TAC VDRfl/fl and VDRCM-KO mice (n = 6 VDRfl/fl sham; n = 6 VDRCM-KO sham; n = 5 VDRfl/fl TAC; n = 6 VDRCM-KO TAC). (D) Serum calcium and phosphate (n = 6 VDRfl/fl sham; n = 6 VDRCM-KO sham; n = 5 VDRfl/fl TAC; n = 5 VDRCM-KO TAC), and (E) aldosterone concentrations in sham-operated and TAC VDRfl/fl and VDRCM-KO mice (n = 6 VDRfl/fl sham; n = 5 VDRCM-KO sham; n = 4 VDRfl/fl TAC; n = 6 VDRCM-KO TAC). (F) Relative cardiac Fgf23 mRNA expression (n = 6 VDRfl/fl sham; n = 6 VDRCM-KO sham; n = 5 VDRfl/fl TAC; n = 5 VDRCM-KO TAC) and (G) histological images of cardiac paraffin sections stained with picrosirius red (PSR) and quantification of PSR-stained area (n = 6 VDRfl/fl sham; n = 5 VDRCM-KO sham; n = 5 VDRfl/fl TAC; n = 7 VDRCM-KO TAC) as well as relative cardiac collagen 1 (Col1a1) mRNA expression in sham-operated and TAC VDRfl/fl and VDRCM-KO mice (n = 5 VDRfl/fl sham; n = 4 VDRCM-KO sham; n = 5 VDRfl/fl TAC; n = 5 VDRCM-KO TAC). Scale bar =100 µm. Data are given as bar dot plots with SEM. * p < 0.05, ** p < 0.01, **** p < 0.0001 by one-way ANOVA followed by Student–Newman–Keuls multiple comparison test; ns, not significant.
2.3. Lack of VDR Signaling in Cardiomyocytes Aggravates TAC-Induced LV Functional Impairment
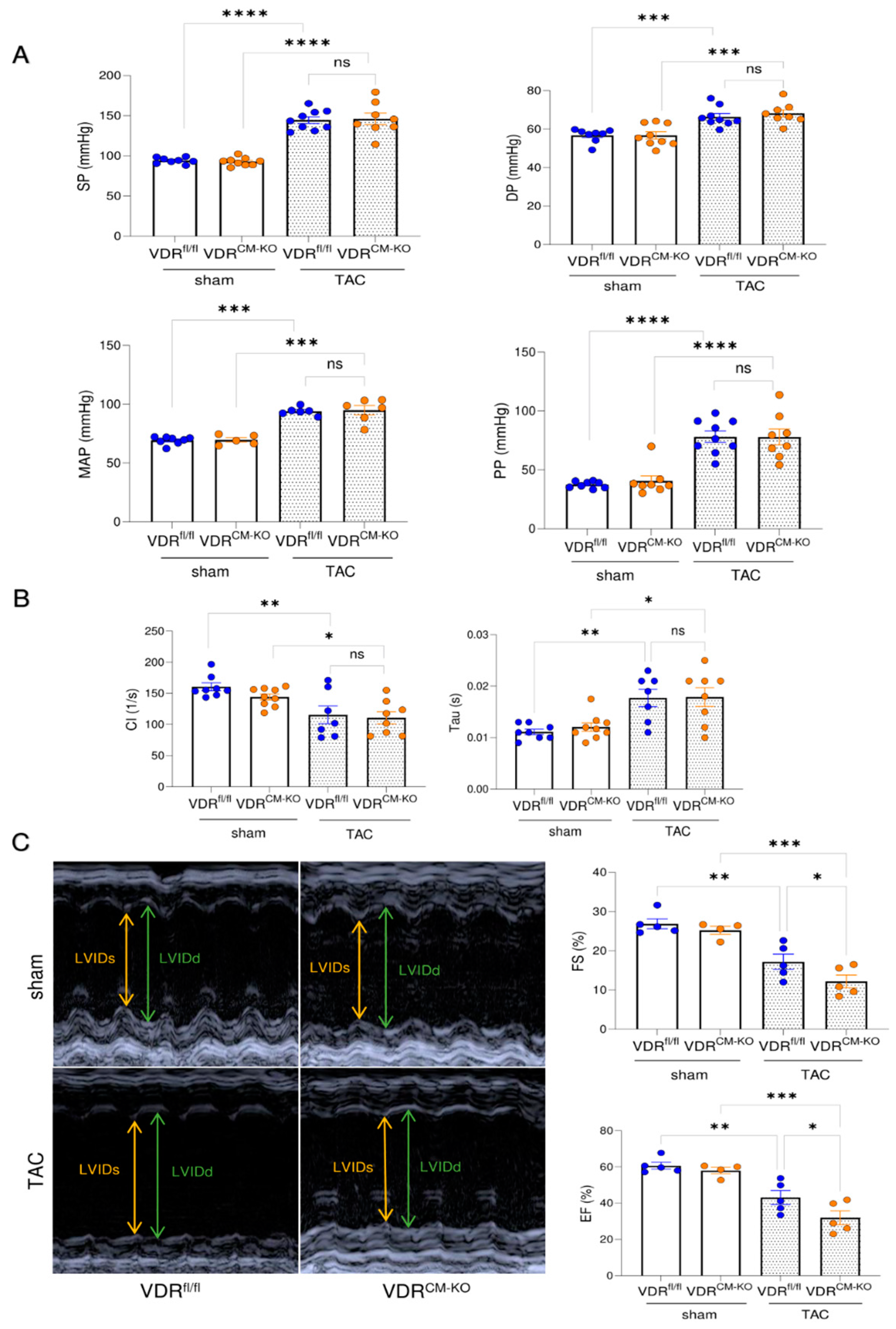
To assess cardiac functionality in our experimental model, we performed intra-arterial and intra-cardiac catheterization as well as echocardiography. TAC results in hypertension upstream of the constriction site, thereby increasing LV afterload. Lack of VDR in cardiomyocytes did not influence the TAC-induced increase in arterial systolic pressure (SP), diastolic pressure (DP), mean arterial pressure (MAP), and pulse pressure (PP) (Figure 3A). In addition, both TAC VDRfl/fl and VDRCM-KO mice developed elevated end diastolic pressure (EDP) relative to sham controls (Supplementary Figure S2A). Left ventricular catheterization showed a comparable decline in cardiac contractility and increase of relaxation time constant (Tau) in both TAC VDRfl/fl and VDRCM-KO mice, relative to sham controls (Figure 3B). In contrast, echocardiography revealed a further reduction of ejection fraction (EF) and fractional shortening (FS) in TAC VDRCM-KO mice, relative to TAC VDRfl/fl controls, suggesting a more pronounced development of HFrEF in mice lacking VDR in cardiomyocytes (Figure 3C). LV internal diameter in diastole and systole (LVIDd and LVIDs) tended to be elevated in TAC VDRCM-KO mice in comparison with TAC VDRfl/fl mice (Supplementary Figure S2B). However, the difference did not reach statistical significance. Taken together, these data suggest that the absence of VDR signaling in cardiomyocytes augments the TAC-induced reduction in cardiac functionality.
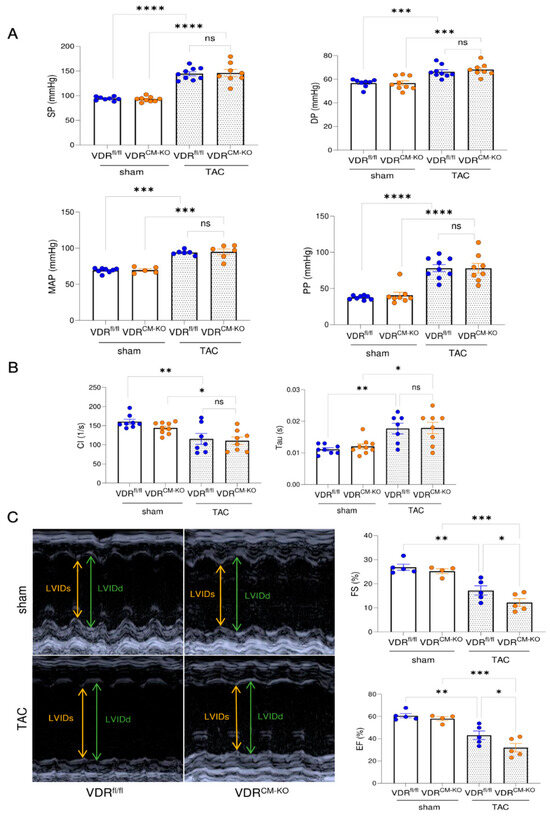
Figure 3.
