Analysis of Oral and Gut Microbiome Composition and Its Impact in Patients with Oral Squamous Cell Carcinoma
Abstract
1. Introduction
2. Results
2.1. Baseline Characteristics
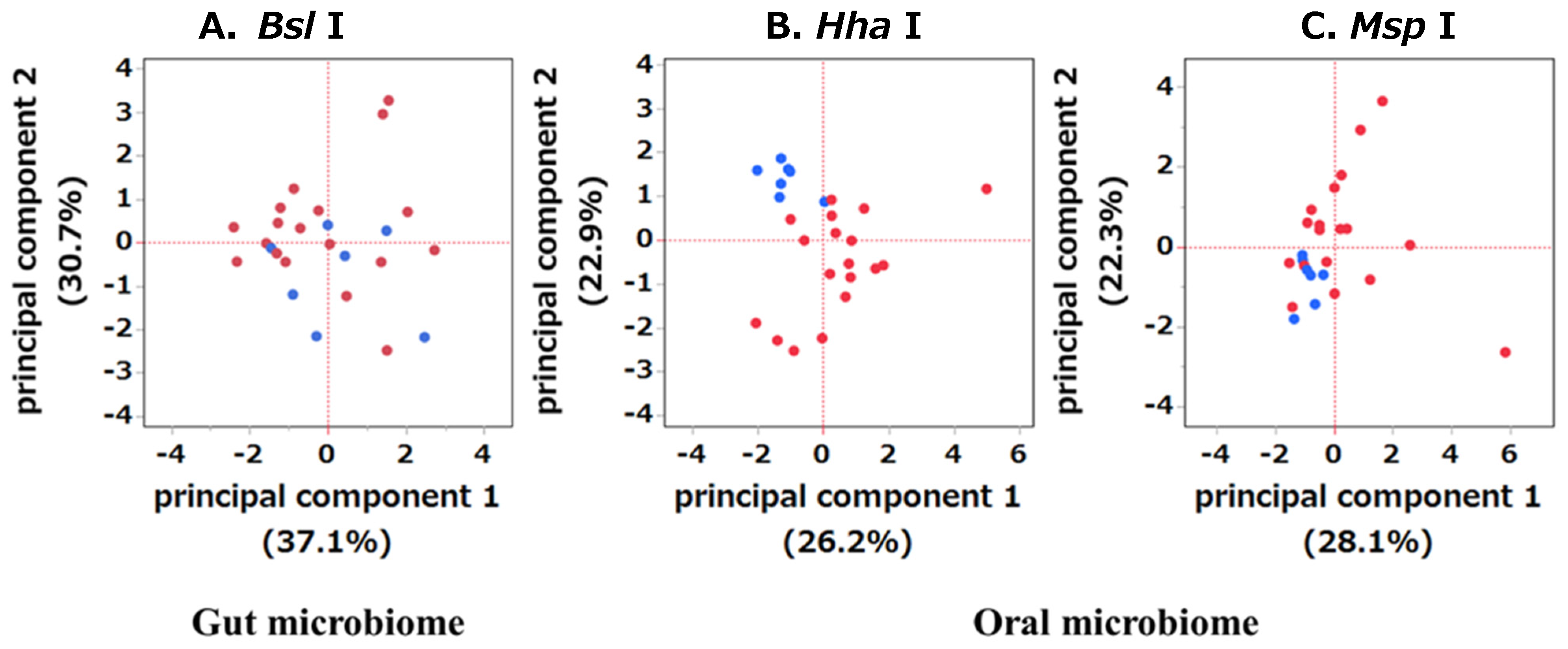
2.2. Comparison of Diversity between the OSCC and HC Groups in Gut and Oral Microbiomes
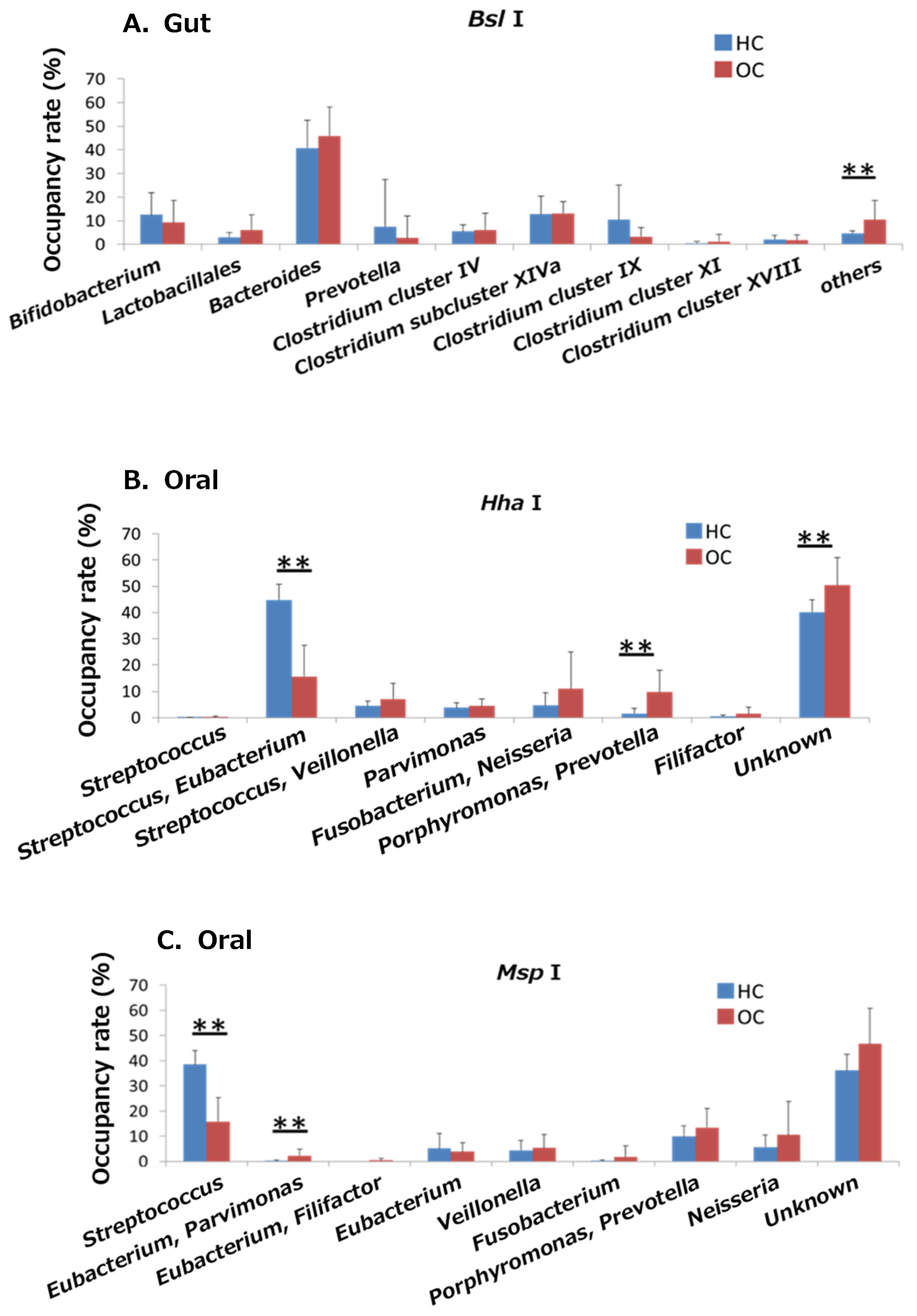
2.3. Comparison of Bacterial Abundances between the Gut and Oral Microbiomes of the OSCC and HC Groups
2.4. Comparison of Clinicopathological Factors between the PD-L1-Positive and -Negative Groups and the PD-1-Positive and -Negative Groups
2.5. Comparison of the Prognosis between the PD-L1-Positive and -Negative Groups and the PD-1-Positive and -Negative Groups
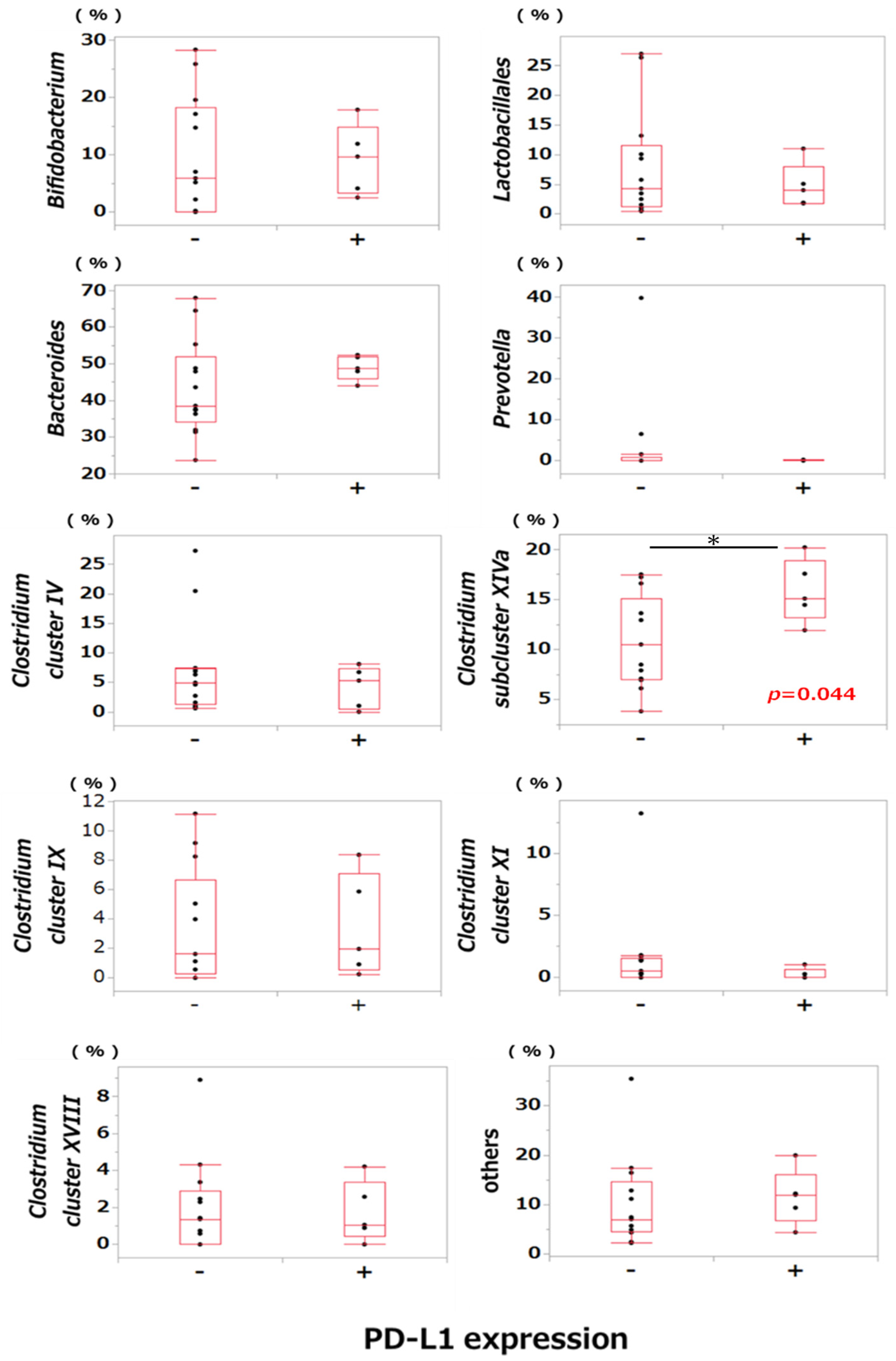
2.6. Comparison of Bacterial Occupancy between the PD-L1-Positive and PD-L1-Negative Groups
3. Discussion
4. Materials and Methods
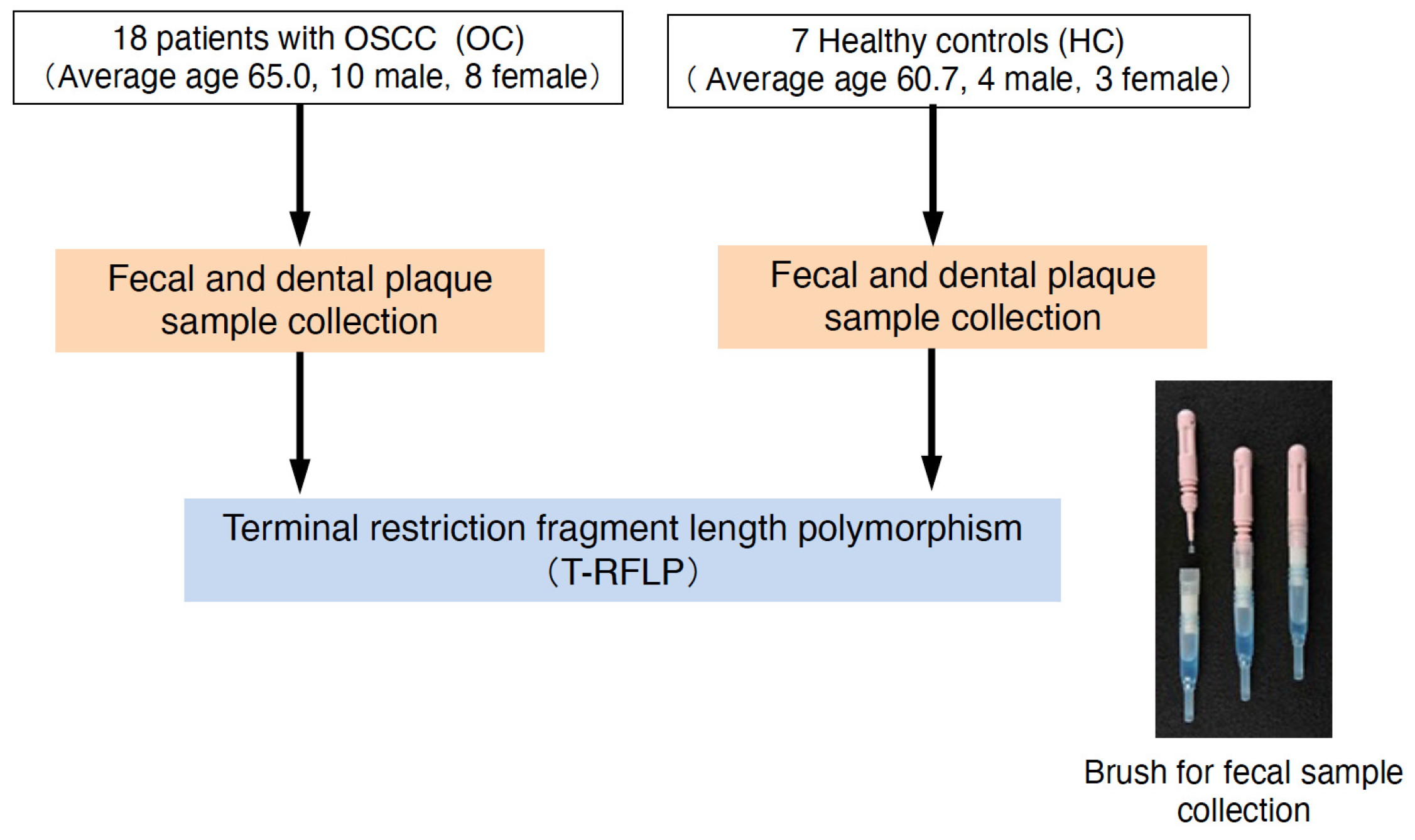
4.1. Participants
4.2. Collection of Fecal Matter and Dental Plaque
4.3. DNA Extraction
4.4. T-RFLP Analysis
4.5. Immunohistochemical Analysis
4.6. Statistical Analysis
4.7. Ethical Considerations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lederberg, J. Infectious history. Science 2000, 288, 287–293. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef]
- Noval Rivas, M.; Burton, O.T.; Wise, P.; Zhang, Y.Q.; Hobson, S.A.; Garcia Lloret, M.; Chehoud, C.; Kuczynski, J.; DeSantis, T.; Warrington, J.; et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol. 2013, 131, 201–212. [Google Scholar] [CrossRef]
- Shanahan, F.; Quigley, E.M. Manipulation of the microbiota for treatment of IBS and IBD-challenges and controversies. Gastroenterology 2014, 146, 1554–1563. [Google Scholar] [CrossRef] [PubMed]
