Deciphering Abnormal Platelet Subpopulations in COVID-19, Sepsis and Systemic Lupus Erythematosus through Machine Learning and Single-Cell Transcriptomics
Abstract
1. Introduction
2. Results
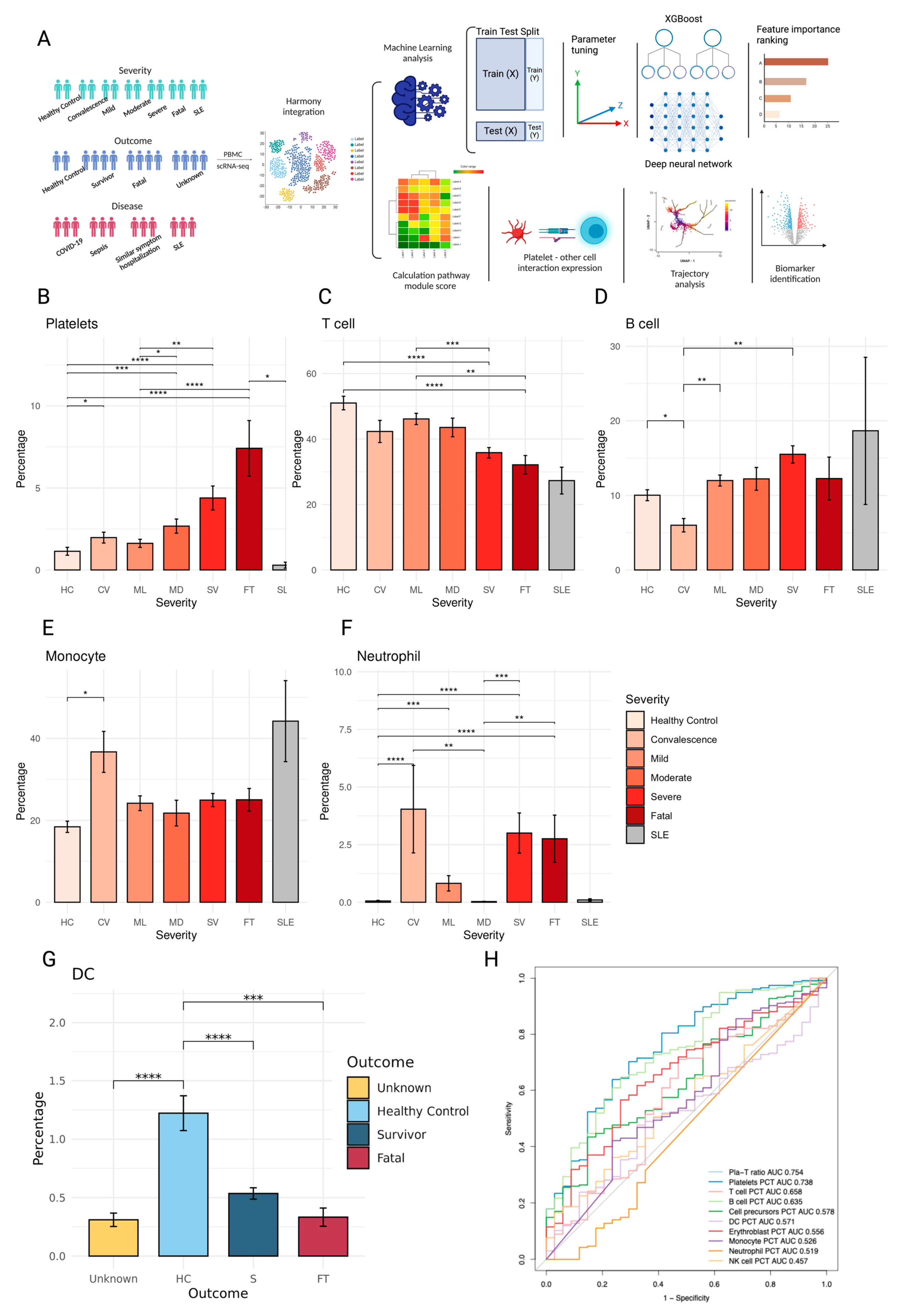
2.1. PBMC Composition Changes with Patient Disease Severity and Outcome
2.2. XGBoost and Deep Neural Network Modeling Identifies Biomarkers of Survival and Fatal Platelets
2.3. Platelets Amplify Endotheliopathy and Disseminated Intravascular Coagulation in Fatal Patients
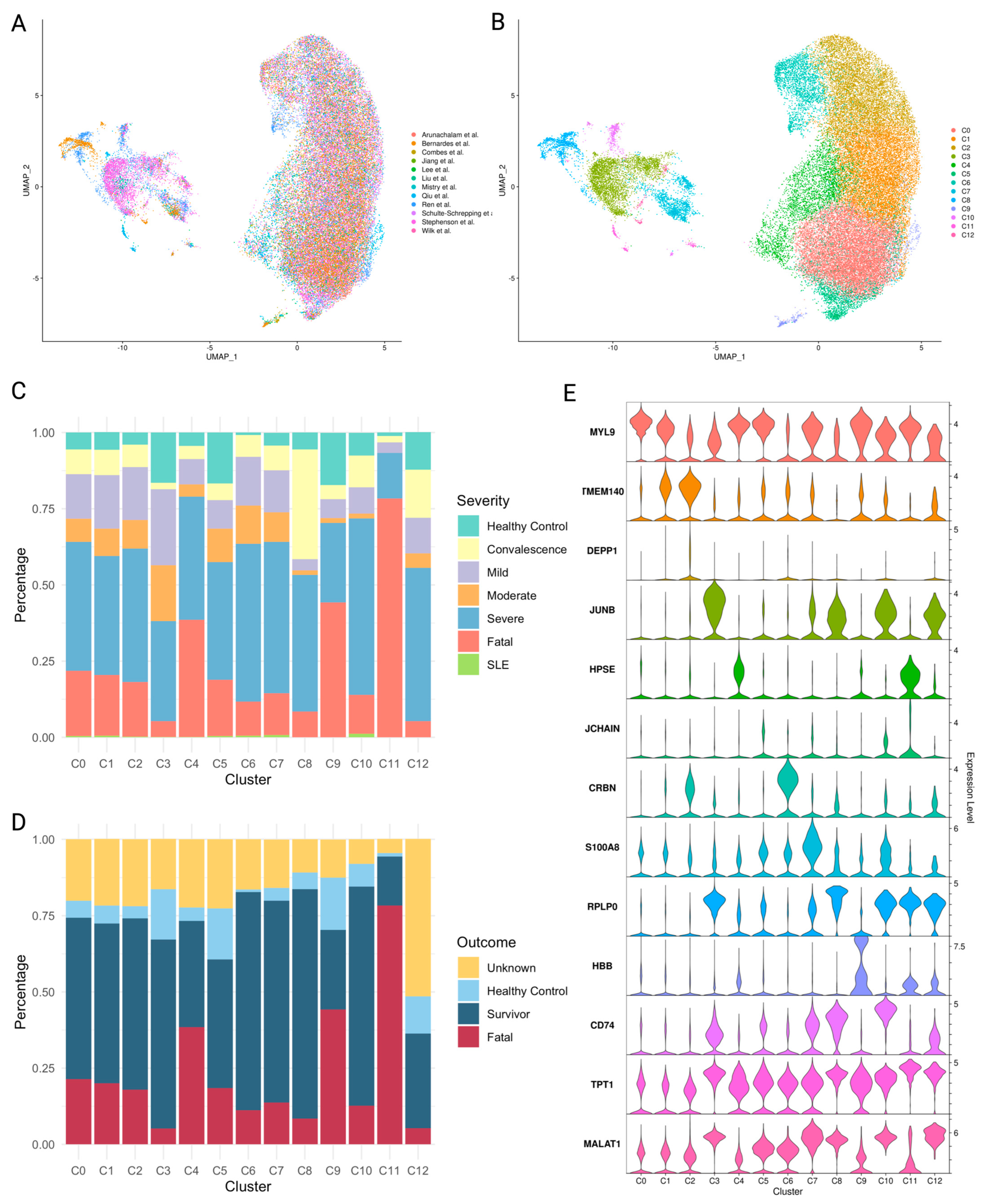
2.4. Platelet Subpopulations Associated with Disease Severity
2.5. Characteristics of Platelet Subpopulations
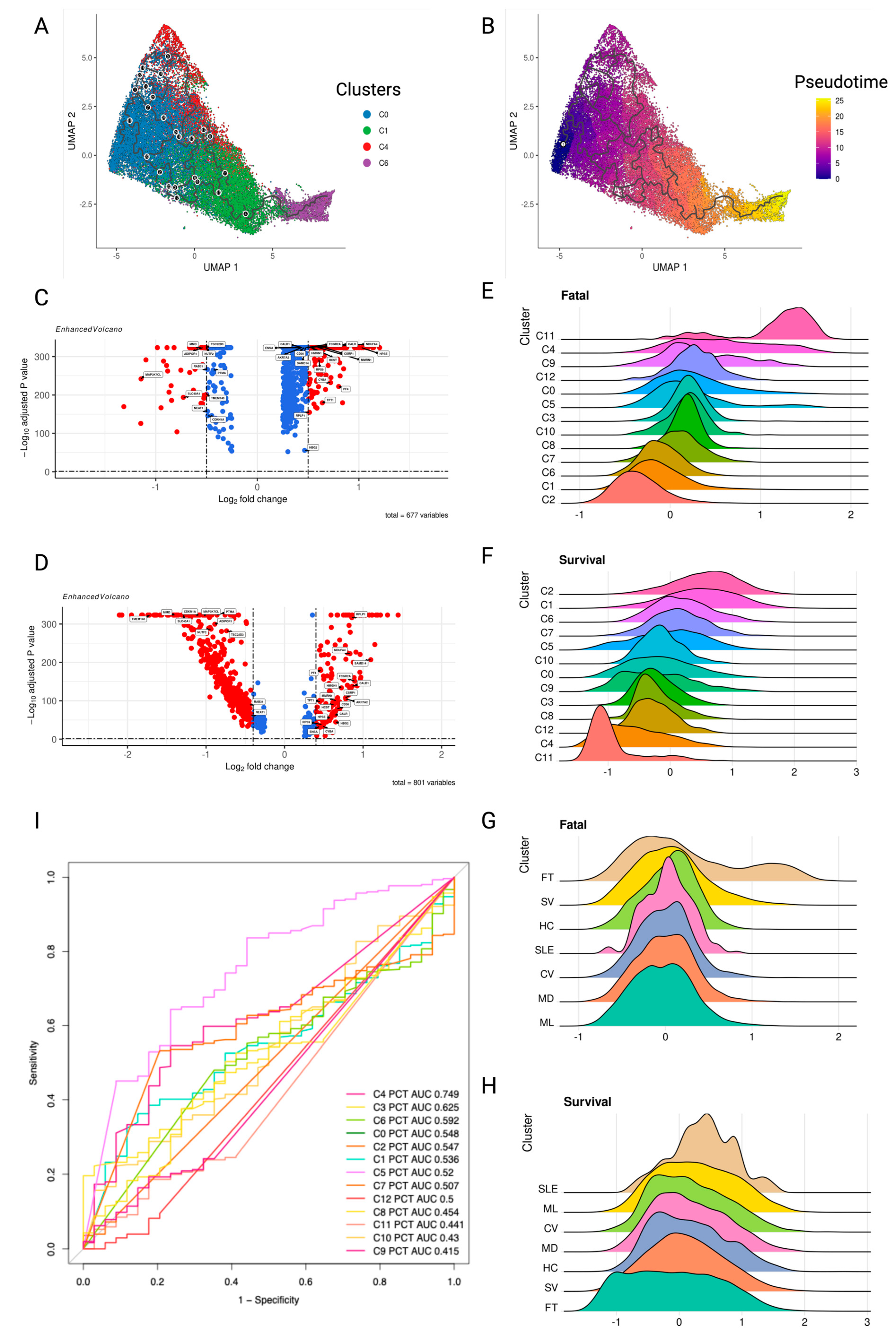
2.6. Pseudotime Trajectory Analysis Identifies Platelet Signature Dynamics in Survival or Fatal Disease Outcomes
2.7. Unique and Shared Gene Expression Changes in Platelets from COVID-19, SSH, Sepsis, and SLE Samples
2.8. Pathway Enrichment Related to Disease Severity in Platelets
2.9. The Platelets’ Crosstalk with Monocytes and Lymphocytes
3. Discussion
4. Materials and Methods
4.1. Integrated Single-Cell Transcriptome Atlas of Peripheral Blood Mononuclear Cells (PBMCs) from COVID-19, Sepsis, and Systemic Lupus Erythematosus Patients
4.2. Removal of Doublets in scRNA-Seq Data and Identifying the Cell Types
4.3. Integrating the Datasets
4.4. Differential Expression Analysis
4.5. Machine Learning Models
4.6. Comparison of Module Scores
4.7. Pathway Enrichment Analysis
4.8. Calculation of Ligand-Receptor Interaction Scores between Platelets and Other Cell Types
- ei,j = expression gene j in cell i
- ncelltypeA = number of cells in the cell type A
4.9. Trajectory Inference of Transition in Platelets Subclusters
4.10. Data and Code Availability Statement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koupenova, M.; Clancy, L.; Corkrey, H.A.; Freedman, J.E. Circulating Platelets as Mediators of Immunity, Inflammation, and Thrombosis. Circ. Res. 2018, 122, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Maouia, A.; Rebetz, J.; Kapur, R.; Semple, J.W. The Immune Nature of Platelets Revisited. Transfus. Med. Rev. 2020, 34, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Yu, S.; Song, Z.; Zhu, X.; Wang, C.; Yan, J.; Wu, F.; Nanda, A.; Granger, D.N.; Li, G. Soluble CD40 ligand stimulates CD40-dependent activation of the β2 integrin Mac-1 and protein kinase C zeda (PKCζ) in neutrophils: Implications for neutrophil-platelet interactions and neutrophil oxidative burst. PLoS ONE 2013, 8, e64631. [Google Scholar] [CrossRef] [PubMed]