Mice lacking VDR in cardiomyocytes display a more pronounced reduction in cardiac functionality after TAC. (A) Systolic pressure (SP), diastolic pressure (DP), mean arterial pressure (MAP), and pulse pressure (PP) are comparably increased in VDRfl/fl and VDRCM-KO mice following TAC (SP, DP, PP: n = 8 VDRfl/fl sham; n = 9 VDRCM-KO sham; n = 9 VDRfl/fl TAC; n = 8 VDRCM-KO TAC; MAP: n = 8 VDRfl/fl sham; n = 5 VDRCM-KO sham; n = 6 VDRfl/fl TAC; n = 6 VDRCM-KO TAC). (B) LV contractility index (CI) and relaxation time (Tau) does not differ between TAC VDRCM-KO and VDRfl/fl mice (n = 8 VDRfl/fl sham; n = 9 VDRCM-KO sham; n = 7 VDRfl/fl TAC; n = 8 VDRCM-KO TAC). (C) Representative original echocardiograms in parasternal short axis M-mode (left). Parameters of cardiac functionality fractional shortening (FS) and ejection fraction (EF) (right) are significantly reduced in TAC VDRCM-KO mice in comparison to TAC VDRfl/fl controls (n = 5 VDRfl/fl sham; n = 4 VDRCM-KO sham; n = 5 VDRfl/fl TAC; n = 5 VDRCM-KO TAC). Data are given as bar dot plots with SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 by one-way ANOVA followed by Student–Newman–Keuls post-hoc test. LVIDd and LVIDs, left ventricular internal diameter in diastole and systole; ns, not significant.
2.4. Spatial Transcriptomics Reveal More Pronounced Pro-Inflammatory and Pro-Fibrotic Gene Regulatory Networks in Hypertrophic Cardiac Tissue Niches of TAC VDRCM-KO Mice
To gain further insight into the molecular changes induced by lacking VDR signaling in cardiomyocytes in our TAC model, we performed spatial transcriptomics on heart cryosections, focusing on the comparison between TAC VDRfl/fl and VDRCM-KO mice. The cross-sections were taken in the middle region of the ventricle. In agreement with profoundly reduced VDR expression in cardiomyocytes of VDRCM-KO mice, we found only one spot with detectable VDR expression in the VDRCM-KO mouse, whereas 44 spots were detected in the VDRfL/fl mouse (Supplementary Figure S3). In all spots with VDR expression, we also detected cardiomyocyte-specific Myl2 expression in the ventricle of the VDRfl/fl mouse. Using the SPIN algorithm [29], we identified two distinct region clusters across TAC VDRfl/fl and VDRCM-KO samples, based on unique gene marker patterns that define specific tissue regions (Figure 4A). This spatial integration revealed two primary clusters corresponding to inner and outer tissue regions (cluster 0 representing the outer region and cluster 1 the inner region), with an even distribution of spots among them (0_fl (1649), 0_KO (1641), 1_fl (1414), and 1_KO (1327) (Figure 4B). The ‘outer’ region includes the outer LV wall and the right ventricle, whereas the ‘inner’ region includes the LV inner wall and septum (Figure 4B). For simplicity, we will refer to the two clusters as ‘inner’ and ‘outer’ regions henceforth.
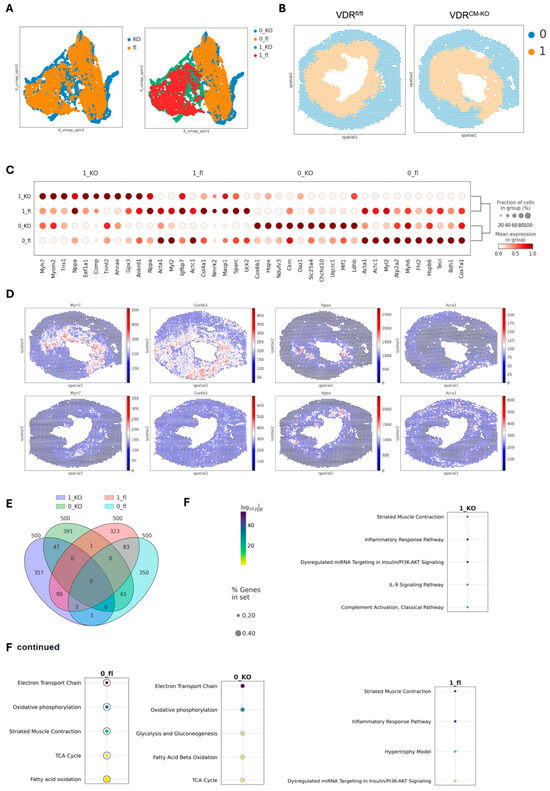
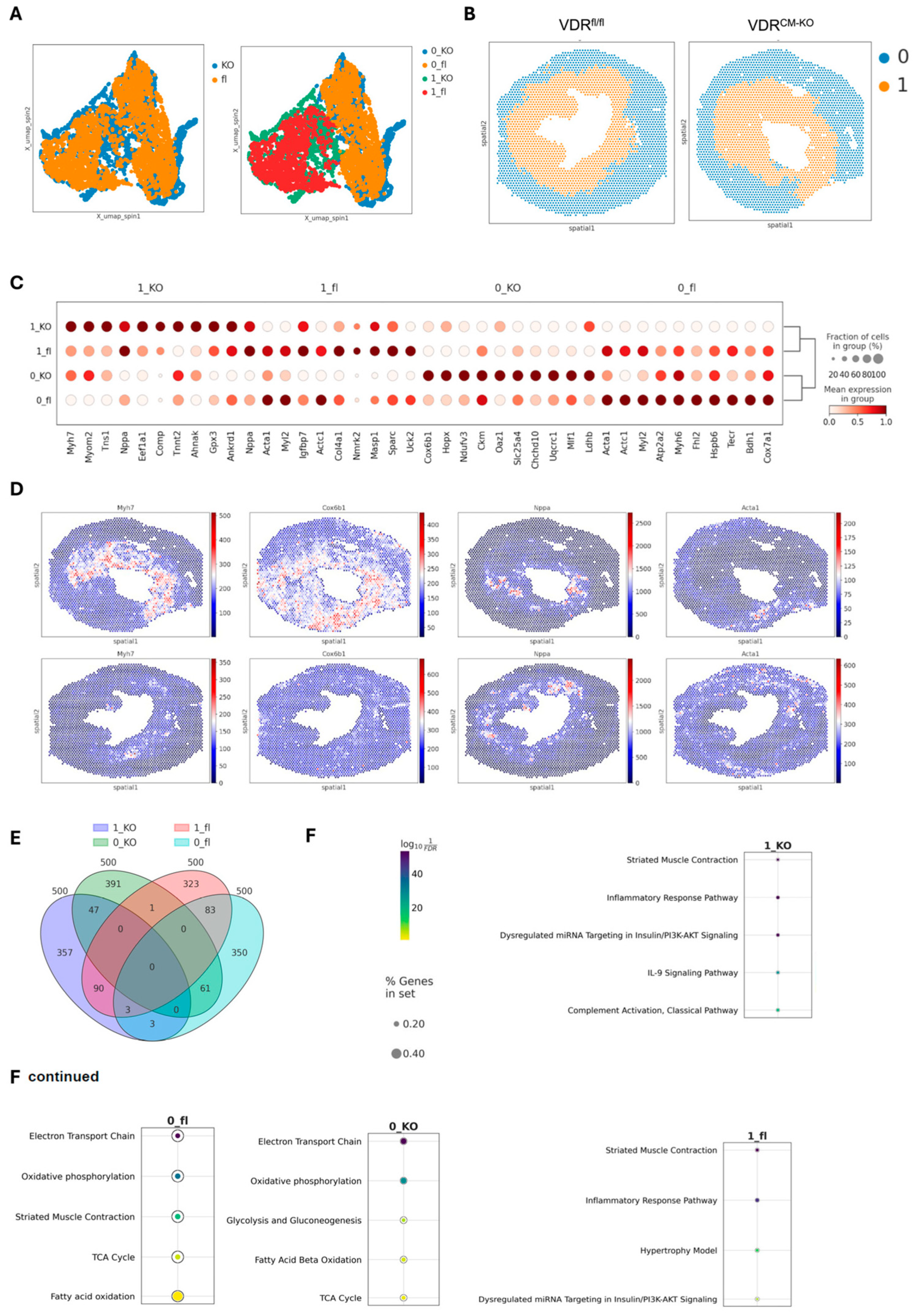
Figure 4.