- Miyake, S.; Kim, S.; Suda, W.; Oshima, K.; Nakamura, M.; Matsuoka, T.; Chihara, N.; Tomita, A.; Sato, W.; Kim, S.W.; et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef]
- Ding, T.; Schloss, P.D. Dynamics and associations of microbial community types across the human body. Nature 2014, 509, 357–360. [Google Scholar] [CrossRef]
- Lauridsen, M.M.; Grønkjær, L.L.; Khraibut, S.; Patel, N.; Deeb, J.G.; Bajaj, J.S. The Multi-dimensional Challenge of Poor Oral Health in Cirrhosis-Disparities and Solutions. Gastroenterology 2024, 166, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819867354. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.A.; Rhoads, T.W.; Merrill, A.E.; Ye, Z.; Westphall, M.S.; Acharya, A.; Shukla, S.K.; Coon, J.J. Proteomics, Lipidomics, Metabolomics, and 16S DNA Sequencing of Dental Plaque From Patients With Diabetes and Periodontal Disease. Mol. Cell. Proteom. 2021, 20, 100126. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef]
- Santacroce, L.; Passarelli, P.C.; Azzolino, D.; Bottalico, L.; Charitos, I.A.; Cazzolla, A.P.; Colella, M.; Topi, S.; Godoy, F.G.; D’Addona, A. Oral microbiota in human health and disease: A perspective. Exp. Biol. Med. 2023, 248, 1288–1301. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef]
- Okazaki, T.; Honjo, T. PD-1 and PD-1 ligands: From discovery to clinical application. Int. Immunol. 2007, 19, 813–824. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Tani, R.; Tokumaru, K. Hepatic Dysfunction Prophylaxis. Japan Patent JP 5823695, 16 October 2015. [Google Scholar]
- Okamoto, T.; Tani, R.; Tokumaru, K. Cancer Treatment Survival Enhancer. Japan Patent JP 2017038961, 27 October 2017. [Google Scholar]
