Identification of a Novel miR-195-5p/PNN Axis in Colorectal Cancer
Abstract
1. Introduction
2. Results
2.1. PNN as a Target of miR-195-5p
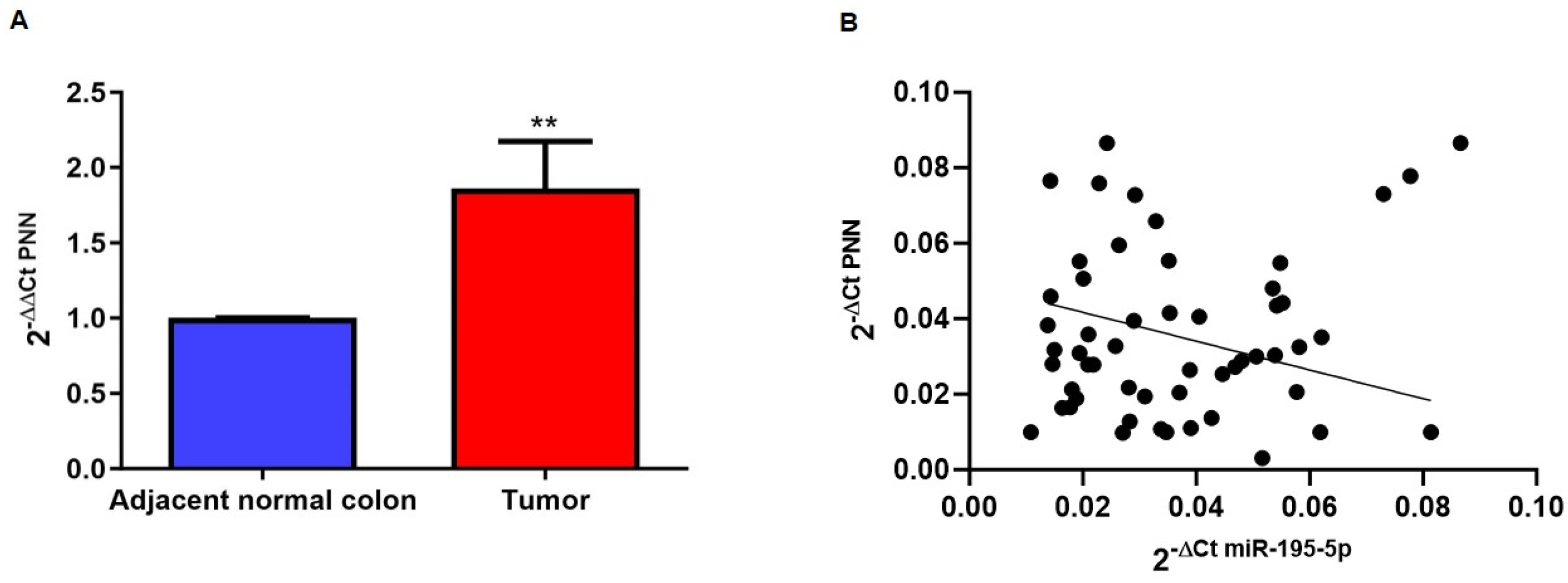
2.2. PNN Expression in CRC Patients
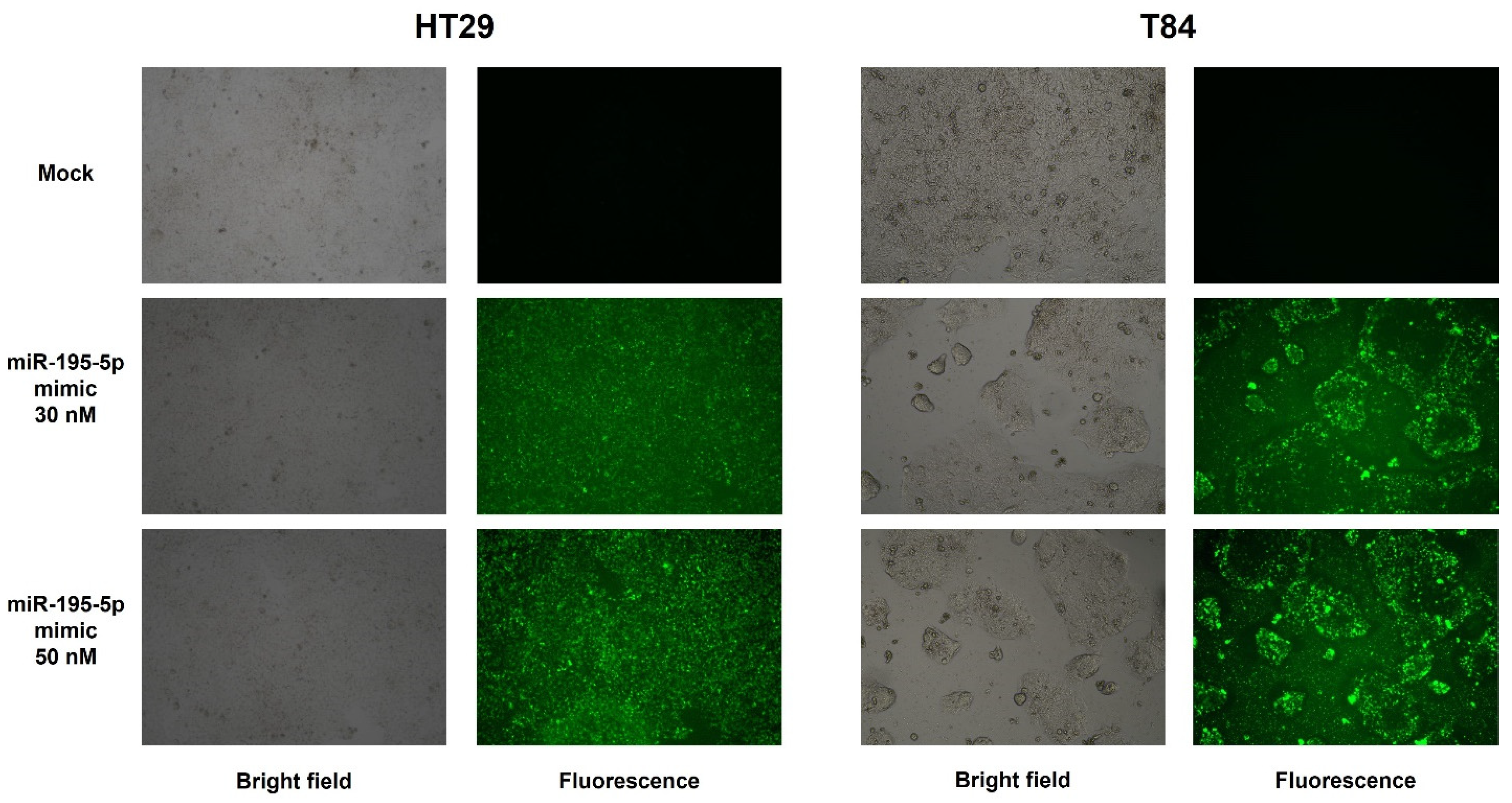
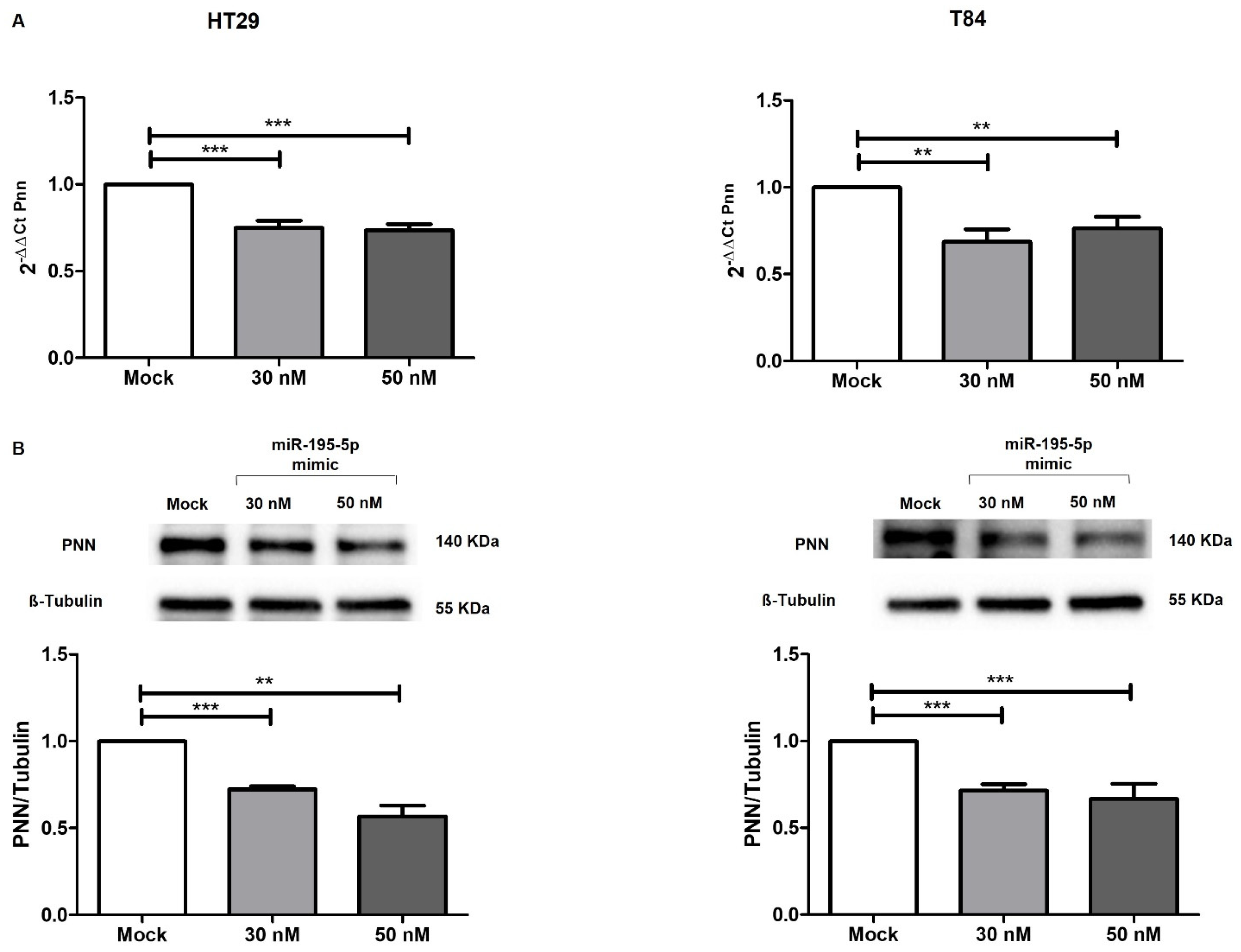
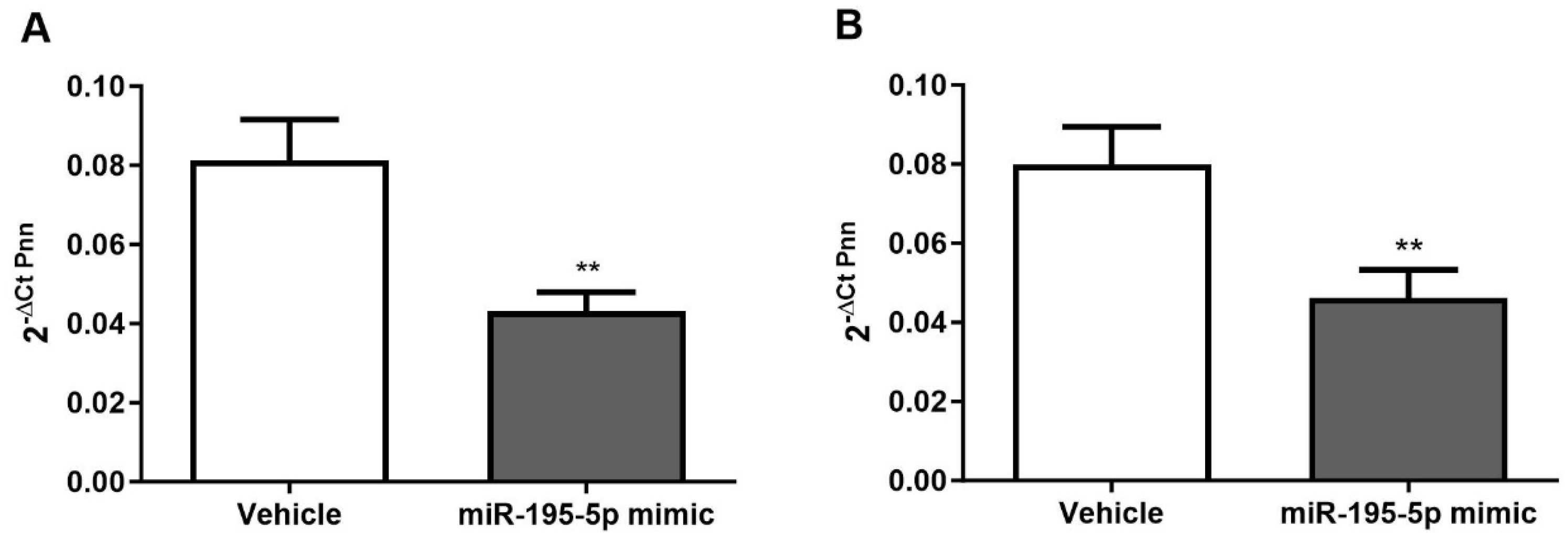
2.3. miR-195-5p Is an Effective Regulator of PNN Expression
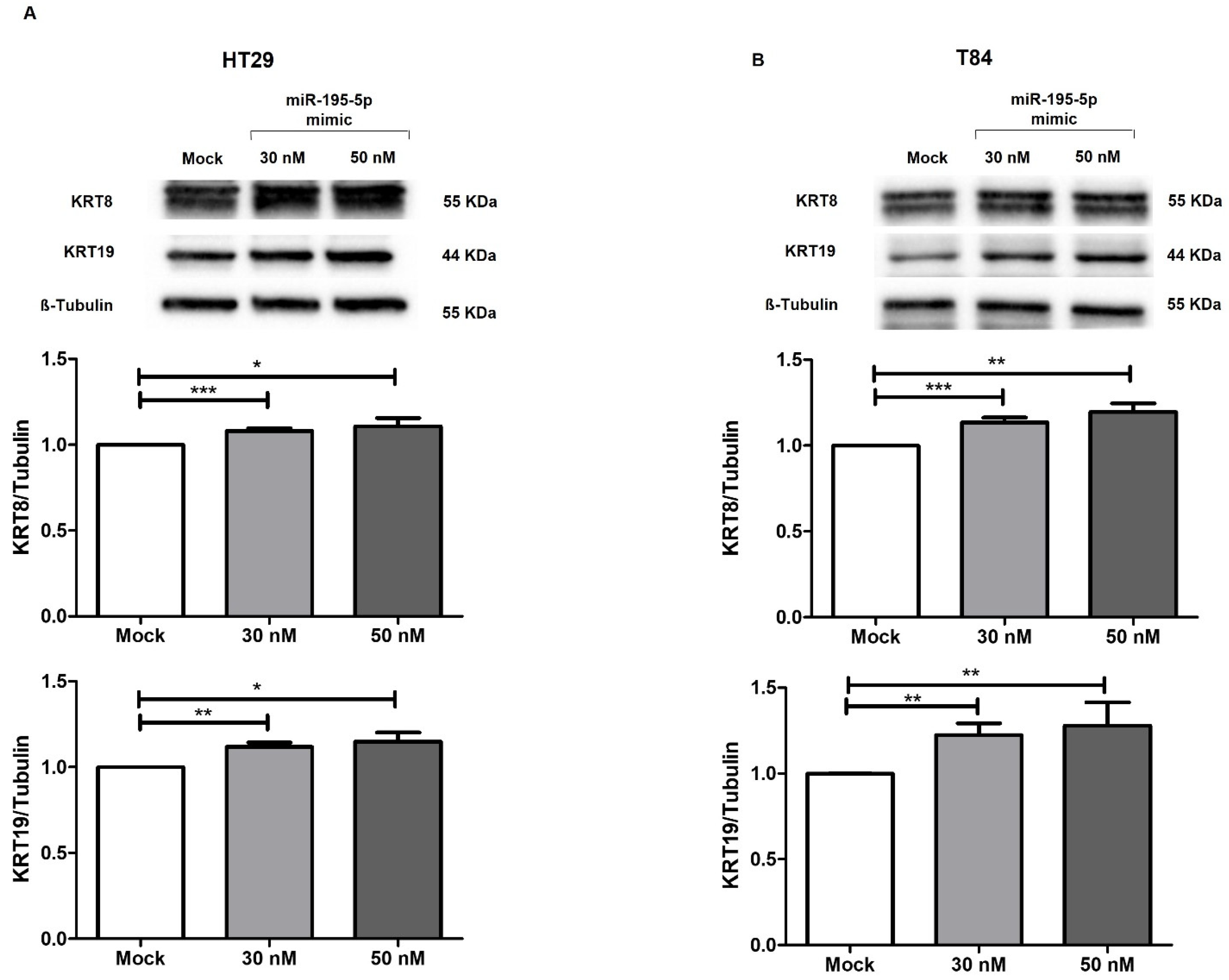
2.4. miR-195-5p Indirectly Modulates the Expression of Cytokeratin-8 and Cytokeratin-19
2.5. PNN Determination after miR-195-5p Mimic Treatment in a Mouse Model of AOM/DSS-Induced CRC
3. Discussion
4. Materials and Methods
4.1. Human Tissues
4.2. RNA Extraction and Real-Time PCR of FFPE Tissue
4.3. Immunohistochemistry (IHC)
4.4. Cell Culture and In Vitro Transfection
4.5. RNA Extraction and Real-Time PCR
4.6. Protein Isolation and Western Blot Analysis
4.7. Immunofluorescence
4.8. Bioinformatic and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rusu, A.D.; Georgiou, M. The multifarious regulation of the apical junctional complex. Open Biol. 2020, 10, 190278. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Cell adhesion in development: A complex signaling network. Curr. Opin. Genet. Dev. 2003, 13, 365–371. [Google Scholar] [CrossRef]
- Garrod, D.; Chidgey, M. Desmosome structure, composition and function. Biochim. Biophys. Acta 2008, 1778, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Brooke, M.A.; Nitoiu, D.; Kelsell, D.P. Cell–cell connectivity: Desmosomes and disease. J. Pathol. 2012, 226, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Brückner, B.R.; Janshoff, A. Importance of integrity of cell-cell junctions for the mechanics of confluent MDCK II cells. Sci. Rep. 2018, 8, 14117. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Yang, L.; Gray, A.; Srivastava, A.K.; Li, C.; Zhang, G.; Cui, T. The role of desmosomes in carcinogenesis. OncoTargets Ther. 2017, 10, 4059–4063. [Google Scholar] [CrossRef]