Spatial transcriptomic analysis in the TAC-induced heart failure model in VDRCM-KO and VDRfl/fl mice. (A) UMAP visualization of the integrated transcriptomic spots from cryosections of VDRCM-KO and VDRfl/fl TAC mice, showing similarities and differences in gene expression profiles. (B) Spatial mapping of clusters in heart sections of VDRCM-KO and VDRfl/fl TAC mice, revealing the cluster distribution. (C) Dot plot of the top 10 differentially expressed genes (DEG) identified across the combined clusters and conditions, highlighting key genes driving the distinction between clusters. (D) Visualization of spatial expression patterns for the most significant DEG identified per cluster (1_KO, 0_KO, 1_fl, 0_fl). (E) Venn diagram summarizing the overlap and uniqueness among the top 500 DEG across all combined regions and conditions. (F) GO (Gene Ontology) enrichment analysis for the top 500 DEGs, examining enriched pathways across the four identified clusters. KO, VDRCM-KO; fl, VDRfl/fl.
The differential gene expression analysis identified the top ten signature genes distinguishing these clusters, with hierarchical clustering showing more similarities within regions than between genotypes (Figure 4C). The outer regions, characterized by genes such as Myh6, Cox6b1, Ckm, and Acta1, reflect typical heart function, underscoring their roles in maintaining cardiac physiology [30,31,32] (Figure 4C). Conversely, the inner regions showed elevated levels of myosin heavy chain-7 (Myh7) and ANP (Nppa), markers linked to cardiac stress and hypertrophy, similar to findings from single-cell RNA-seq studies in the same heart failure TAC model [33,34] (Figure 4D). The upregulation of Myh7 has been linked to a transition from hypertrophy to heart failure, marked by an increase in Myh7 mRNA and protein levels, particularly pronounced in severe cardiac stress [33]. Moreover, the variability in Myh7 and Nppa expression among cardiomyocytes indicated a heterogeneous cellular response to pressure overload [34]. Noteworthy is the differential expression pattern of Myh7 and Myh6 encoding for β-myosin heavy chain (MHC) and α-MHC, respectively, representing distinct MHC isoforms critical for cardiac function [35,36,37]. In the inner, more stressed heart regions in TAC mice, Myh7’s presence correlates with its slower, more energy-efficient contractions, a beneficial adaptation during cardiac stress [36]. Conversely, Myh6 predominates in the outer, more healthy areas, facilitating rapid contractions and higher cardiac output, though at greater energy expense [35]. This differential expression highlights the heart’s adaptability, with Myh7 optimizing energy use under stress and Myh6 enabling swift responses in normal conditions.
A detailed examination of the top 500 differentially expressed genes revealed distinct gene sets for each cluster, with minimal overlap, indicating unique molecular profiles (Figure 4E). GO (Gene Ontology) enrichment analysis further differentiated these regions, associating outer regions with normal heart function pathways, while inner regions showed enrichment for inflammatory pathways, with the VDRfl/fl heart identified as the hypertrophy model (Figure 4F). The IL-9 and complement pathways specific for the inner region in the VDRCM-KO heart hints at vitamin D’s regulatory impact on immune responses [38] in the hypertrophic heart (Figure 4F).
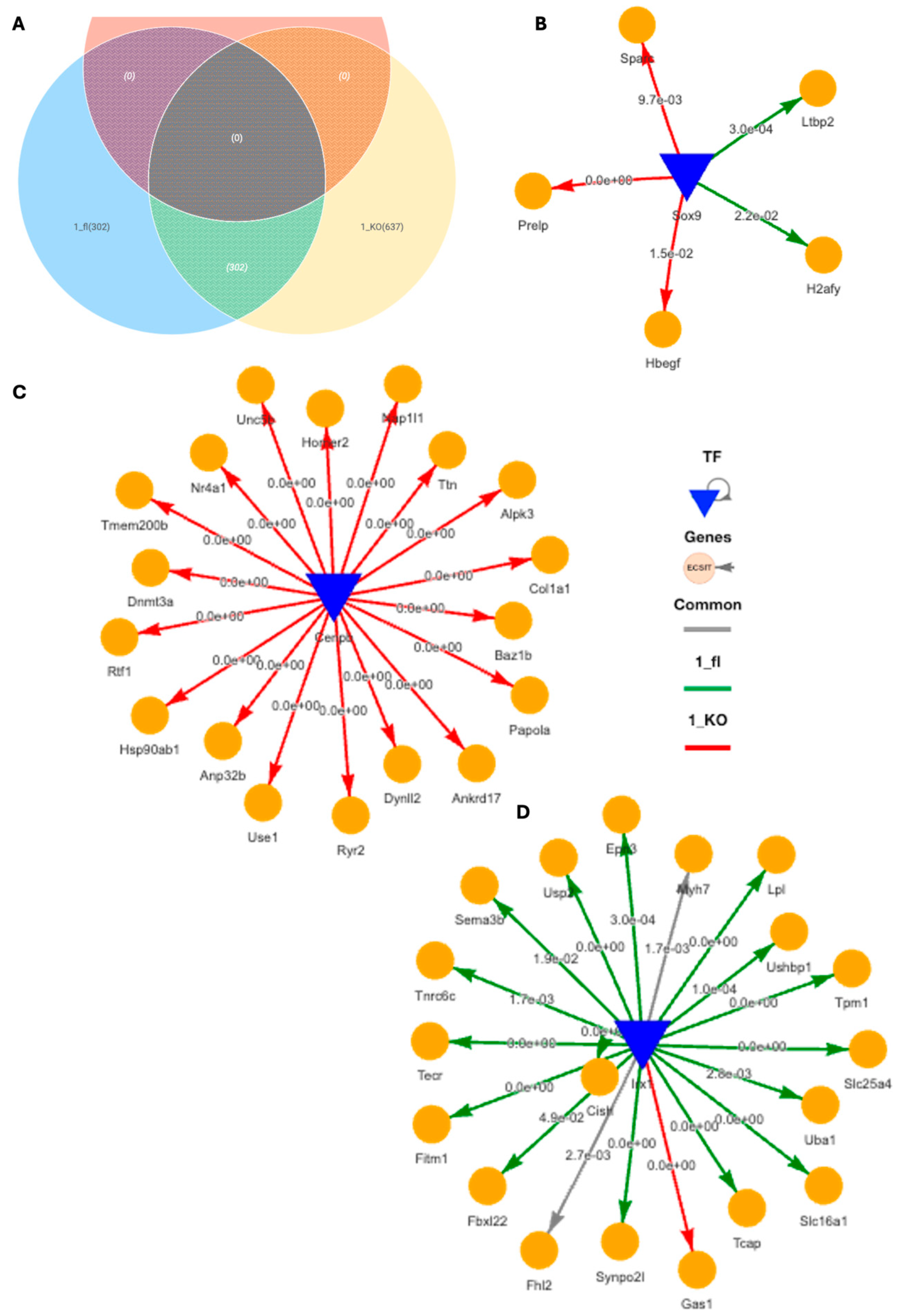
Using DiNiro [39] to analyze differential regulatory disease mechanisms between VDRCM-KO (1_KO) and VDRfl/fl TAC mice (1_fl) in the inner, more stressed hypertrophic tissue niche, we uncovered 302 shared edges, with an additional 302 unique to 1_fl and 637 unique to 1_KO (Figure 5A). Among the significantly highly ranked gene regulatory networks (GRNs), the Sox9-driven network stands out due to its impact in cardiomyocyte-driven hypertrophic cascades [40]. The target genes of Sox9 for the 1_KO are associated with extracellular matrix (ECM) and growth factor signaling (Sparc Prelp, Hbegf) indicative of fibrotic responses, while in 1_fl the targets (H2afy, Ltbp2) hint at mechanisms potentially protective against excessive fibrosis, involving genes in chromatin remodeling and TGF-beta regulation (Figure 5B). CEBPB-driven GRN exclusively regulate genes in 1_KO, such as Col1a1 (fibrosis) and Hsp90ab1 (stress response), as well as dynl2 and dbn3a (cytoskeletal changes), suggesting a focus on structural and stress adaptations (Figure 5C). Additionally, IRX1, a transcription factor associated with anti-fibrotic functions in myocardial health [41], predominantly regulates genes in 1_fl including Fhl2 (modulating cardiac hypertrophy) [42], Lpl (crucial for lipid metabolism and energy utilization) [43], Uba1 (key in protein degradation and cardiac stress management) [44], and Usp2 (regulation of overload-induced cardiac remodeling) [45] (Figure 5D). IRX1 is also the potential mechanistic regulator behind Myh7 for both 1_KO and 1_fl in the inner tissue niche (Figure 5D).