- Inoue, D.; Kimura, I.; Wakabayashi, M.; Tsumoto, H.; Ozawa, K.; Hara, T.; Takei, Y.; Hirasawa, A.; Ishihama, Y.; Tsujimoto, G. Short-chain fatty acid receptor GPR41-mediated activation of sympathetic neurons involves synapsin 2b phosphorylation. FEBS Lett. 2012, 586, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T.; et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef] [PubMed]
- Higashimura, Y.; Naito, Y.; Takagi, T.; Uchiyama, K.; Mizushima, K.; Yoshikawa, T. Propionate Promotes Fatty Acid Oxidation through the Up-Regulation of Peroxisome Proliferator-Activated Receptor α in Intestinal Epithelial Cells. J. Nutr. Sci. Vitaminol. 2015, 61, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.R.M.; Gattuso, G.; Pedullà, E.; Rapisarda, E.; Nicolosi, D.; Salmeri, M. Association of oral dysbiosis with oral cancer development. Oncol. Lett. 2020, 19, 3045–3058. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.F.; Huang, H.D.; Fan, W.L.; Jong, Y.J.; Chen, M.K.; Huang, C.N.; Chuang, C.Y.; Kuo, Y.L.; Chung, W.H.; Su, S.C. Compositional and functional variations of oral microbiota associated with the mutational changes in oral cancer. Oral Oncol. 2018, 77, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef]
- Li, R.; Xiao, L.; Gong, T.; Liu, J.; Li, Y.; Zhou, X.; Li, Y.; Zheng, X. Role of oral microbiome in oral oncogenesis, tumor progression, and metastasis. Mol. Oral Microbiol. 2023, 38, 9–22. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, H.; Tian, Y.; Li, J.; Xin, Y.; Jiang, X. Mendelian randomization analysis to investigate the gut microbiome in oral and oropharyngeal cancer. Front. Cell. Infect. Microbiol. 2023, 13, 1210807. [Google Scholar] [CrossRef] [PubMed]
