Role of iRhom2 in Olfaction: Implications for Odorant Receptor Regulation and Activity-Dependent Adaptation
Abstract
1. Introduction
2. Results
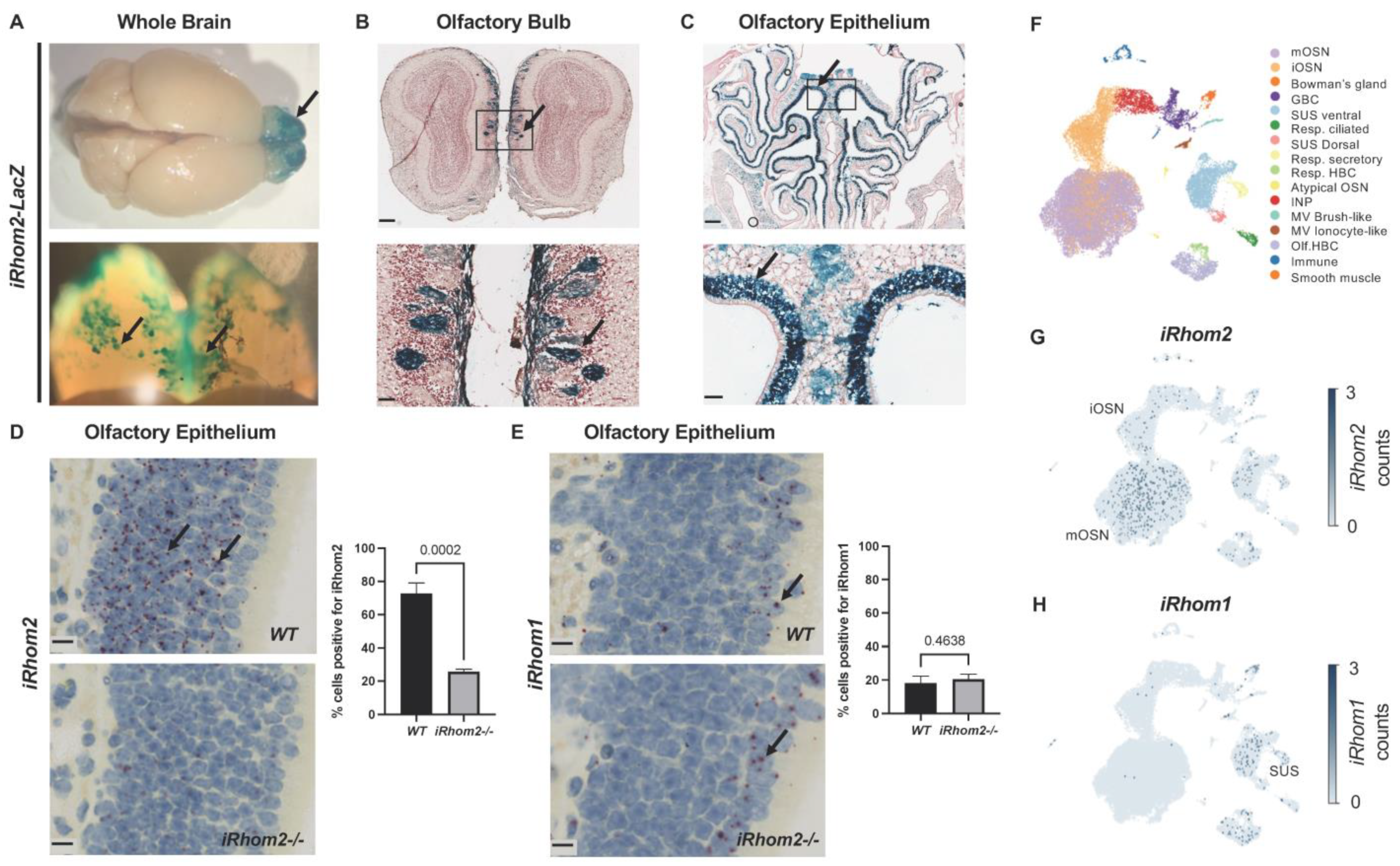
2.1. iRhom2 Is Prominently Expressed in the Olfactory Sensory Neurons
2.2. iRhom2-/- Olfactory Epithelium Shows No Histopathologic Defects
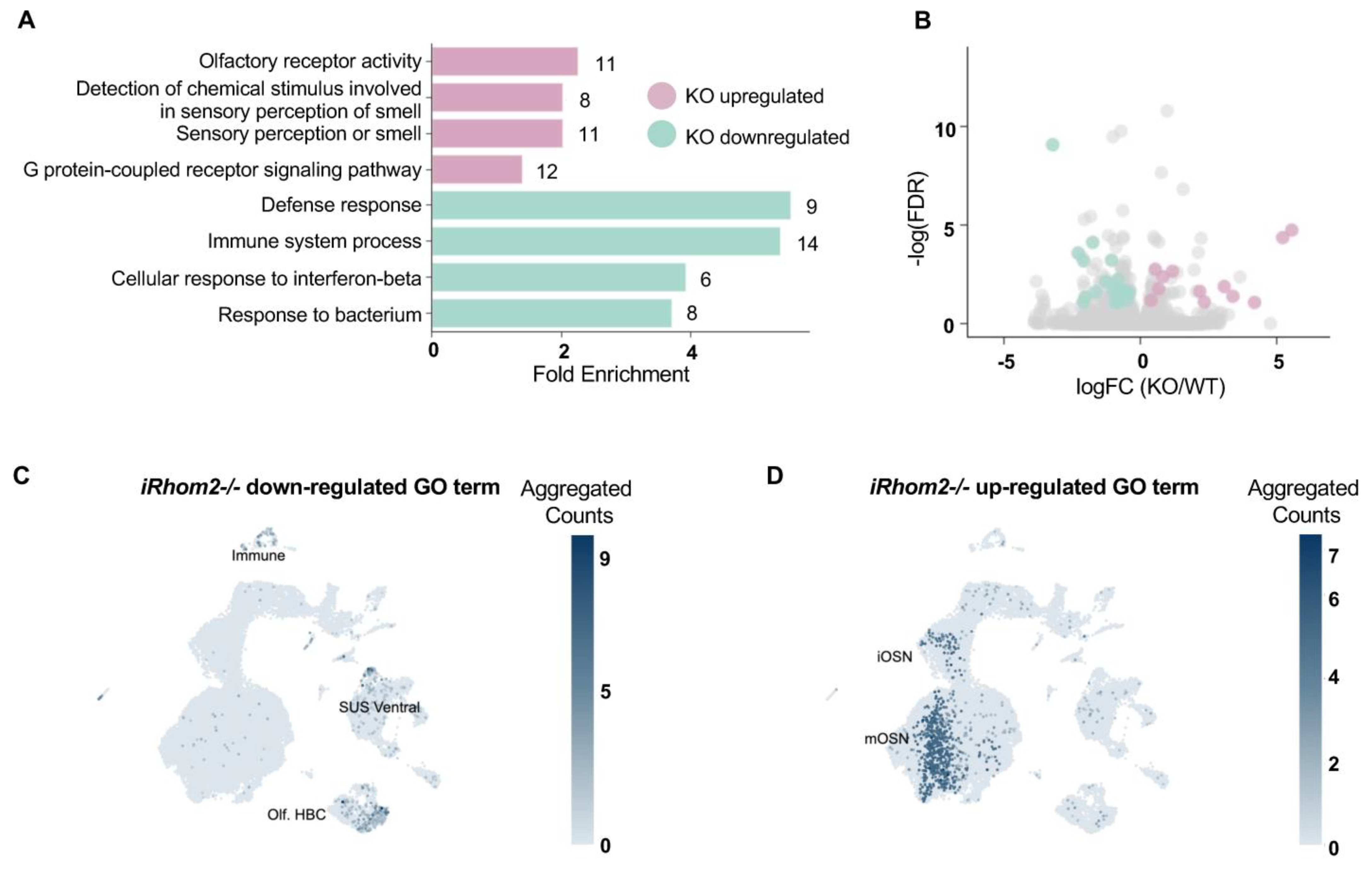
2.3. RNAseq GO Analysis
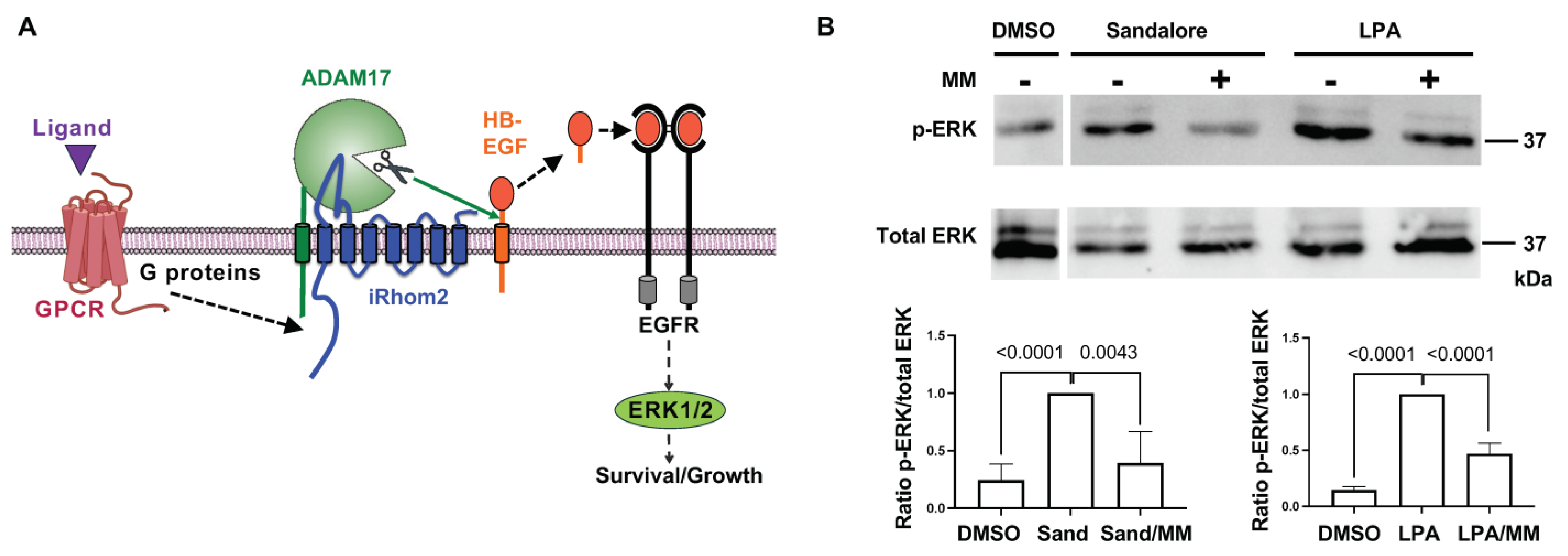
2.4. Activation of iRhom2/ADAM17 by OR Signaling in a Non-OSN Cell Type
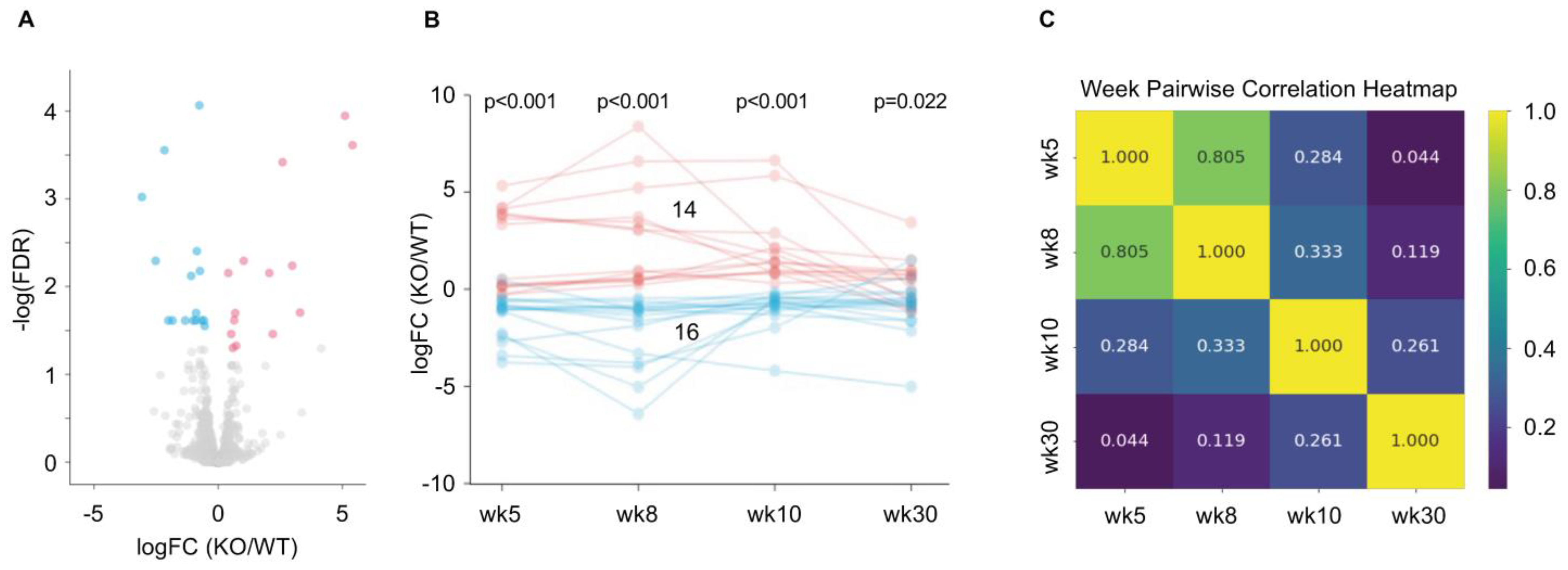
2.5. Change in Differential Expression of ORs in iRhom2-/- over Time
2.6. iRhom2 Expression Negatively Correlates with Neuronal Activity
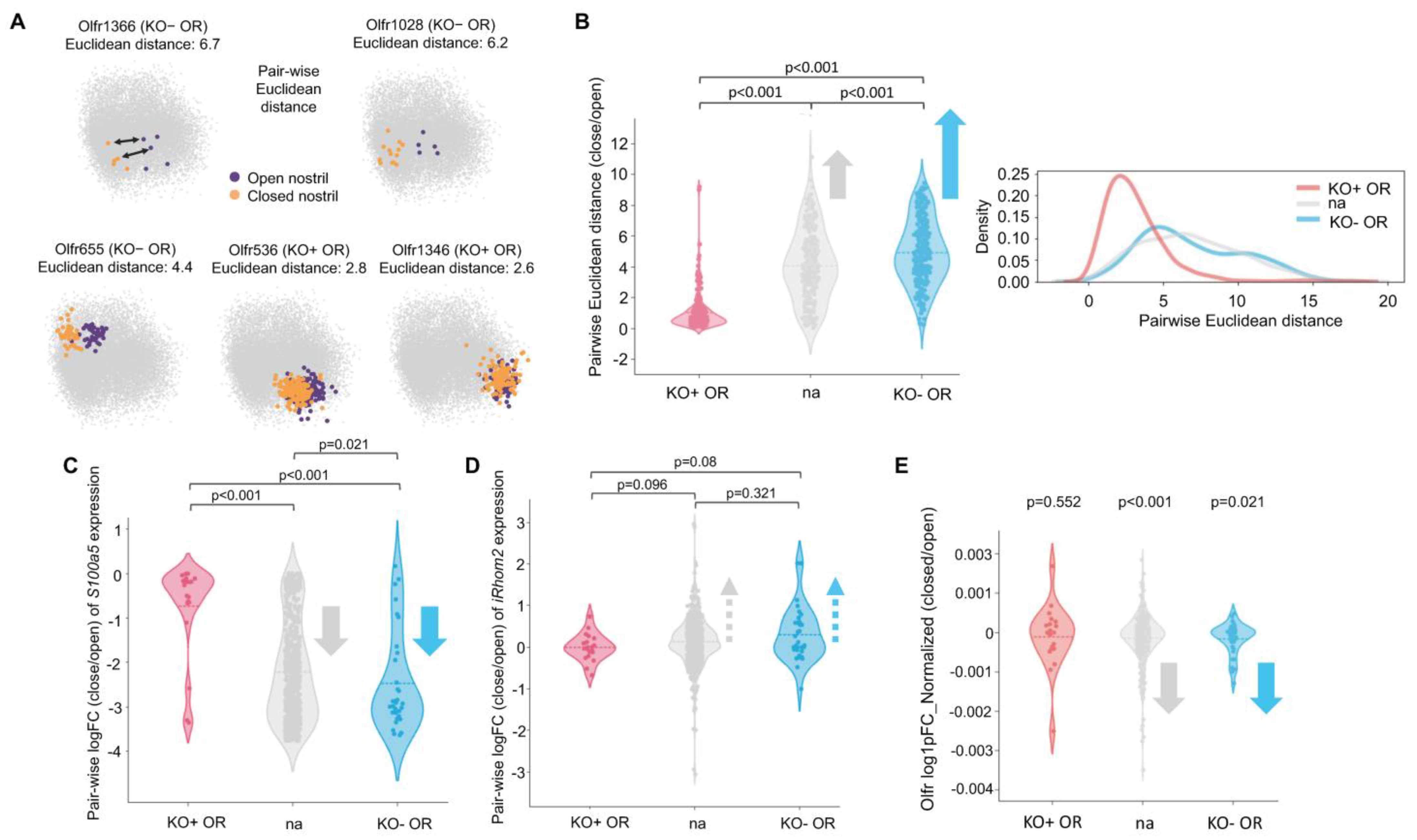
2.7. OSNs Expressing ORs Upregulated in iRhom2-/- Maintain Unusually Stable Gene Expression Amid Environmental Odor Changes
2.8. Gene Expression Changes in iRhom2-/- OE Mimic Changes Occuring in Naris Occlusion
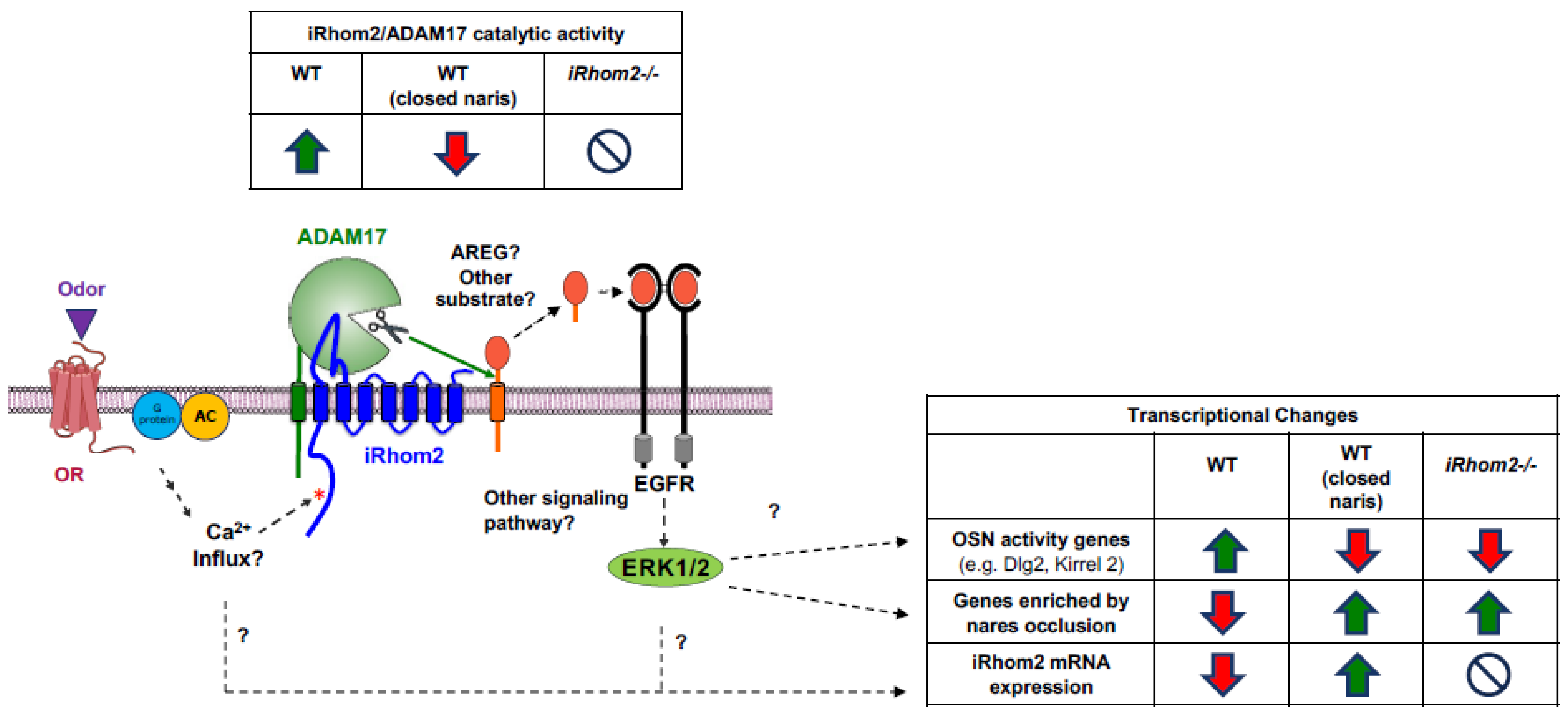
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. X-Gal Staining
4.3. RNA In Situ Hybridization
4.4. Treatment of Keratinocytes with Sandalore and Phospho-ERK Western Blot
4.5. Immunohistochemistry
4.6. Slide Scanning and Cellular/Subcellular Quantification
4.7. BrdU Injection, Tissue Harvest and Staining
4.8. Western Blot Densitometry