- Gudbrandsdottir, S.; Hasselbalch, H.C.; Nielsen, C.H. Activated platelets enhance IL-10 secretion and reduce TNF-α secretion by monocytes. J. Immunol. 2013, 191, 4059–4067. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.A.; Wuescher, L.M.; Worth, R.G. Platelets: Essential components of the immune system. Curr. Trends Immunol. 2015, 16, 65–78. [Google Scholar] [PubMed]
- Chatterjee, M.; von Ungern-Sternberg, S.N.; Seizer, P.; Schlegel, F.; Büttcher, M.; Sindhu, N.A.; Müller, S.; Mack, A.; Gawaz, M. Platelet-derived CXCL12 regulates monocyte function, survival, differentiation into macrophages and foam cells through differential involvement of CXCR4-CXCR7. Cell Death Dis. 2015, 6, e1989. [Google Scholar] [CrossRef] [PubMed]
- Portier, I.; Campbell, R.A. Role of Platelets in Detection and Regulation of Infection. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Xiang, B.; Zhang, G.; Guo, L.; Li, X.A.; Morris, A.J.; Daugherty, A.; Whiteheart, S.W.; Smyth, S.S.; Li, Z. Platelets protect from septic shock by inhibiting macrophage-dependent inflammation via the cyclooxygenase 1 signalling pathway. Nat. Commun. 2013, 4, 2657. [Google Scholar] [CrossRef] [PubMed]
- Duffau, P.; Seneschal, J.; Nicco, C.; Richez, C.; Lazaro, E.; Douchet, I.; Bordes, C.; Viallard, J.F.; Goulvestre, C.; Pellegrin, J.L.; et al. Platelet CD154 potentiates interferon-alpha secretion by plasmacytoid dendritic cells in systemic lupus erythematosus. Sci. Transl. Med. 2010, 2, 47ra63. [Google Scholar] [CrossRef] [PubMed]
- Gorog, D.A.; Storey, R.F.; Gurbel, P.A.; Tantry, U.S.; Berger, J.S.; Chan, M.Y.; Duerschmied, D.; Smyth, S.S.; Parker, W.A.E.; Ajjan, R.A.; et al. Current and novel biomarkers of thrombotic risk in COVID-19: A Consensus Statement from the International COVID-19 Thrombosis Biomarkers Colloquium. Nat. Rev. Cardiol. 2022, 19, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Assinger, A.; Schrottmaier, W.C.; Salzmann, M.; Rayes, J. Platelets in Sepsis: An Update on Experimental Models and Clinical Data. Front. Immunol. 2019, 10, 1687. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Li, J.; Bonenfant, J.; Jaroszewski, L.; Mittal, A.; Klein, W.; Godzik, A.; Nair, M.G. Dynamic changes in human single-cell transcriptional signatures during fatal sepsis. J. Leukoc. Biol. 2021, 110, 1253–1268. [Google Scholar] [CrossRef] [PubMed]
- Linge, P.; Fortin, P.R.; Lood, C.; Bengtsson, A.A.; Boilard, E. The non-haemostatic role of platelets in systemic lupus erythematosus. Nat. Rev. Rheumatol. 2018, 14, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Shannon, O. The role of platelets in sepsis. Res. Pract. Thromb. Haemost. 2021, 5, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Tauseef, A.; Zafar, M.; Arshad, W.; Thirumalareddy, J.; Sood, A.; Farooque, U.; Nair, S.; Mirza, M. Role of immature platelet fraction (IPF) in sepsis patients: A systematic review. J. Fam. Med. Prim. Care 2021, 10, 2148–2152. [Google Scholar] [CrossRef] [PubMed]