- Baunwall, S.M.D.; Andreasen, S.E.; Hansen, M.M.; Kelsen, J.; Høyer, K.L.; Rågård, N.; Eriksen, L.L.; Støy, S.; Rubak, T.; Damsgaard, E.M.S.; et al. Faecal microbiota transplantation for first or second Clostridioides difficile infection (Early FMT): A randomised, double-blind, placebo-controlled trial. Lancet Gastroenterol. Hepatol. 2022, 7, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Rancich, M.; Roman, C. Updated guidelines for diagnosing and managing Clostridium difficile. J. Am. Acad. Physician Assist. 2019, 32, 48–50. [Google Scholar] [CrossRef]
- Campbell, B.R.; Sanders, C.B.; Netterville, J.L.; Sinard, R.J.; Rohde, S.L.; Langerman, A.; Mannion, K.; Kim, Y.J.; Murphy, B.A.; Lewis, J.S., Jr.; et al. Early onset oral tongue squamous cell carcinoma: Associated factors and patient outcomes. Head Neck 2019, 41, 1952–1960. [Google Scholar] [CrossRef]
- Ju, X.; Mejia, G.; Chrisopoulos, S.; Luzzi, L.; Jamieson, L.M. A longitudinal assessment of chronic periodontitis in Australian adults. J. Clin. Periodontol. 2023, 50, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Printz, C. Study adds evidence to link between gum disease and cancer risk: Researchers observe connection with gastric, esophageal cancer. Cancer 2021, 127, 495–496. [Google Scholar] [CrossRef] [PubMed]
- John, A.A.; Naresh, K.C.; Ranganath, V.; Subramaniam, M.R.; Patil, A.S.; Jumani, P.N. Relationship between the nutritional status and antimicrobial protein levels with the periodontal condition in untreated head and neck cancer patients. J. Fam. Med. Prim. Care 2019, 8, 3325–3333. [Google Scholar] [CrossRef]
- Olsen, I.; Yilmaz, Ö. Possible role of Porphyromonas gingivalis in orodigestive cancers. J. Oral Microbiol. 2019, 11, 1563410. [Google Scholar] [CrossRef]
- Yilmaz, O.; Yao, L.; Maeda, K.; Rose, T.M.; Lewis, E.L.; Duman, M.; Lamont, R.J.; Ojcius, D.M. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cell. Microbiol. 2008, 10, 863–875. [Google Scholar] [CrossRef]