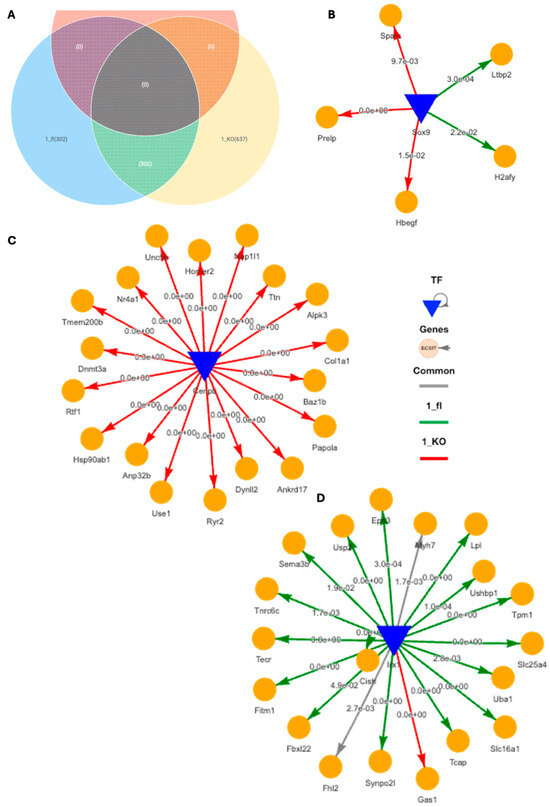
Figure 5.
Differential gene regulatory networks in cardiac hypertrophic tissue niche in TAC VDRCM-KO and VDRfl/fl mice. (A) Venn diagram of shared and unique regulatory edges within the inner, stressed heart tissue region in TAC VDRfl/fl and VDRCM-KO mice. (B) SOX9-driven gene regulatory network (GRN) with target genes specifically in the VDRCM-KO (1_KO, red) or in the VDRfl/fl (1_fl, green) mouse. (C) CEBPB-driven GRN exclusively in 1_KO. (D) IRX1-driven GRN primarily in 1_fl. TF, transcription factor.
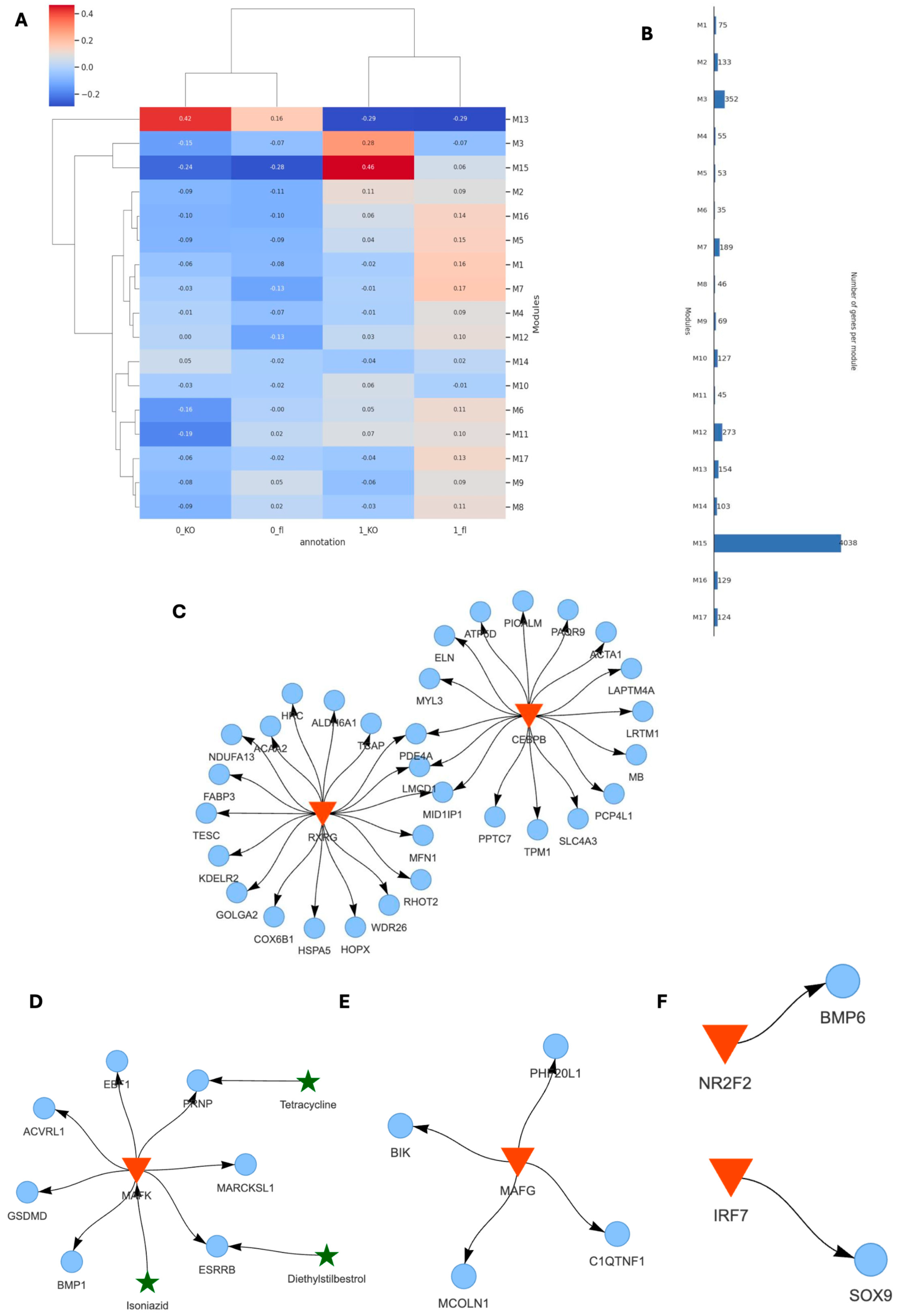
Using SCANet, we analyzed co-expression gene modules across inner and outer tissue regions of TAC VDRCM-KO and VDRfl/fl mice (0_fl, 0_KO, 1_fl, 1_KO) (Figure 6A). This approach enabled de novo identification of genes exhibiting synchronized expression patterns, thus facilitating an unbiased examination of distinct behavioral patterns and their functional implications within specific tissue niches. Notably, modules M13 and M15 showed a stronger correlation in 0_KO and 1_KO, respectively (Figure 6A). Module M13 contains 154 genes, whereas M15 consists of 4038 genes (Figure 6B). Within the 0_KO outer niche, the M13 module featured the major transcription factors RXRG and CEBPB, which targeted the outer signature gene markers (Cox6b1, Acta1) (Figure 4C and Figure 6C). Conversely, the M15 module in the inner niche of 1_KO revealed smaller GRNs, driven by, among others, the pro-inflammatory transcription factors Mafk and Mafg, targeting genes such as C1qtnf1 (complement factor), Acvrl1 (TGF-beta signaling for vascular integrity and fibrosis), Bmp1 (ECM organization, impacting fibrosis and tissue repair), Bik (pro-apoptotic), Mcoln1 (lysosomal function and autophagy), Esrrb (energy metabolism and mitochondrial function), and Prnnp (protective against oxidative stress) (Figure 6D,E). In addition, the transcription factor NR2F2 was connected to BMP6. Given that NR2F2 binding is very flexible, including normally regulating VDR through competitive binding [46], our finding of NR2F2 regulating BMP6 in VDRCM-KO tissue may be a strategic shift in response to VDR’s absence (Figure 6E). Finally, Sox9, the potential regulator of cardiomyocyte-driven hypertrophic pathways (Figure 5B), is regulated by the pro-inflammatory transcription factor IR7 in KO_1 (Figure 6F).
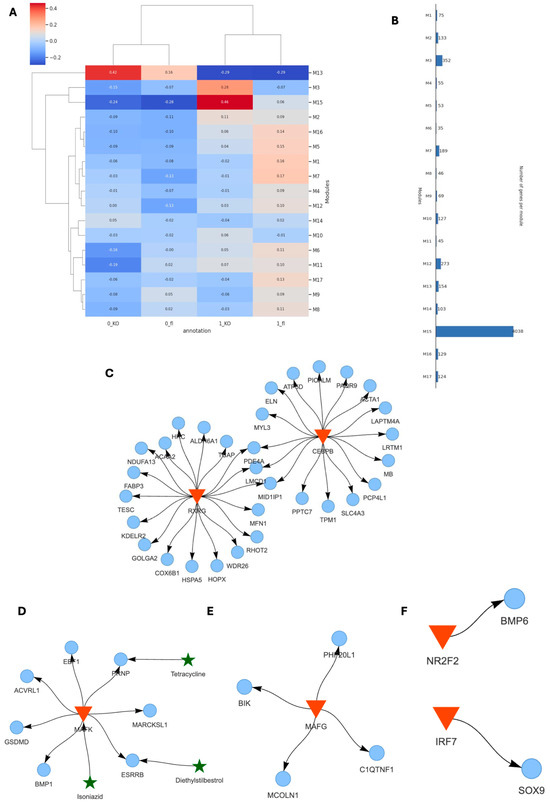
Figure 6.