- Enz Hubert, R.M.; Rodrigues, M.V.; Andreguetto, B.D.; Santos, T.M.; de Fatima Pereira Gilberti, M.; de Castro, V.; Annichino-Bizzacchi, J.M.; Dragosavac, D.; Carvalho-Filho, M.A.; De Paula, E.V. Association of the immature platelet fraction with sepsis diagnosis and severity. Sci. Rep. 2015, 5, 8019. [Google Scholar] [CrossRef] [PubMed]
- Bergersen, K.V.; Pham, K.; Li, J.; Ulrich, M.T.; Merrill, P.; He, Y.; Alaama, S.; Qiu, X.; Harahap-Carrillo, I.S.; Ichii, K.; et al. Health disparities in COVID-19: Immune and vascular changes are linked to disease severity and persist in a high-risk population in Riverside County, California. BMC Public Health 2023, 23, 1584. [Google Scholar] [CrossRef] [PubMed]
- Wool, G.D.; Miller, J.L. The Impact of COVID-19 Disease on Platelets and Coagulation. Pathobiology 2021, 88, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Puhm, F.; Allaeys, I.; Lacasse, E.; Dubuc, I.; Galipeau, Y.; Zaid, Y.; Khalki, L.; Belleannee, C.; Durocher, Y.; Brisson, A.R.; et al. Platelet activation by SARS-CoV-2 implicates the release of active tissue factor by infected cells. Blood Adv. 2022, 6, 3593–3605. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhou, Q.; Xu, J. Mechanism of thrombocytopenia in COVID-19 patients. Ann. Hematol. 2020, 99, 1205–1208. [Google Scholar] [CrossRef] [PubMed]
- Vils, S.R.; Troldborg, A.; Hvas, A.-M.; Thiel, S. Platelets and the Lectin Pathway of Complement Activation in Patients with Systemic Lupus Erythematosus or Antiphospholipid Syndrome. TH Open 2023, 7, e155–e167. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Joo, J.Y.; Kang, J.; Yu, Y.; Kim, Y.H.; Park, H.R. Single-cell analysis of platelets from patients with periodontitis and diabetes. Res. Pract. Thromb. Haemost. 2023, 7, 100099. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Li, H.Y.; Gao, Y.; Xie, G.Y.; Chi, J.H.; Li, G.L.; Zeng, S.Q.; Xiong, X.M.; Liu, J.H.; Shi, L.L.; et al. Platelet RNA signature independently predicts ovarian cancer prognosis by deep learning neural network model. Protein Cell 2023, 14, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Garma, L.D.; Deng, H.; Goldschmidt, E. Integrated analysis of transcriptomic data reveals the platelet response in COVID-19 disease. Sci. Rep. 2022, 12, 6851. [Google Scholar] [CrossRef] [PubMed]