- Inaba, H.; Sugita, H.; Kuboniwa, M.; Iwai, S.; Hamada, M.; Noda, T.; Morisaki, I.; Lamont, R.J.; Amano, A. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell. Microbiol. 2014, 16, 131–145. [Google Scholar] [CrossRef]
- Baba, Y.; Hara, Y.; Toihata, T.; Kosumi, K.; Iwatsuki, M.; Iwagami, S.; Miyamoto, Y.; Yoshida, N.; Komohara, Y.; Baba, H. Relationship between gut microbiome Fusobacterium nucleatum and LINE-1 methylation level in esophageal cancer. Esophagus 2023, 20, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Hora, S.S.; Patil, S.K. Oral Microflora in the Background of Oral Cancer: A Review. Cureus 2022, 14, e33129. [Google Scholar] [CrossRef] [PubMed]
- Mattox, A.K.; Lee, J.; Westra, W.H.; Pierce, R.H.; Ghossein, R.; Faquin, W.C.; Diefenbach, T.J.; Morris, L.G.; Lin, D.T.; Wirth, L.J.; et al. PD-1 Expression in Head and Neck Squamous Cell Carcinomas Derives Primarily from Functionally Anergic CD4(+) TILs in the Presence of PD-L1(+) TAMs. Cancer Res. 2017, 77, 6365–6374. [Google Scholar] [CrossRef] [PubMed]
- de Vicente, J.C.; Rodríguez-Santamarta, T.; Rodrigo, J.P.; Blanco-Lorenzo, V.; Allonca, E.; García-Pedrero, J.M. PD-L1 Expression in Tumor Cells Is an Independent Unfavorable Prognostic Factor in Oral Squamous Cell Carcinoma. Cancer Epidemiol. Biomark. Prev. 2019, 28, 546–554. [Google Scholar] [CrossRef]
- Blachier, F.; Andriamihaja, M.; Larraufie, P.; Ahn, E.; Lan, A.; Kim, E. Production of hydrogen sulfide by the intestinal microbiota and epithelial cells and consequences for the colonic and rectal mucosa. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G125–G135. [Google Scholar] [CrossRef] [PubMed]