Gene regulatory networks in different cardiac tissue niches in the absence of VDR. (A) Overview of co-expression gene modules identified in VDRCM-KO and VDRfl/fl models across inner and outer heart regions, illustrating distinct co-expression patterns. (B) Comparison of co-expression gene module sizes. (C) GRN of module M13 in the outer tissue niche in the VDRCM-KO heart (0_KO) including transcription factors (red triangles) and their target genes (blue circles). (D–F) Small GRNs in the inner hypertrophic tissue niche in the VDRCM-KO heart (1_KO) with interacting repurposing drugs (green stars).
3. Discussion
Taken together, our data indicate that vitamin D signaling in cardiomyocytes is dispensable for heart function under physiological conditions in 5-month-old mice. Additionally, we found that the development of cardiomyocyte hypertrophy in response to chronic pressure overload is independent of VDR expression in cardiomyocytes. However, lack of vitamin D signaling in cardiomyocytes aggravated TAC-induced LV functional impairment. Spatial transcriptomics of heart cryosections revealed a more pronounced inflammatory and pro-fibrotic phenotype in the inner, more stressed tissue niche of VDRCM-KO compared with VDRfl/fl mice after TAC.
Our finding that cardiomyocyte-specific VDR deletion does not result in LVH under basal conditions contrasts with the findings reported by Chen et al. [21]. The latter authors suggested that ablation of the VDR in cardiomyocytes results in LVH, both at baseline and after isoproterenol infusion in comparison to WT controls. Given that both studies employed the same Cre driver mouse line, the difference cannot be explained by specificity of Cre expression. The Cre driver mice used are a knock-in model in which parts of the Mlc2v gene were replaced by a Cre cassette [25]. In line with prior studies [26], our data show that Mlc2v (Myl2) haploinsufficiency does not result in a cardiac phenotype. In contrast, homozygous Mlc2v mutant mice develop a severe phenotype and die before birth [25]. The discrepancies between our findings and those of Chen et al. [21] might stem from the use of different VDR floxed mice: our model targets exon 3 [24], whereas theirs targets exon 4 of the VDR gene [21]. Nonetheless, both gene targeting strategies are expected to result in complete VDR inactivation. A more plausible explanation for the discrepancies could be differences in knockout approach: Chen et al. [21] used Mlc2v Cre-expressing VDRfl/- mice as tissue-specific knockout model, i.e., VDR floxed mice on a heterozygous global VDR deficient background. This is a major difference from our study, and likely explains the more severe phenotype in their study [21] due to the interaction between cardiomyocyte-specific VDR deletion and global VDR haploinsufficiency. In light of our finding of an altered inflammatory cardiac phenotype in TAC mice lacking VDR in cardiomyocytes, VDR haploinsufficiency in immune cells may have a major modulating influence in these experiments.
In addition, we found germline deletion in our Mlc2v Cre model, a frequent problem encountered with Cre driver mice, which can make tissue-specific conditional knockout experiments uninterpretable [47]. Mating of Mlc2v Cre-positive VDRfl/wt females with male VDRfl/wt mice frequently resulted in germline VDR deletion. Germline deletion was not observed when male Cre-positive VDRfl/wt mice were crossed with female VDRfl/wt mice. Therefore, we solely utilized paternal inheritance of the Cre allele for the present study. Germline activation of Cre results in global deletion of the floxed allele. Hence, depending on the breeding strategy, this problem can result in heterozygous or even homozygous global deletion of the floxed gene, which in turn may bias the results.
Our finding that mice with a cardiomyocyte-specific deletion of the VDR exhibited more pronounced LV functional impairment after TAC than controls aligns with a post-hoc analysis of the EVITA (Effect of VITamin D on All-cause mortality in heart failure) trial. The latter trial investigated the impact of daily vitamin D supplementation for up to 3 years on various cardiac functional parameters in patients with advanced heart failure. Although the trial did not demonstrate any significant correlation between treatment duration and all examined functional parameters, a noteworthy improvement in cardiac function was noted in aged patients, indicated by a modest, yet statistically significant, increase in EF [48].
In our spatial transcriptomics analysis, we first applied the SPIN algorithm [29] to address autocorrelation issues between neighboring spots, enhancing the resolution of spatial patterns and improving cluster identification between hypertrophic and healthy heart tissues. This precision allowed for a detailed scrutiny of gene regulatory network (GRN). The analysis differentiated heart regions into clusters with distinct gene expressions indicative of normal heart function (outer regions) or stress-induced hypertrophy (inner regions). Notably, Myh6 was prevalent in the outer, while Myh7 was upregulated in the inner regions as adaptation to cardiac stress. In the inner region of the VDRCM-KO TAC heart, a more inflammatory profile was evident compared to the VDRfl/fl TAC heart, characterized by small regulatory differences and activation of unique complement and IL-9 pathways through de novo mechanistic GRNs driven by Mafg, Mafk, and Irf7.
VDR deficiency in the inner, stressed region also correlated with fibrosis-promoting regulatory networks. This finding is in agreement with the upregulation of Col1a1 transcription assessed by qRT-PCR (Figure 2G). Hence, the more pronounced fibrosis-promoting regulatory networks in the inner region of VDRCM-KO TAC hearts may translate into augmented cardiac fibrosis at later time points than the 4 weeks post-TAC used in the current study. CEBPB-driven GRNs appeared to be characteristic of the VDRCM-KO TAC heart across the tissue niches. Yet, the specific genes targeted by these GRNs differed, likely reflecting the diverse microenvironmental influences. Given that CEBPB is influenced by the vitamin D pathway [49], the absence of VDR may explain its aberrant regulatory network. Conversely, the VDRfl/fl mouse had more protective regulatory patterns in the inner, stressed region. We hypothesize that the LV functional impairment associated with VDR deficiency in cardiomyocytes after TAC is the consequence of the more inflammatory phenotype in the inner, stressed regions of the hypertrophic heart. This inflammatory response, coupled with fibrosis-driving GRNs, could potentially explain the observed exacerbation of HFrEF in VDRCM-KO TAC mice. It is conceivable that the loss of VDR signaling in cardiomyocytes may lead to a more uncontrolled inflammatory response, which in turn may worsen cardiac function. It is interesting to note in this context that CEBPB expression in leukocytes has been linked to muscle function in observational studies [50], corroborating the association between inflammatory processes and muscle function.
A limitation of the current study is that we could compare only one heart cryosection of VDRCM-KO and VDRfl/fl TAC mice each in spatial transcriptomics analysis. Nevertheless, our study has uncovered that conditional ablation of the VDR in cardiomyocytes leads to a more pronounced pro-inflammatory and pro-fibrotic gene regulatory phenotype in the inner, more stressed tissue niche in the TAC-induced LVH model. Future studies will be needed to fully unravel the underlying molecular mechanisms of action. A better understanding of the interaction between vitamin D signaling and heart function may eventually lead to new treatment approaches in patients with heart failure.
4. Material and Methods
4.1. Animals
All animal procedures were approved by the Animal Welfare Committee of the Austrian Federal Ministry of Education, Science and Research and were undertaken in accordance with prevailing guidelines for animal care and welfare (BMWFW-68.205/0188-WF/V/3b/2017). All studies were carried out in male mice at 4–5 months of age. Genotype was assessed by PCR on genomic DNA isolated from mouse ear punches. Throughout the experiments, mice were housed at 24 °C with a 12 h light-dark cycle. They were fed a commercial rodent chow (Sniff, Soest, Germany), and had free access to food pellets and tap water. At necropsy, mice were euthanized by exsanguination from the abdominal vena cava under general anesthesia with ketamine/medetomidine (100/0.25 mg/kg) for serum collection.