4.9. WOE Dissection, RNA Extraction and Library Prep
4.10. RNAseq Alignment, Quantification and Differential Expression Analysis
4.11. Principal Component Analysis
4.12. Visualization via UMAP
4.13. Identification of the OR Expressed in Each OSN
4.14. OR Pair-Wise Comparison
4.15. OR Population Comparison
4.16. RNAseq Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McIlwain, D.R.; Lang, P.A.; Maretzky, T.; Hamada, K.; Ohishi, K.; Maney, S.K.; Berger, T.; Murthy, A.; Duncan, G.; Xu, H.C.; et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science 2012, 335, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Adrain, C.; Zettl, M.; Christova, Y.; Taylor, N.; Freeman, M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science 2012, 335, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Siggs, O.M.; Xiao, N.; Wang, Y.; Shi, H.; Tomisato, W.; Li, X.; Xia, Y.; Beutler, B. iRhom2 is required for the secretion of mouse TNFalpha. Blood 2012, 119, 5769–5771. [Google Scholar] [CrossRef] [PubMed]
- Christova, Y.; Adrain, C.; Bambrough, P.; Ibrahim, A.; Freeman, M. Mammalian iRhoms have distinct physiological functions including an essential role in TACE regulation. EMBO Rep. 2013, 14, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Maretzky, T.; Weskamp, G.; Monette, S.; Qing, X.; Issuree, P.D.; Crawford, H.C.; McIlwain, D.R.; Mak, T.W.; Salmon, J.E.; et