- Kamakura, R.; Raza, G.S.; Sodum, N.; Lehto, V.P.; Kovalainen, M.; Herzig, K.H. Colonic Delivery of Nutrients for Sustained and Prolonged Release of Gut Peptides: A Novel Strategy for Appetite Management. Mol. Nutr. Food Res. 2022, 66, e2200192. [Google Scholar] [CrossRef] [PubMed]
- Rysz, J.; Franczyk, B.; Ławiński, J.; Olszewski, R.; Ciałkowska-Rysz, A.; Gluba-Brzózka, A. The Impact of CKD on Uremic Toxins and Gut Microbiota. Toxins 2021, 13, 252. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- Nagashima, K.; Hisada, T.; Sato, M.; Mochizuki, J. Application of new primer-enzyme combinations to terminal restriction fragment length polymorphism profiling of bacterial populations in human feces. Appl. Environ. Microbiol. 2003, 69, 1251–1262. [Google Scholar] [CrossRef]
- Nagashima, K.; Mochizuki, J.; Hisada, T.; Suzuki, S.; Shimomura, K. Phylogenetic analysis of 16S ribosomal RNA gene sequences from human fecal microbiota and improved utility of terminal restriction fragment length polymorphism profiling. Biosci. Microflora 2006, 25, 99–107. [Google Scholar] [CrossRef]
- Sakamoto, M.; Huang, Y.; Ohnishi, M.; Umeda, M.; Ishikawa, I.; Benno, Y. Changes in oral microbial profiles after periodontal treatment as determined by molecular analysis of 16S rRNA genes. J. Med. Microbiol. 2004, 53, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Hiraishi, A.; Kamagata, Y.; Nakamura, K. Polymerase chain reaction amplification and restriction fragment length polymorphism analysis of 16S rRNA genes from methanogens. J. Ferment. Bioeng. 1995, 79, 523–529. [Google Scholar] [CrossRef]