- Chidgey, M.; Dawson, C. Desmosomes: A role in cancer? Br. J. Cancer 2007, 96, 1783–1787. [Google Scholar] [CrossRef]
- Al-Amoudi, A.; Castaño-Diez, D.; Devos, D.P.; Russell, R.B.; Johnson, G.T.; Frangakis, A.S. The three-dimensional molecular structure of the desmosomal plaque. Proc. Natl. Acad. Sci. USA 2011, 108, 6480–6485. [Google Scholar] [CrossRef]
- Broussard, J.A.; Getsios, S.; Green, K.J. Desmosome regulation and signaling in disease. Cell Tissue Res. 2015, 360, 501–512. [Google Scholar] [CrossRef]
- Müller, L.; Hatzfeld, M.; Keil, R. Desmosomes as signaling hubs in the regulation of cell behavior. Front. Cell Dev. Biol. 2021, 9, 745670. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-Z.; Ren, Y.-Y.; Wang, K.-J.; Chen, J.-F.; Su, R.; Jiang, J.-H.; Wang, P.; Ma, Q. Pinin promotes tumor progression via activating CREB through PI3K/AKT and ERK/MAPK pathway in prostate cancer. Am. J. Cancer Res. 2021, 11, 1286–1303. [Google Scholar] [PubMed]
- Ouyang, P.; Sugrue, S.P. Characterization of pinin, a novel protein associated with the desmosome-intermediate filament complex. J. Cell Biol. 1996, 135, 1027–1042. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tabesh, M.; Sugrue, S.P. Role of cell Adhesion–Associated protein, pinin (DRS/memA), in corneal epithelial migration. Investig. Ophthalmol. Vis. Sci. 2000, 41, 1337–1345. [Google Scholar]
- Shi, J.; Sugrue, S.P. Dissection of protein linkage between keratins and pinin, a protein with dual location at desmosome-intermediate filament complex and in the nucleus. J. Biol. Chem. 2000, 275, 14910–14915. [Google Scholar] [CrossRef] [PubMed]