al. iRhoms 1 and 2 are essential upstream regulators of ADAM17-dependent EGFR signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 6080–6085. [Google Scholar] [CrossRef]
- Grieve, A.G.; Xu, H.; Kunzel, U.; Bambrough, P.; Sieber, B.; Freeman, M. Phosphorylation of iRhom2 at the plasma membrane controls mammalian TACE-dependent inflammatory and growth factor signalling. eLife 2017, 6, e23968. [Google Scholar] [CrossRef] [PubMed]
- Badenes, M.; Burbridge, E.; Oikonomidi, I.; Amin, A.; de Carvalho, E.; Kosack, L.; Domingos, P.; Faisca, P.; Adrain, C. The ADAM17 sheddase complex regulator iTAP modulates inflammation, epithelial repair, and tumor growth. bioRxiv 2022. [Google Scholar] [CrossRef]
- Cavadas, M.; Oikonomidi, I.; Gaspar, C.J.; Burbridge, E.; Badenes, M.; Felix, I.; Bolado, A.; Hu, T.; Bileck, A.; Gerner, C.; et al. Phosphorylation of iRhom2 Controls Stimulated Proteolytic Shedding by the Metalloprotease ADAM17/TACE. Cell Rep. 2017, 21, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Oikonomidi, I.; Burbridge, E.; Cavadas, M.; Sullivan, G.; Collis, B.; Naegele, H.; Clancy, D.; Brezinova, J.; Hu, T.; Bileck, A.; et al. iTAP, a novel iRhom interactor, controls TNF secretion by policing the stability of iRhom/TACE. eLife 2018, 7, e35032. [Google Scholar] [CrossRef]