- Begum, S.; Zhang, Y.; Shintani, T.; Toratani, S.; Sato, J.D.; Okamoto, T. Immunohistochemical expression of heparin-binding protein 17/fibroblast growth factor-binding protein-1 (HBp17/FGFBP-1) as an angiogenic factor in head and neck tumorigenesis. Oncol. Rep. 2007, 17, 591–596. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ito, N.; Yamasaki, S.; Shintani, T.; Matsui, K.; Obayashi, F.; Koizumi, K.; Tani, R.; Yanamoto, S.; Okamoto, T. Tumor-Infiltrating CD45RO+ Memory Cells Are Associated with Favorable Prognosis in Oral Squamous Cell Carcinoma Patients. Cancers 2023, 15, 2221. [Google Scholar] [CrossRef] [PubMed]

| Category | OC (n = 18) | HC (n = 7) | |
|---|---|---|---|
| Sex | (male/female) | 10/8 | 4/3 |
| Age | average (range) | 65.0 (32–84) | 60.7 (32–94) |
| T | 1 | 1 | |
| 2 | 9 | ||
| 3 | 3 | ||
| 4 | 5 | ||
| N | negative | 14 | |
| positive | 4 | ||
| Stage | Ⅰ | 1 | |
| Ⅱ | 8 | ||
| Ⅲ | 4 | ||
| Ⅳ | 5 | ||
| Gut Microbiome | Oral Microbiome | |
|---|---|---|
| <restriction enzyme: Bsl I> | <restriction enzyme: Hha I> | <restriction enzyme: Msp I> |
| Bifidobacterium | Streptococcus | Streptococcus |
| Lactobacillales | Streptococcus, Eubacterium | Eubacterium, Parvimonas |
| Bacteroides | Streptococcus, Veillonella | Eubacterium, Filifactor |
| Prevotella | Parvimonas | Eubacterium |
| Clostridium cluster IV | Fusobacterium, Neisseria | Veillonella |
| Clostridium cluster IX | Porphyromonas, Prevotella | Fusobacterium |
| Clostridium cluster XI | Filifactor | Porphyromonas, Prevotella |
| Clostridium cluster XIVa | others | Neisseria |
| Clostridium cluster XVIII | others | |
| others | ||
| Category | PD-L1 | p-Value | PD-1 | p-Value | |||
|---|---|---|---|---|---|---|---|
| Positive (n = 5) | Negative (n = 13) | Positive (n = 6) | Negative (n = 12) | ||||
| Sex | (male/female) | 4/1 | 5/8 | 0.291 1 | 2/4 | 7/5 | 0.621 1 |
| Age | median (IQR) | 62.4 (40–68) | 69.1 (40–84) | 0.372 2 | 71.8 (60–82) | 64.9 (40–84) | 0.332 2 |