- Horton, L.E.; Cross, R.W.; Hartnett, J.N.; Engel, E.J.; Sakabe, S.; Goba, A.; Momoh, M.; Sandi, J.D.; Geisbert, T.W.; Garry, R.F.; et al. Endotheliopathy and Platelet Dysfunction as Hallmarks of Fatal Lassa Fever. Emerg. Infect. Dis. 2020, 26, 2625–2637. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.A.; Larsen, J.B.; Hvas, A.M. Platelet function in disseminated intravascular coagulation: A systematic review. Platelets 2018, 29, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Popescu, N.I.; Lupu, C.; Lupu, F. Disseminated intravascular coagulation and its immune mechanisms. Blood 2022, 139, 1973–1986. [Google Scholar] [CrossRef] [PubMed]
- Coenen, D.M.; Mastenbroek, T.G.; Cosemans, J.M.E.M. Platelet interaction with activated endothelium: Mechanistic insights from microfluidics. Blood 2017, 130, 2819–2828. [Google Scholar] [CrossRef] [PubMed]
- Boral, B.M.; Williams, D.J.; Boral, L.I. Disseminated Intravascular Coagulation. Am. J. Clin. Pathol. 2016, 146, 670–680. [Google Scholar] [CrossRef] [PubMed]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martínez-Colón, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Park, S.; Jeong, H.W.; Ahn, J.Y.; Choi, S.J.; Lee, H.; Choi, B.; Nam, S.K.; Sa, M.; Kwon, J.S.; et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020, 5, eabd1554. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, 1419–1440.e23. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tak-Yin Tsang, O.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Wen, W.; Fan, X.; Hou, W.; Su, B.; Cai, P.; Li, J.; Liu, Y.; Tang, F.; Zhang, F.; et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 2021, 184, 1895–1913.e19. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Martins, A.J.; Lau, W.W.; Rachmaninoff, N.; Chen, J.; Imberti, L.; Mostaghimi, D.; Fink, D.L.; Burbelo, P.D.; Dobbs, K.; et al. Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19. Cell 2021, 184, 1836–1857.e22. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, E.; Reynolds, G.; Botting, R.A.; Calero-Nieto, F.J.; Morgan, M.D.; Tuong, Z.K.; Bach, K.; Sungnak, W.; Worlock, K.B.; Yoshida, M.; et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021, 27, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, J.P.; Mishra, N.; Tran, F.; Bahmer, T.; Best, L.; Blase, J.I.; Bordoni, D.; Franzenburg, J.; Geisen, U.; Josephs-Spaulding, J.; et al. Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity 2020, 53, 1296–1314.e9. [Google Scholar] [CrossRef] [PubMed]
- Combes, A.J.; Courau, T.; Kuhn, N.F.; Hu, K.H.; Ray, A.; Chen, W.S.; Chew, N.W.; Cleary, S.J.; Kushnoor, D.; Reeder, G.C.; et al. Global absence and targeting of protective immune states in severe COVID-19. Nature 2021, 591, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Rosborough, B.R.; Chen, J.; Das, S.; Kitsios, G.D.; McVerry, B.J.; Mallampalli, R.K.; Lee, J.S.; Ray, A.; Chen, W.; et al. Single cell RNA sequencing identifies an early monocyte gene signature in acute respiratory distress syndrome. JCI Insight 2020, 5, e135678. [Google Scholar] [CrossRef] [PubMed]