- Brandner, J.M.; Reidenbach, S.; Franke, W.W. Evidence that “pinin”, reportedly a differentiation-specific desmosomal protein, is actually a widespread nuclear protein. Differentiation 1997, 62, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Akin, D.; Newman, J.R.; McIntyre, L.M.; Sugrue, S.P. RNA-seq analysis of impact of PNN on gene expression and alternative splicing in corneal epithelial cells. Mol. Vis. 2016, 22, 40–60. [Google Scholar]
- Joo, J.-H.; Correia, G.P.; Li, J.-L.; Lopez, M.-C.; Baker, H.V.; Sugrue, S.P. Transcriptomic analysis of PNN-and ESRP1-regulated alternative pre-mRNA splicing in human corneal epithelial cells. Investig. Ophthalmol. Vis. Sci. 2013, 54, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.H.; Ryu, D.; Peng, Q.; Sugrue, S.P. Role of Pnn in alternative splicing of a specific subset of lncRNAs of the corneal epithelium. Mol. Vis. 2014, 20, 1629–1642. [Google Scholar]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Scalavino, V.; Piccinno, E.; Bianco, G.; Schena, N.; Armentano, R.; Giannelli, G.; Serino, G. The increase of miR-195-5p reduces intestinal permeability in ulcerative colitis, modulating tight junctions’ expression. Int. J. Mol. Sci. 2022, 23, 5840. [Google Scholar] [CrossRef]
- Scalavino, V.; Piccinno, E.; Valentini, A.M.; Schena, N.; Armentano, R.; Giannelli, G.; Serino, G. miR-369-3p Modulates Intestinal Inflammatory Response via BRCC3/NLRP3 Inflammasome Axis. Cells 2023, 12, 2184. [Google Scholar] [CrossRef] [PubMed]
- Scalavino, V.; Piccinno, E.; Valentini, A.M.; Mastronardi, M.; Armentano, R.; Giannelli, G.; Serino, G. A novel mechanism of immunoproteasome regulation via miR-369-3p in intestinal inflammatory response. Int. J. Mol. Sci. 2022, 23, 13771. [Google Scholar] [CrossRef] [PubMed]