| T | 1 | 0 | 1 | 0.411 1 | 1 | 0 | 0.451 1 |
| 2 | 2 | 7 | 2 | 7 | |||
| 3 | 2 | 1 | 1 | 2 | |||
| 4 | 1 | 4 | 2 | 3 | |||
| N | negative | 4 | 10 | 0.671 1 | 6 | 8 | 0.111 1 |
| positive | 1 | 3 | 0 | 4 | |||
| Stage | Ⅰ | 0 | 1 | 0.61 1 | 1 | 0 | 0.481 1 |
| Ⅱ | 2 | 6 | 2 | 6 | |||
| Ⅲ | 2 | 2 | 1 | 6 | |||
| Ⅳ | 1 | 4 | 2 | 3 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsui, K.; Tani, R.; Yamasaki, S.; Ito, N.; Hamada, A.; Shintani, T.; Otomo, T.; Tokumaru, K.; Yanamoto, S.; Okamoto, T. Analysis of Oral and Gut Microbiome Composition and Its Impact in Patients with Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2024, 25, 6077. https://doi.org/10.3390/ijms25116077
Matsui K, Tani R, Yamasaki S, Ito N, Hamada A, Shintani T, Otomo T, Tokumaru K, Yanamoto S, Okamoto T. Analysis of Oral and Gut Microbiome Composition and Its Impact in Patients with Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2024; 25(11):6077. https://doi.org/10.3390/ijms25116077
Chicago/Turabian StyleMatsui, Kensaku, Ryouji Tani, Sachiko Yamasaki, Nanako Ito, Atsuko Hamada, Tomoaki Shintani, Takeshi Otomo, Koichiro Tokumaru, Souichi Yanamoto, and Tetsuji Okamoto. 2024. "Analysis of Oral and Gut Microbiome Composition and Its Impact in Patients with Oral Squamous Cell Carcinoma" International Journal of Molecular Sciences 25, no. 11: 6077. https://doi.org/10.3390/ijms25116077
APA StyleMatsui, K., Tani, R., Yamasaki, S., Ito, N., Hamada, A., Shintani, T., Otomo, T., Tokumaru, K., Yanamoto, S., & Okamoto, T. (2024). Analysis of Oral and Gut Microbiome Composition and Its Impact in Patients with Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences, 25(11), 6077. https://doi.org/10.3390/ijms25116077