- Mistry, P.; Nakabo, S.; O’Neil, L.; Goel, R.R.; Jiang, K.; Carmona-Rivera, C.; Gupta, S.; Chan, D.W.; Carlucci, P.M.; Wang, X.; et al. Transcriptomic, epigenetic, and functional analyses implicate neutrophil diversity in the pathogenesis of systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 2019, 116, 25222–25228. [Google Scholar] [CrossRef] [PubMed]
- Nakazato, T.; Bono, H.; Matsuda, H.; Takagi, T. Gendoo: Functional profiling of gene and disease features using MeSH vocabulary. Nucleic Acids Res. 2009, 37, W166–W169. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Silva-Filho, J.L.; Caruso-Neves, C.; Pinheiro, A.A.S. IL-4: An important cytokine in determining the fate of T cells. Biophys. Rev. 2014, 6, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Sayols, S. rrvgo: A Bioconductor package for interpreting lists of Gene Ontology terms. Micropublication Biol. 2023, 2023. [Google Scholar] [CrossRef]
- Xu, E.; Xie, Y.; Al-Aly, Z. Long-term neurologic outcomes of COVID-19. Nat. Med. 2022, 28, 2406–2415. [Google Scholar] [CrossRef] [PubMed]
- Widmann, C.N.; Heneka, M.T. Long-term cerebral consequences of sepsis. Lancet Neurol. 2014, 13, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Shen, S.; Rowley, J.W.; Tolley, N.D.; Jia, W.; Manne, B.K.; McComas, K.N.; Bolingbroke, B.; Kosaka, Y.; Krauel, K.; et al. Platelet MHC class I mediates CD8+ T-cell suppression during sepsis. Blood 2021, 138, 401–416. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.; Cardoso, A.P.; Gonçalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef] [PubMed]
- Germann, T.; Gately, M.K.; Schoenhaut, D.S.; Lohoff, M.; Mattner, F.; Fischer, S.; Jin, S.C.; Schmitt, E.; Rüde, E. Interleukin-12/T cell stimulating factor, a cytokine with multiple effects on T helper type 1 (Th1) but not on Th2 cells. Eur. J. Immunol. 1993, 23, 1762–1770. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Bencurova, E.; Psota, E.; Neurgaonkar, P.; Prelog, M.; Scheller, C.; Dandekar, T. Population-Predicted MHC Class II Epitope Presentation of SARS-CoV-2 Structural Proteins Correlates to the Case Fatality Rates of COVID-19 in Different Countries. Int. J. Mol. Sci. 2021, 22, 2630. [Google Scholar] [CrossRef] [PubMed]
- Vardon-Bounes, F.; Ruiz, S.; Gratacap, M.P.; Garcia, C.; Payrastre, B.; Minville, V. Platelets Are Critical Key Players in Sepsis. Int. J. Mol. Sci. 2019, 20, 3494. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, R.I.; Evtugina, N.G.; Peshkova, A.D.; Safiullina, S.I.; Andrianova, I.A.; Khabirova, A.I.; Nagaswami, C.; Khismatullin, R.R.; Sannikova, S.S.; Weisel, J.W. Altered platelet and coagulation function in moderate-to-severe COVID-19. Sci. Rep. 2021, 11, 16290. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.T.; Doherty, G.; Fallon, P.G.; Cummins, E.P. Hypoxia-dependent regulation of inflammatory pathways in immune cells. J. Clin. Investig. 2016, 126, 3716–3724. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, M.D.; Kinning, K.T.; Sullivan, K.D.; Araya, P.; Smith, K.P.; Granrath, R.E.; Shaw, J.R.; Baxter, R.; Jordan, K.R.; Russell, S.; et al. Specialized interferon action in COVID-19. Proc. Natl. Acad. Sci. USA 2022, 119, e2116730119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 2020, 27, 883–890.e2. [Google Scholar] [CrossRef] [PubMed]
- de Lusignan, S.; Hoang, U.; Liyanage, H.; Tripathy, M.; Yonova, I.; Byford, R.; Ferreira, F.; Diez-Domingo, J.; Clark, T. Integrating molecular point-of-care testing for influenza into primary care: A mixed-methods feasibility study. Br. J. Gen. Pract. 2020, 70, e555–e562. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Galani, I.E.; Rovina, N.; Lampropoulou, V.; Triantafyllia, V.; Manioudaki, M.; Pavlos, E.; Koukaki, E.; Fragkou, P.C.; Panou, V.; Rapti, V.; et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021, 22, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, R.; Zhang, S.; Song, S.; Jiang, C.; Han, G.; Wang, M.; Ajani, J.; Futreal, A.; Wang, L. iTALK: An R Package to characterize and illustrate intercellular communication. bioRxiv 2019, 507871. [Google Scholar] [CrossRef]
- Fantuzzi, L.; Tagliamonte, M.; Gauzzi, M.C.; Lopalco, L. Dual CCR5/CCR2 targeting: Opportunities for the cure of complex disorders. Cell. Mol. Life Sci. 2019, 76, 4869–4886. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.E.; Paust, H.J.; Bennstein, S.B.; Bramke, P.; Krebs, C.; Steinmetz, O.M.; Velden, J.; Haag, F.; Stahl, R.A.; Panzer, U. Protective role for CCR5 in murine lupus nephritis. Am. J. Physiol. Renal Physiol. 2012, 302, F1503–F1515. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, M.K.; Armitage, R.J.; Strockbine, L.; Clifford, K.N.; Macduff, B.M.; Sato, T.A.; Maliszewski, C.R.; Fanslow, W.C. Recombinant human CD40 ligand stimulates B cell proliferation and immunoglobulin E secretion. J. Exp. Med. 1992, 176, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Roller, J.; Zhang, S.; Syk, I.; Menger, M.D.; Jeppsson, B.; Thorlacius, H. Metalloproteinases regulate CD40L shedding from platelets and pulmonary recruitment of neutrophils in abdominal sepsis. Inflamm. Res. 2012, 61, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.X.; Tyagi, T.; Jain, K.; Gu, V.W.; Lee, S.H.; Hwa, J.M.; Kwan, J.M.; Krause, D.S.; Lee, A.I.; Halene, S.; et al. Thrombocytopathy and endotheliopathy: Crucial contributors to COVID-19 thromboinflammation. Nat. Rev. Cardiol. 2021, 18, 194–209. [Google Scholar] [CrossRef] [PubMed]