- Kunzel, U.; Grieve, A.G.; Meng, Y.; Sieber, B.; Cowley, S.A.; Freeman, M. FRMD8 promotes inflammatory and growth factor signalling by stabilising the iRhom/ADAM17 sheddase complex. eLife 2018, 7, e35012. [Google Scholar] [CrossRef]
- Geesala, R.; Issuree, P.D.; Maretzky, T. Novel functions of inactive rhomboid proteins in immunity and disease. J. Leukoc. Biol. 2019, 106, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Issuree, P.D.; Maretzky, T.; McIlwain, D.R.; Monette, S.; Qing, X.; Lang, P.A.; Swendeman, S.L.; Park-Min, K.H.; Binder, N.; Kalliolias, G.D.; et al. iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. J. Clin. Investig. 2013, 123, 928–932. [Google Scholar] [CrossRef] [PubMed]
- Black, R.; Rauch, C.T.; Kozlosky, C.J.; Peschon, J.J.; Slack, J.L.; Wolfson, M.F.; Castner, B.J.; Stocking, K.L.; Reddy, P.; Srinivasan, S.; et al. A metalloprotease disintegrin that releases tumour-necrosis factor-a from cells. Nature 1997, 385, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Moss, M.L.; Jin, S.-L.C.; Milla, M.E.; Burkhart, W.; Cartner, H.L.; Chen, W.-J.; Clay, W.C.; Didsbury, J.R.; Hassler, D.; Hoffman, C.R.; et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-a. Nature 1997, 385, 733–736. [Google Scholar] [CrossRef]