- Scalavino, V.; Piccinno, E.; Lacalamita, A.; Tafaro, A.; Armentano, R.; Giannelli, G.; Serino, G. miR-195-5p Regulates Tight Junctions Expression via Claudin-2 Downregulation in Ulcerative Colitis. Biomedicines 2022, 10, 919. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhang, Q.; Deng, M.; Miao, J.; Guo, Y.; Gao, W.; Cui, Q. An analysis of human microRNA and disease associations. PLoS ONE 2008, 3, e3420. [Google Scholar] [CrossRef] [PubMed]
- Piccinno, E.; Scalavino, V.; Armentano, R.; Giannelli, G.; Serino, G. miR-195-5p as Regulator of γ-Catenin and Desmosome Junctions in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 17084. [Google Scholar] [CrossRef] [PubMed]
- Piccinno, E.; Scalavino, V.; Labarile, N.; Bianco, G.; Savino, M.T.; Armentano, R.; Giannelli, G.; Serino, G. Downregulation of γ-Catenin by miR-195-5p Inhibits Colon Cancer Progression, Regulating Desmosome Function. Int. J. Mol. Sci. 2023, 25, 494. [Google Scholar] [CrossRef]
- Slaby, O.; Svoboda, M.; Fabian, P.; Smerdova, T.; Knoflickova, D.; Bednarikova, M.; Nenutil, R.; Vyzula, R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 2008, 72, 397–402. [Google Scholar] [CrossRef]
- Pidíkova, P.; Reis, R.; Herichova, I. miRNA clusters with down-regulated expression in human colorectal cancer and their regulation. Int. J. Mol. Sci. 2020, 21, 4633. [Google Scholar] [CrossRef]
- Rübsam, M.; Broussard, J.A.; Wickström, S.A.; Nekrasova, O.; Green, K.J.; Niessen, C.M. Adherens junctions and desmosomes coordinate mechanics and signaling to orchestrate tissue morphogenesis and function: An evolutionary perspective. Cold Spring Harb. Perspect. Biol. 2018, 10, a029207. [Google Scholar] [CrossRef]