- Songdej, N.; Rao, A.K. Hematopoietic transcription factor mutations: Important players in inherited platelet defects. Blood 2017, 129, 2873–2881. [Google Scholar] [CrossRef] [PubMed]
- Eustes, A.S.; Campbell, R.A.; Middleton, E.A.; Tolley, N.D.; Manne, B.K.; Montenont, E.; Rowley, J.W.; Krauel, K.; Blair, A.; Guo, L.; et al. Heparanase expression and activity are increased in platelets during clinical sepsis. J. Thromb. Haemost. 2021, 19, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Buijsers, B.; Yanginlar, C.; de Nooijer, A.; Grondman, I.; Maciej-Hulme, M.L.; Jonkman, I.; Janssen, N.A.F.; Rother, N.; de Graaf, M.; Pickkers, P.; et al. Increased Plasma Heparanase Activity in COVID-19 Patients. Front. Immunol. 2020, 11, 575047. [Google Scholar] [CrossRef] [PubMed]
- Goshua, G.; Pine, A.B.; Meizlish, M.L.; Chang, C.H.; Zhang, H.; Bahel, P.; Baluha, A.; Bar, N.; Bona, R.D.; Burns, A.J.; et al. Endotheliopathy in COVID-19-associated coagulopathy: Evidence from a single-centre, cross-sectional study. Lancet Haematol. 2020, 7, e575–e582. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.J.; Cornwell, M.; Myndzar, K.; Rolling, C.C.; Xia, Y.; Drenkova, K.; Biebuyck, A.; Fields, A.T.; Tawil, M.; Luttrell-Williams, E.; et al. Platelets amplify endotheliopathy in COVID-19. Sci. Adv. 2021, 7, eabh2434. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C. Sepsis and septic shock: Endothelial molecular pathogenesis associated with vascular microthrombotic disease. Thromb. J. 2019, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Rondina, M.T.; Carlisle, M.; Fraughton, T.; Brown, S.M.; Miller, R.R.; Harris, E.S.; Weyrich, A.S.; Zimmerman, G.A.; Supiano, M.A.; Grissom, C.K. Platelet-monocyte aggregate formation and mortality risk in older patients with severe sepsis and septic shock. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Engel, J.; Teichmann, S.A.; Lonnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Abdelnabi, J.; Al-Dulaimi, R.; Aggarwal, A.; Ramos, M.; Davis, S.; Riester, M.; Waldron, L. HGNChelper: Identification and correction of invalid gene symbols for human and mouse. F1000Research 2020, 9, 1493. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Finak, G.; McDavid, A.; Yajima, M.; Deng, J.; Gersuk, V.; Shalek, A.K.; Slichter, C.K.; Miller, H.W.; McElrath, M.J.; Prlic, M.; et al. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015, 16, 278. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Carlson, M. GO.db: A Set of Annotation Maps Describing the Entire Gene Ontology, R Package Version 3.8.2; Bioconductor: Boston, MA, USA, 2019.