- Peschon, J.J.; Slack, J.L.; Reddy, P.; Stocking, K.L.; Sunnarborg, S.W.; Lee, D.C.; Russel, W.E.; Castner, B.J.; Johnson, R.S.; Fitzner, J.N.; et al. An essential role for ectodomain shedding in mammalian development. Science 1998, 282, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Blobel, C.P. ADAMs: Key players in EGFR-signaling, development and disease. Nat. Rev. Mol. Cell Biol. 2005, 6, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.I.; Jenkins, B.J. The protease ADAM17 at the crossroads of disease: Revisiting its significance in inflammation, cancer, and beyond. FEBS J. 2024, 291, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Tushaus, J.; Muller, S.A.; Shrouder, J.; Arends, M.; Simons, M.; Plesnila, N.; Blobel, C.P.; Lichtenthaler, S.F. The pseudoprotease iRhom1 controls ectodomain shedding of membrane proteins in the nervous system. FASEB J. 2021, 35, e21962. [Google Scholar] [CrossRef] [PubMed]
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef]
- Zhang, X.; Firestein, S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002, 5, 124–133. [Google Scholar] [CrossRef]
- Shykind, B.M.; Rohani, S.C.; O’Donnell, S.; Nemes, A.; Mendelsohn, M.; Sun, Y.; Axel, R.; Barnea, G. Gene switching and the stability of odorant receptor gene choice. Cell 2004, 117, 801–815. [Google Scholar] [CrossRef]