4.2. Transverse Aortic Constriction (TAC)
The TAC procedure was performed to induce pressure overload and cardiac hypertrophy, as previously described [22]. Briefly, mice were anesthetized with ketamine/medetomidine (100/0.25 mg/kg i.p.), intubated, and ventilated using a small animal ventilator (Model 845, Harvard Apparatus, Holliston, MA, USA). The chest was opened via a midline sternotomy, and the transverse aorta was ligated using a 6-0 silk suture tied around a 27-gauge needle. The needle was then promptly removed, creating a stenosis in the aortic arch. Sham-operated animals underwent the same surgical procedure except that the ligature was not tied. After the TAC or sham procedure, the chest was closed in layers and the animals were allowed to recover on a warming pad. Buprenorphine (0.25 mg/kg s.c.) was administered subcutaneously every 24 h for four days to alleviate pain and enrofloxacin (10 mg/kg s.c.) for five days to prevent infection. Four weeks after the surgery, mice were sacrificed as stated above.
4.3. Doppler Echocardiography
Transthoracic in vivo echocardiography was performed to assess cardiac function using a linear transducer system (Accuson s2000tm, Siemens, Munich, Germany) equipped with a 14-MHz probe, as previously described [22]. Briefly, mice were anesthetized with 1.5% isoflurane and positioned in the supine position on a heating pad. M-mode images were obtained from the parasternal short-axis view at the level of the papillary muscles to measure left ventricular (LV) wall thickness and chamber dimensions (LVIDd, LVIDs, LVAW, and LVPW thickness). Fractional shortening (FS) and ejection fraction (EF) were calculated from these dimensions. At least five cardiac cycles were averaged for each measured parameter.
4.4. Central Arterial and Cardiac Pressure Measurements
Central arterial pressure was measured by inserting a SPR-671 micro-tip catheter (1.4 Millar Instruments, Houston, TX, USA) into the ascending aorta via the right carotid artery under 1.5% isoflurane anesthesia. The catheter was then further advanced into the left ventricle for cardiac pressure measurements. Pressure waveforms were recorded for at least three minutes after stable hemodynamic parameters were achieved and analyzed via LabChart7 software (V. 7.3.8, ADInstruments, Dunedin, New Zealand). Mean arterial blood pressure was calculated as 2/3 diastolic pressure plus 1/3 systolic blood pressure.
4.5. Biochemical Analysis
Serum levels of phosphate and calcium were measured using a Cobas c111 analyzer (Roche, Mannheim, Germany). Serum was extracted with diethylether and resuspended in steroid-free serum (DRG Diagnostics, Marburg, Germany) for the aldosterone ELISA (DRG Diagnostics).
4.6. Tissue Harvesting and Histological Analysis
Hearts were harvested and fixed in 4% formalin. Paraffin-embedded sections at 5-μm thickness were stained with FITC-conjugated wheat germ agglutinin (WGA) to evaluate cardiomyocyte size and picrosirius red (PSR) to assess collagen deposition and fibrosis in the heart as described previously [51]. Images were taken using a Zeiss LSM 880 Airyscan confocal microscope. Cardiomyocyte size and fibrotic areas were assessed with ImageJ (National Institutes of Health) with the investigator blinded for the experimental conditions.
4.7. RNA Isolation and Quantitative Real-Time PCR
Hearts were harvested from mice and snap-frozen in liquid nitrogen. Total RNA was extracted from heart tissue using a TRI Reagent solution (Applied Biosystems, Thermo Fischer Scientific, Waltham, MA, USA) as described previously [51,52]. The purity and integrity of the extracted RNA were assessed using electrophoresis (Agilent Tapestation, Santa Clara, CA, USA). Purified RNA (2 µg) was synthesized into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific, USA). Quantitative RT-PCR was performed on the QTower device (Analytic Jena, Jena, Germany) using Fast Eva Green Kit (Biotium, Fremont, CA, USA) or by TaqMan probe, depending on the assay. All samples were measured in triplicate and normalized to either one reference gene (Dpm1) for Fgf23, Col1a1, and Bnp expression, or two reference genes (Txnl4a and Dpm1) for VDR expression. The 2−ΔΔCt standard method was employed to calculate the relative expression level of genes.
4.8. Statistical Analysis of Phenotyping Data
All phenotyping data are expressed as mean ± standard error of the mean (SEM). Sample sizes were determined by analysis based on data collected by our laboratory in published studies [22,51,52]. Statistical analysis of the data was carried out using GraphPad Prism software (GraphPad Software 8.3.0, San Diego, CA, USA). Comparison between groups was made using one-way analysis of variance (ANOVA) followed by Student–Newman–Keuls test for multiple comparisons. Differences were considered statistically significant at values of p < 0.05.
4.9. Spatial Transcriptomics and Bioinformatic Analysis
Heart cryosections were air-dried, fixed in methanol at −20 °C, and stained with hematoxylin-eosin (HE) according to standard 10× Genomics protocols. Prior to hybridization, high resolution images of the HE-stained sections were taken. Spatial transcriptomics were performed according to standard procedures at the Genomics Core Facility of the Medical University Vienna, using the mouse 10× Genomics Fresh-frozen v2 kit on a CytAssist machine.
4.9.1. Spatial Transcriptomics Data Preprocessing
Spatially resolved transcriptomic datasets from heart sections of TAC VDRCM-KO and VDRfl/fl mice were processed using Scanpy [53]. We performed quality control by filtering out spatial spots with low and extreme total RNA counts (<2500 and >20,000 counts) and genes detected in fewer than 20 spots (Supplementary Figure S4).
4.9.2. Data Integration
The SPIN algorithm [29] integrated TAC VDRCM-KO and VDRfl/fl datasets, addressing batch effects while retaining spatial information. Dimensionality reduction via PCA and spatial pattern visualization through UMAP facilitated the identification of unique expression domains through Leiden clustering.
4.9.3. Differential Expression and Marker Identification
We employed Scanpy’s differential expression analysis to find genes differentiating spatial tissue niches across VDRCM-KO and VDRfl/fl conditions. Selected markers were visualized across tissue sections, linking gene expression patterns to spatial regions.
4.9.4. Functional Enrichment Analysis
GO (Gene Ontology) enrichment analysis, conducted with GSEApy [54] against different gene sets, such as comprehensive biological process, cellular component, and molecular function databases, elucidated the functional landscapes of spatially variable genes, identifying potential biological functions of observed spatial expression patterns.
4.9.5. Differential Gene Regulatory Network Analysis
We conducted a comparative analysis using DiNiro [39] to examine the differences in gene regulatory networks between spots from the 1_KO region (region 1 in VDRCM-KO) and those from 1_fl (region 1 in VDRfl/fl). DiNiro is a computational tool designed to unravel regulatory mechanisms from single-cell data, providing valuable insights into gene expression patterns. Our analysis utilized the following parameters: number of subsamples = 10, sub-sampling size (%) = 80, occurrence threshold (%) = 100, significance cutoff = 0.05. For further details regarding parameter selection and their implications, we refer to the DiNiro publication [39].
4.9.6. Co-Expression Modules and Gene-Regulatory Networks (GRN)
We employed SCANet [55] to investigate differences in gene co-expression networks (GCNs) among various spatial regions in the integrated dataset using the 6000 highly variable genes. SCANet inferred de novo GCN modules from the spatial data and analyzed region associations, identifying modules with altered expression in specific regions. These GCNs were further converted to gene regulatory networks (GRNs), and we conducted a drug repositioning analysis based on these networks.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25115929/s1.
Author Contributions
A.Z., N.L., J.B., M.L.E. and R.G.E. conceived and designed the study; A.Z., N.L., M.O., A.R. and M.L.E. performed experiments, and analyzed and interpreted the data; G.C. and J.B. provided important tools; A.Z., M.O., M.L.E. and R.G.E. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Austrian Science Fund (FWF P26534-B13) to R.G.E., by resources of the University of Veterinary Medicine Vienna and of the Genomics Core Facility of the University of Veterinary Medicine Vienna, and by Open Access Funding of the University of Veterinary Medicine Vienna. The work of M.O. and J.B. is part of the NetMap project and is funded by the German Federal Ministry of Education and Research (BMBF) under grant number (031L0309B). M.L.E. was supported by Lundbeckfonden (R347-2020-2454).
Institutional Review Board Statement
All animal procedures were approved by the Animal Welfare Committee of the Austrian Federal Ministry of Education, Science and Research and were undertaken in accordance with prevailing guidelines for animal care and welfare (BMWFW-68.205/0188-WF/V/3b/2017).
Informed Consent Statement
Not applicable.
Data Availability Statement
All mouse phenotyping data generated or analyzed in this study are included in this article (and its Supplementary Information Files). The spatial transcriptomics raw data that support the findings of this study are available at https://github.com/oubounyt/heart_visium (accessed on 23 May 2024).