- Frederick, M.; Skinner, H.D.; Kazi, S.A.; Sikora, A.G.; Sandulache, V.C. High expression of oxidative phosphorylation genes predicts improved survival in squamous cell carcinomas of the head and neck and lung. Sci. Rep. 2020, 10, 6380. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Yan, G.R.; He, Q.Y. DOSE: An R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 2015, 31, 608–609. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.P.; Du, J.; Lagoudas, G.; Jiao, Y.; Sawyer, A.; Drummond, D.C.; Lauffenburger, D.A.; Raue, A. Analysis of Single-Cell RNA-Seq Identifies Cell-Cell Communication Associated with Tumor Characteristics. Cell Rep. 2018, 25, 1458–1468.e4. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979. [Google Scholar] [CrossRef] [PubMed]
- Haghverdi, L.; Lun, A.T.L.; Morgan, M.D.; Marioni, J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018, 36, 421–427. [Google Scholar] [CrossRef] [PubMed]





| Pathway Module | Healthy Control | Convalescence | Mild | Moderate | Severe | Fatal | SLE |
|---|---|---|---|---|---|---|---|
| Glycolysis | 0.0221 | 0.0585 | 0.0151 | 0.0065 | 0.0131 | 0.0736 | −0.0675 |
| MHC ClassII | −0.0319 | 0.0672 | −0.0318 | −0.0438 | −0.0575 | −0.1183 | −0.0519 |
| OXPHOS | 0.0323 | 0.0419 | 0.0191 | 0.047 | 0.0474 | 0.0801 | 0.0469 |
| Coagulation | 0.3007 | 0.2432 | 0.3034 | 0.2642 | 0.3243 | 0.3643 | 0.2209 |
| Response to Interferon Beta | −0.1375 | −0.0454 | −0.0335 | 0.0277 | −0.0373 | −0.0263 | −0.0674 |
| Response to Interferon Gamma | 0.094 | 0.1052 | 0.1109 | 0.1293 | 0.1024 | 0.1091 | 0.0603 |
| Response to Type I Interferon | −0.0947 | −0.0314 | −0.0091 | 0.0364 | −0.0245 | −0.0201 | −0.0045 |
| Translational Initiation | 0.0877 | 0.0778 | 0.0653 | 0.0841 | 0.0549 | 0.0535 | 0.058 |
| Group | Platelets |
|---|---|
| Healthy Controls (HC) | 3205 |
| Convalescence (CV) | 3695 |
| Mild (ML) | 7359 |
| Moderate (MD) | 4330 |
| Severe (SV) | 19,805 |
| Fatal (FT) | 9414 |
| Systemic Lupus Erythematosus (SLE) | 169 |
| Outcomes | |
| Fatal (FT) | 9414 |
| Survivors (S) | 25,750 |
| Unknown | 9608 |
| Diseases | |
| COVID-19 | 38,673 |
| Hospitalized Patients with Similar Symptoms (SSH) | 2508 |
| Sepsis | 3422 |
| Systemic Lupus Erythematosus (SLE) | 169 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, X.; Nair, M.G.; Jaroszewski, L.; Godzik, A. Deciphering Abnormal Platelet Subpopulations in COVID-19, Sepsis and Systemic Lupus Erythematosus through Machine Learning and Single-Cell Transcriptomics. Int. J. Mol. Sci. 2024, 25, 5941. https://doi.org/10.3390/ijms25115941
Qiu X, Nair MG, Jaroszewski L, Godzik A. Deciphering Abnormal Platelet Subpopulations in COVID-19, Sepsis and Systemic Lupus Erythematosus through Machine Learning and Single-Cell Transcriptomics. International Journal of Molecular Sciences. 2024; 25(11):5941. https://doi.org/10.3390/ijms25115941
Chicago/Turabian StyleQiu, Xinru, Meera G. Nair, Lukasz Jaroszewski, and Adam Godzik. 2024. "Deciphering Abnormal Platelet Subpopulations in COVID-19, Sepsis and Systemic Lupus Erythematosus through Machine Learning and Single-Cell Transcriptomics" International Journal of Molecular Sciences 25, no. 11: 5941. https://doi.org/10.3390/ijms25115941
APA StyleQiu, X., Nair, M. G., Jaroszewski, L., & Godzik, A. (2024). Deciphering Abnormal Platelet Subpopulations in COVID-19, Sepsis and Systemic Lupus Erythematosus through Machine Learning and Single-Cell Transcriptomics. International Journal of Molecular Sciences, 25(11), 5941. https://doi.org/10.3390/ijms25115941