- Mombaerts, P. Odorant receptor gene choice in olfactory sensory neurons: The one receptor-one neuron hypothesis revisited. Curr. Opin. Neurobiol. 2004, 14, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Van der Linden, C.J.; Gupta, P.; Bhuiya, A.I.; Riddick, K.R.; Hossain, K.; Santoro, S.W. Olfactory Stimulation Regulates the Birth of Neurons That Express Specific Odorant Receptors. Cell Rep. 2020, 33, 108210. [Google Scholar] [CrossRef]
- Santoro, S.W.; Dulac, C. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. eLife 2012, 1, e00070. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, T.A.; Cleland, T.A. The anatomical logic of smell. Trends Neurosci. 2005, 28, 620–627. [Google Scholar] [CrossRef]
- Brann, D.H.; Tsukahara, T.; Weinreb, C.; Lipovsek, M.; Van den Berge, K.; Gong, B.; Chance, R.; Macaulay, I.C.; Chou, H.J.; Fletcher, R.B.; et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020, 6, eabc5801. [Google Scholar] [CrossRef]
- Maretzky, T.; McIlwain, D.R.; Issuree, P.D.; Li, X.; Malapeira, J.; Amin, S.; Lang, P.A.; Mak, T.W.; Blobel, C.P. iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. Proc. Natl. Acad. Sci. USA 2013, 110, 11433–11438. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Ishiguro, J.; Kitamura, H.; Arima, N.; Okutani, M.; Shuto, A.; Higashiyama, S.; Ohwada, T.; Arai, H.; Makide, K.; et al. TGFalpha shedding assay: An accurate and versatile method for detecting GPCR activation. Nat. Methods 2012, 9, 1021–1029. [Google Scholar] [CrossRef]
- Maretzky, T.; Evers, A.; Zhou, W.; Swendeman, S.L.; Wong, P.M.; Rafii, S.; Reiss, K.; Blobel, C.P. Migration of growth factor-stimulated epithelial and endothelial cells depends on EGFR transactivation by ADAM17. Nat. Commun. 2011, 2, 229. [Google Scholar] [CrossRef]
- Ohtsu, H.; Dempsey, P.J.; Frank, G.D.; Brailoiu, E.; Higuchi, S.; Suzuki, H.; Nakashima, H.; Eguchi, K.; Eguchi, S. ADAM17 mediates epidermal growth factor receptor transactivation and vascular smooth muscle cell hypertrophy induced by angiotensin II. Arterioscler. Thromb. Vasc. Biol. 2006, 26, e133–e137. [Google Scholar] [CrossRef]
- Forrester, S.J.; Kawai, T.; O’Brien, S.; Thomas, W.; Harris, R.C.; Eguchi, S. Epidermal Growth Factor Receptor Transactivation: Mechanisms, Pathophysiology, and Potential Therapies in the Cardiovascular System. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 627–653. [Google Scholar] [CrossRef] [PubMed]
- Gschwind, A.; Hart, S.; Fischer, O.M.; Ullrich, A. TACE cleavage of proamphiregulin regulates GPCR-induced proliferation and motility of cancer cells. EMBO J. 2003, 22, 2411–2421. [Google Scholar] [CrossRef] [PubMed]