- Green, K.J.; Jaiganesh, A.; Broussard, J.A. Desmosomes: Essential contributors to an integrated intercellular junction network. F1000Research 2019, 8, 2150. [Google Scholar] [CrossRef]
- Lim, H.Y.G.; Plachta, N. Cytoskeletal control of early mammalian development. Nat. Rev. Mol. Cell Biol. 2021, 22, 548–562. [Google Scholar] [CrossRef]
- Jaalouk, D.E.; Lammerding, J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009, 10, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Luciano, M.; Versaevel, M.; Vercruysse, E.; Procès, A.; Kalukula, Y.; Remson, A.; Deridoux, A.; Gabriele, S. Appreciating the role of cell shape changes in the mechanobiology of epithelial tissues. Biophys. Rev. 2022, 3, 011305. [Google Scholar] [CrossRef]
- Delva, E.; Tucker, D.K.; Kowalczyk, A.P. The desmosome. Cold Spring Harb. Perspect. Biol. 2009, 1, a002543. [Google Scholar] [CrossRef] [PubMed]
- Nekrasova, O.; Green, K.J. Desmosome assembly and dynamics. Trends Cell Biol. 2013, 23, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Ma, W.; Qi, X.; Zhu, X.; Wang, Y.; Xu, Z.; Luo, J.; Wang, D.; Guo, W.; Li, X.; et al. Pinin facilitated proliferation and metastasis of colorectal cancer through activating EGFR/ERK signaling pathway. Oncotarget 2016, 7, 29429–29439. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lin, R.-I.; Lai, M.-C.; Ouyang, P.; Tarn, W.-Y. Nuclear Pnn/DRS protein binds to spliced mRNPs and participates in mRNA processing and export via interaction with RNPS1. Mol. Cell. Biol. 2003, 23, 763–7376. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lou, P.-J.; Leu, S.; Ouyang, P. Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochem. Biophys. Res. Commun. 2002, 294, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Alpatov, R.; Munguba, G.C.; Caton, P.; Joo, J.H.; Shi, Y.; Shi, Y.; Hunt, M.E.; Sugrue, S.P. Nuclear speckle-associated protein Pnn/DRS binds to the transcriptional corepressor CtBP and relieves CtBP-mediated repression of the E-cadherin gene. Mol. Cell. Biol. 2004, 24, 10223–10235. [Google Scholar] [CrossRef]