Acknowledgments
The authors cordially thank Alexandra Petric, Claudia Bergow, Nikole Ginner, and Christiane Schüler for their excellent technical support and Ute Zeitz for help with animal breeding and husbandry. In addition, the authors acknowledge the Core Facilities of the Medical University of Vienna, a member of VLSI.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schocken, D.D.; Benjamin, E.J.; Fonarow, G.C.; Krumholz, H.M.; Levy, D.; Mensah, G.A.; Narula, J.; Shor, E.S.; Young, J.B.; Hong, Y. Prevention of heart failure: A scientific statement from the American Heart Association Councils on Epidemiology and Prevention, Clinical Cardiology, Cardiovascular Nursing, and High Blood Pressure Research; Quality of Care and Outcomes Research Interdisciplinary Working Group; and Functional Genomics and Translational Biology Interdisciplinary Working Group. Circulation 2008, 117, 2544–2565. [Google Scholar] [PubMed]
- Cheng, S.; Massaro, J.M.; Fox, C.S.; Larson, M.G.; Keyes, M.J.; McCabe, E.L.; Robins, S.J.; O’Donnell, C.J.; Hoffmann, U.; Jacques, P.F.; et al. Adiposity, cardiometabolic risk, and vitamin D status: The Framingham Heart Study. Diabetes 2010, 59, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Melamed, M.L.; Muntner, P.; Michos, E.D.; Uribarri, J.; Weber, C.; Sharma, J.; Raggi, P. Serum 25-hydroxyvitamin D levels and the prevalence of peripheral arterial disease: Results from NHANES 2001 to 2004. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Meng, X.; Tian, Q.; Cao, W.; Fan, X.; Wu, L.; Song, M.; Meng, Q.; Wang, W.; Wang, Y. Vitamin D and Multiple Health Outcomes: An Umbrella Review of Observational Studies, Randomized Controlled Trials, and Mendelian Randomization Studies. Adv. Nutr. 2022, 13, 1044–1062. [Google Scholar] [CrossRef] [PubMed]
- Scragg, R.; Stewart, A.W.; Waayer, D.; Lawes, C.M.M.; Toop, L.; Sluyter, J.; Murphy, J.; Khaw, K.T.; Camargo, C.A., Jr. Effect of Monthly High-Dose Vitamin D Supplementation on Cardiovascular Disease in the Vitamin D Assessment Study: A Randomized Clinical Trial. JAMA Cardiol. 2017, 2, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Manson, J.E.; Cook, N.R.; Lee, I.M.; Christen, W.; Bassuk, S.S.; Mora, S.; Gibson, H.; Gordon, D.; Copeland, T.; D’Agostino, D.; et al. Vitamin D Supplements and Prevention of Cancer and Cardiovascular Disease. N. Engl. J. Med. 2019, 380, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Djousse, L.; Cook, N.R.; Kim, E.; Bodar, V.; Walter, J.; Bubes, V.; Luttmann-Gibson, H.; Mora, S.; Joseph, J.; Lee, I.M.; et al. Supplementation With Vitamin D and Omega-3 Fatty Acids and Incidence of Heart Failure Hospitalization: VITAL-Heart Failure. Circulation 2020, 141, 784–786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Naughton, D.P. Vitamin D in health and disease: Current perspectives. Nutr. J. 2010, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Cauley, J.A.; Lacroix, A.Z.; Wu, L.; Horwitz, M.; Danielson, M.E.; Bauer, D.C.; Lee, J.S.; Jackson, R.D.; Robbins, J.A.; Wu, C.; et al. Serum 25-hydroxyvitamin D concentrations and risk for hip fractures. Ann. Intern. Med. 2008, 149, 242–250. [Google Scholar] [CrossRef]
- Bouillon, R.; Carmeliet, G.; Verlinden, L.; van Etten, E.; Verstuyf, A.; Luderer, H.F.; Lieben, L.; Mathieu, C.; Demay, M. Vitamin D and human health: Lessons from vitamin D receptor null mice. Endocr. Rev. 2008, 29, 726–776. [Google Scholar] [CrossRef]
- Al Mheid, I.; Patel, R.; Murrow, J.; Morris, A.; Rahman, A.; Fike, L.; Kavtaradze, N.; Uphoff, I.; Hooper, C.; Tangpricha, V.; et al. Vitamin D status is associated with arterial stiffness and vascular dysfunction in healthy humans. J. Am. Coll. Cardiol. 2011, 58, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Tishkoff, D.X.; Nibbelink, K.A.; Holmberg, K.H.; Dandu, L.; Simpson, R.U. Functional vitamin D receptor (VDR) in the t-tubules of cardiac myocytes: VDR knockout cardiomyocyte contractility. Endocrinology 2008, 149, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Glenn, D.J.; Ni, W.; Grigsby, C.L.; Olsen, K.; Nishimoto, M.; Law, C.S.; Gardner, D.G. Expression of the vitamin d receptor is increased in the hypertrophic heart. Hypertension 2008, 52, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Gardner, D.G.; Chen, S.; Glenn, D.J. Vitamin D and the heart. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R969–R977. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; You, Y.; Swier, V.; Tang, L.; Radwan, M.M.; Pandya, A.N.; Agrawal, D.K. Vitamin D Protects Against Atherosclerosis via Regulation of Cholesterol Efflux and Macrophage Polarization in Hypercholesterolemic Swine. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 2432–2442. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Pan, W.; Kong, J.; Zheng, W.; Szeto, F.L.; Wong, K.E.; Cohen, R.; Klopot, A.; Zhang, Z.; Li, Y.C. 1,25-dihydroxyvitamin D3 suppresses renin gene transcription by blocking the activity of the cyclic AMP response element in the renin gene promoter. J. Biol. Chem. 2007, 282, 29821–29830. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Watts, S.W.; Ng, M.; Chen, S.; Glenn, D.J.; Gardner, D.G. Elimination of vitamin D receptor in vascular endothelial cells alters vascular function. Hypertension 2014, 64, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Riek, A.E.; Darwech, I.; Funai, K.; Shao, J.; Chin, K.; Sierra, O.L.; Carmeliet, G.; Ostlund, R.E., Jr.; Bernal-Mizrachi, C. Deletion of macrophage Vitamin D receptor promotes insulin resistance and monocyte cholesterol transport to accelerate atherosclerosis in mice. Cell Rep. 2015, 10, 1872–1886. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Kong, J.; Wei, M.; Chen, Z.F.; Liu, S.Q.; Cao, L.P. 1,25-Dihydroxyvitamin D3 is a negative endocrine regulator of the renin-angiotensin system. J. Clin. Investig. 2002, 110, 229–238. [Google Scholar] [CrossRef]
- Andrukhova, O.; Slavic, S.; Zeitz, U.; Riesen, S.C.; Heppelmann, M.S.; Ambrisko, T.D.; Markovic, M.; Kuebler, W.M.; Erben, R.G. Vitamin D is a regulator of endothelial nitric oxide synthase and arterial stiffness in mice. Mol. Endocrinol. 2014, 28, 53–64. [Google Scholar] [CrossRef]
- Chen, S.; Law, C.S.; Grigsby, C.L.; Olsen, K.; Hong, T.T.; Zhang, Y.; Yeghiazarians, Y.; Gardner, D.G. Cardiomyocyte-specific deletion of the vitamin D receptor gene results in cardiac hypertrophy. Circulation 2011, 124, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Slavic, S.; Ford, K.; Modert, M.; Becirovic, A.; Handschuh, S.; Baierl, A.; Katica, N.; Zeitz, U.; Erben, R.G.; Andrukhova, O. Genetic Ablation of Fgf23 or Klotho Does not Modulate Experimental Heart Hypertrophy Induced by Pressure Overload. Sci. Rep. 2017, 7, 11298. [Google Scholar] [CrossRef] [PubMed]
- Ford, K.; Latic, N.; Slavic, S.; Zeitz, U.; Dolezal, M.; Andrukhov, O.; Erben, R.G.; Andrukhova, O. Lack of vitamin D signalling per se does not aggravate cardiac functional impairment induced by myocardial infarction in mice. PLoS ONE 2018, 13, e0204803. [Google Scholar] [CrossRef] [PubMed]
- Meir, T.; Levi, R.; Lieben, L.; Libutti, S.; Carmeliet, G.; Bouillon, R.; Silver, J.; Naveh-Many, T. Deletion of the vitamin D receptor specifically in the parathyroid demonstrates a limited role for the receptor in parathyroid physiology. Am. J. Physiol. Renal Physiol. 2009, 297, F1192–F1198. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Kubalak, S.W.; Minamisawa, S.; Price, R.L.; Becker, K.D.; Hickey, R.; Ross, J., Jr.; Chien, K.R. Selective requirement of myosin light chain 2v in embryonic heart function. J. Biol. Chem. 1998, 273, 1252–1256. [Google Scholar] [CrossRef] [PubMed]