- Leu, S. The role and regulation of Pnn in proliferative and non-dividing cells: Form embryogenesis to pathogenesis. Biochem. Pharmacol. 2021, 192, 114672. [Google Scholar] [CrossRef]
- Hsu, S.-Y.; Chen, Y.-J.; Ouyang, P. Pnn and SR family proteins are differentially expressed in mouse central nervous system. Histochem. Cell Biol. 2011, 135, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.-H.; Taxter, T.J.; Munguba, G.C.; Kim, Y.H.; Dhaduvai, K.; Dunn, N.W.; Degan, W.J.; Oh, S.P.; Sugrue, S.P. Pinin modulates expression of an intestinal homeobox gene, Cdx2, and plays an essential role for small intestinal morphogenesis. Dev. Biol. 2010, 345, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Li, D.; Liu, W.; Wang, P.; Xiang, Z.; Liu, K. Pinin acts as a poor prognostic indicator for renal cell carcinoma by reducing apoptosis and promoting cell migration and invasion. J. Cell. Mol. Med. 2021, 25, 4340–4348. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Wu, X. Hsa_circ_0032463 acts as the tumor promoter in osteosarcoma by regulating the miR-330-3p/PNN axis. Int. J. Mol. Med. 2021, 47, 92. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sun, D.; Dong, C.; Tian, Y.; Gao, Z.; Wang, L. Pinin associates with prognosis of hepatocellular carcinoma through promoting cell proliferation and suppressing glucose deprivation-induced apoptosis. Oncotarget 2016, 7, 39694–39704. [Google Scholar] [CrossRef]
- Mini, E.; Lapucci, A.; Perrone, G.; D’Aurizio, R.; Napoli, C.; Brugia, M.; Landini, I.; Tassi, R.; Picariello, L.; Simi, L.; et al. RNA sequencing reveals PNN and KCNQ1OT1 as predictive biomarkers of clinical outcome in stage III colorectal cancer patients treated with adjuvant chemotherapy. Int. J. Cancer 2019, 145, 2580–2593. [Google Scholar] [CrossRef] [PubMed]
- Lapucci, A.; Perrone, G.; Di Paolo, A.; Napoli, C.; Landini, I.; Roviello, G.; Calosi, L.; Naccarato, A.G.; Falcone, A.; Bani, D. PNN and KCNQ1OT1 can predict the efficacy of adjuvant fluoropyrimidine-based chemotherapy in colorectal cancer patients. Oncol. Res. 2021, 28, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Soifer, H.S.; Rossi, J.J.; Sætrom, P. MicroRNAs in disease and potential therapeutic applications. Mol. Ther. 2007, 15, 2070–2079. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Ren, L.-L.; Yan, T.-T.; Shen, C.-Q.; Tang, J.-Y.; Kong, X.; Wang, Y.-C.; Chen, J.; Liu, Q.; He, J.; Zhong, M.; et al. The distinct role of strand-specific miR-514b-3p and miR-514b-5p in colorectal cancer metastasis. Cell Death Dis. 2018, 9, 687. [Google Scholar] [CrossRef]
- Li, A.; Yang, P.-M. Overexpression of miR-21-5p in colorectal cancer cells promotes self-assembly of E-cadherin-dependent multicellular tumor spheroids. Tissue Cell 2020, 65, 101365. [Google Scholar] [CrossRef] [PubMed]
- Dino, P.; D’Anna, C.; Sangiorgi, C.; Di Sano, C.; Di Vincenzo, S.; Ferraro, M.; Pace, E. Cigarette smoke extract modulates E-Cadherin, Claudin-1 and miR-21 and promotes cancer invasiveness in human colorectal adenocarcinoma cells. Toxicol. Lett. 2019, 317, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wu, L.; Liu, Z.; Zhang, X.; Han, S.; Zhao, N.; Bao, H.; Yuan, W.; Chen, J.; Ji, J.; et al. MicroRNA-214-3p targets the PLAGL2-MYH9 axis to suppress tumor proliferation and metastasis in human colorectal cancer. Aging 2020, 12, 9633–9657. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Quillet, A.; Saad, C.; Ferry, G.; Anouar, Y.; Vergne, N.; Lecroq, T.; Dubessy, C. Improving bioinformatics prediction of microRNA targets by ranks aggregation. Front. Genet. 2020, 10, 1330. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Mitra, R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics 2009, 25, 2625–2631. [Google Scholar] [CrossRef]
- Cho, S.; Jang, I.; Jun, Y.; Yoon, S.; Ko, M.; Kwon, Y.; Choi, I.; Chang, H.; Ryu, D.; Lee, B.; et al. MiRGator v3. 0: A microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2012, 41, D252–D257. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piccinno, E.; Scalavino, V.; Labarile, N.; De Marinis, L.; Armentano, R.; Giannelli, G.; Serino, G. Identification of a Novel miR-195-5p/PNN Axis in Colorectal Cancer. Int. J. Mol. Sci. 2024, 25, 5980. https://doi.org/10.3390/ijms25115980
Piccinno E, Scalavino V, Labarile N, De Marinis L, Armentano R, Giannelli G, Serino G. Identification of a Novel miR-195-5p/PNN Axis in Colorectal Cancer. International Journal of Molecular Sciences. 2024; 25(11):5980. https://doi.org/10.3390/ijms25115980
Chicago/Turabian StylePiccinno, Emanuele, Viviana Scalavino, Nicoletta Labarile, Lucia De Marinis, Raffaele Armentano, Gianluigi Giannelli, and Grazia Serino. 2024. "Identification of a Novel miR-195-5p/PNN Axis in Colorectal Cancer" International Journal of Molecular Sciences 25, no. 11: 5980. https://doi.org/10.3390/ijms25115980
APA StylePiccinno, E., Scalavino, V., Labarile, N., De Marinis, L., Armentano, R., Giannelli, G., & Serino, G. (2024). Identification of a Novel miR-195-5p/PNN Axis in Colorectal Cancer. International Journal of Molecular Sciences, 25(11), 5980. https://doi.org/10.3390/ijms25115980