- Minamisawa, S.; Gu, Y.; Ross, J., Jr.; Chien, K.R.; Chen, J. A post-transcriptional compensatory pathway in heterozygous ventricular myosin light chain 2-deficient mice results in lack of gene dosage effect during normal cardiac growth or hypertrophy. J. Biol. Chem. 1999, 274, 10066–10070. [Google Scholar] [CrossRef] [PubMed]
- Merino, D.; Gil, A.; Gomez, J.; Ruiz, L.; Llano, M.; Garcia, R.; Hurle, M.A.; Nistal, J.F. Experimental modelling of cardiac pressure overload hypertrophy: Modified technique for precise, reproducible, safe and easy aortic arch banding-debanding in mice. Sci. Rep. 2018, 8, 3167. [Google Scholar] [CrossRef] [PubMed]
- Eitner, F.; Richter, B.; Schwanen, S.; Szaroszyk, M.; Vogt, I.; Grund, A.; Thum, T.; Heineke, J.; Haffner, D.; Leifheit-Nestler, M. Comprehensive Expression Analysis of Cardiac Fibroblast Growth Factor 23 in Health and Pressure-induced Cardiac Hypertrophy. Front. Cell Dev. Biol. 2021, 9, 791479. [Google Scholar] [CrossRef]
- Maher, K.; Wu, M.; Zhou, Y.; Huang, J.; Zhang, Q.; Wang, X. Mitigating autocorrelation during spatially resolved transcriptomics data analysis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Olson, T.M.; Michels, V.V.; Thibodeau, S.N.; Tai, Y.S.; Keating, M.T. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science 1998, 280, 750–752. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Wan, J.; Zhang, P.; Pei, F. COX6B1 relieves hypoxia/reoxygenation injury of neonatal rat cardiomyocytes by regulating mitochondrial function. Biotechnol. Lett. 2019, 41, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Brewster, L.M.; Haan, Y.C.; Zwinderman, A.H.; van den Born, B.J.; van Montfrans, G.A. CK (Creatine Kinase) Is Associated With Cardiovascular Hemodynamics: The HELIUS Study. Hypertension 2020, 76, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Nomura, S.; Harada, M.; Yamaguchi, T.; Ko, T.; Sumida, T.; Toko, H.; Naito, A.T.; Takeda, N.; Tobita, T.; et al. High-throughput single-molecule RNA imaging analysis reveals heterogeneous responses of cardiomyocytes to hemodynamic overload. J. Mol. Cell Cardiol. 2019, 128, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Yu, P.; Li, D.; Li, Z.; Liao, Y.; Wang, Y.; Zhou, B.; Wang, L. Single-Cell Reconstruction of Progression Trajectory Reveals Intervention Principles in Pathological Cardiac Hypertrophy. Circulation 2020, 141, 1704–1719. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, R.E.; Hedges, D.J.; Morales, A. Dilated cardiomyopathy: The complexity of a diverse genetic architecture. Nat. Rev. Cardiol. 2013, 10, 531–547. [Google Scholar] [CrossRef]
- Nag, S.; Trivedi, D.V.; Sarkar, S.S.; Adhikari, A.S.; Sunitha, M.S.; Sutton, S.; Ruppel, K.M.; Spudich, J.A. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 2017, 24, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Barrick, S.K.; Greenberg, M.J. Cardiac myosin contraction and mechanotransduction in health and disease. J. Biol. Chem. 2021, 297, 101297. [Google Scholar] [CrossRef]
- Martens, P.-J.; Gysemans, C.; Verstuyf, A.; Mathieu, C. Vitamin D’s Effect on Immune Function. Nutrients 2020, 12, 1248. [Google Scholar] [CrossRef] [PubMed]
- Oubounyt, M.; Elkjaer, M.L.; Laske, T.; Grønning, A.G.; Moeller, M.J.; Baumbach, J. De-novo reconstruction and identification of transcriptional gene regulatory network modules differentiating single-cell clusters. NAR Genom. Bioinform. 2023, 5, lqad018. [Google Scholar]
- Schauer, A.; Adams, V.; Poitz, D.M.; Barthel, P.; Joachim, D.; Friedrich, J.; Linke, A.; Augstein, A. Loss of Sox9 in cardiomyocytes delays the onset of cardiac hypertrophy and fibrosis. Int. J. Cardiol. 2019, 282, 68–75. [Google Scholar] [CrossRef]
- Zeng, L.; Gu, N.; Chen, J.; Jin, G.; Zheng, Y. IRX1 hypermethylation promotes heart failure by inhibiting CXCL14 expression. Cell Cycle 2019, 18, 3251–3262. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, F.W.; Reischmann, S.; Schwalm, A.; Unger, A.; Ramanujam, D.; Munch, J.; Muller, O.J.; Hengstenberg, C.; Galve, E.; Charron, P.; et al. FHL2 expression and variants in hypertrophic cardiomyopathy. Basic. Res. Cardiol. 2014, 109, 451. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.A.; Kersten, S.; Qi, L. Lipoprotein Lipase and Its Regulators: An Unfolding Story. Trends Endocrinol. Metab. 2021, 32, 48–61. [Google Scholar] [CrossRef]
- Hunt, L.C.; Pagala, V.; Stephan, A.; Xie, B.; Kodali, K.; Kavdia, K.; Wang, Y.-D.; Shirinifard, A.; Curley, M.; Graca, F.A.; et al. An adaptive stress response that confers cellular resilience to decreased ubiquitination. Nat. Commun. 2023, 14, 7348. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Li, P.; Hong, J.; Wang, M.; Liu, Y.; Gao, Y.; Dong, J.; Gu, H.; Li, L. Overexpression of Ubiquitin-Specific Protease 2 (USP2) in the Heart Suppressed Pressure Overload-Induced Cardiac Remodeling. Mediat. Inflamm. 2020, 2020, 4121750. [Google Scholar] [CrossRef] [PubMed]
- Erdos, E.; Balint, B.L. NR2F2 Orphan Nuclear Receptor is Involved in Estrogen Receptor Alpha-Mediated Transcriptional Regulation in Luminal A Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 1910. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, V.; Martin, C.; Dorchies, E.; Vallez, E.; Dehondt, H.; Trabelsi, M.S.; Tailleux, A.; Caron, S.; Staels, B. Screening strategy to generate cell specific recombination: A case report with the RIP-Cre mice. Transgenic Res. 2015, 24, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Zittermann, A.; Ernst, J.B.; Prokop, S.; Fuchs, U.; Dreier, J.; Kuhn, J.; Knabbe, C.; Birschmann, I.; Schulz, U.; Berthold, H.K.; et al. Effect of vitamin D on all-cause mortality in heart failure (EVITA): A 3-year randomized clinical trial with 4000 IU vitamin D daily. Eur. Heart J. 2017, 38, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Koivisto, O.; Hanel, A.; Carlberg, C. Key Vitamin D Target Genes with Functions in the Immune System. Nutrients 2020, 12, 1140. [Google Scholar] [CrossRef]
- Harries, L.W.; Pilling, L.C.; Hernandez, L.D.; Bradley-Smith, R.; Henley, W.; Singleton, A.B.; Guralnik, J.M.; Bandinelli, S.; Ferrucci, L.; Melzer, D. CCAAT-enhancer-binding protein-beta expression in vivo is associated with muscle strength. Aging Cell 2012, 11, 262–268. [Google Scholar] [CrossRef]
- Latic, N.; Zupcic, A.; Frauenstein, D.; Erben, R.G. Activation of RAAS Signaling Contributes to Hypertension in Aged Hyp Mice. Biomedicines 2022, 10, 1691. [Google Scholar] [CrossRef] [PubMed]
- Latic, N.; Peitzsch, M.; Zupcic, A.; Pietzsch, J.; Erben, R.G. Long-Term Excessive Dietary Phosphate Intake Increases Arterial Blood Pressure, Activates the Renin-Angiotensin-Aldosterone System, and Stimulates Sympathetic Tone in Mice. Biomedicines 2022, 10, 2510. [Google Scholar] [CrossRef] [PubMed]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Liu, X.; Peltz, G. GSEApy: A comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 2022, 39, btac757. [Google Scholar] [CrossRef]
- Oubounyt, M.; Adlung, L.; Patroni, F.; Wenke, N.K.; Maier, A.; Hartung, M.; Baumbach, J.; Elkjaer, M.L. Inference of differential key regulatory networks and mechanistic drug repurposing candidates from scRNA-seq data with SCANet. Bioinformatics 2023, 39, btad644. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).