- Mifune, M.; Ohtsu, H.; Suzuki, H.; Nakashima, H.; Brailoiu, E.; Dun, N.J.; Frank, G.D.; Inagami, T.; Higashiyama, S.; Thomas, W.G.; et al. G protein coupling and second messenger generation are indispensable for metalloprotease-dependent, heparin-binding epidermal growth factor shedding through angiotensin II type-1 receptor. J. Biol. Chem. 2005, 280, 26592–26599. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Arima, N.; Ishiguro, J.; Prestwich, G.D.; Arai, H.; Aoki, J. LPA-producing enzyme PA-PLA(1)alpha regulates hair follicle development by modulating EGFR signalling. EMBO J. 2011, 30, 4248–4260. [Google Scholar] [CrossRef] [PubMed]
- Busse, D.; Kudella, P.; Gruning, N.M.; Gisselmann, G.; Stander, S.; Luger, T.; Jacobsen, F.; Steinstrasser, L.; Paus, R.; Gkogkolou, P.; et al. A synthetic sandalwood odorant induces wound-healing processes in human keratinocytes via the olfactory receptor OR2AT4. J. Investig. Dermatol. 2014, 134, 2823–2832. [Google Scholar] [CrossRef]
- Tsukahara, T.; Brann, D.H.; Pashkovski, S.L.; Guitchounts, G.; Bozza, T.; Datta, S.R. A transcriptional rheostat couples past activity to future sensory responses. Cell 2021, 184, 6326–6343.e32. [Google Scholar] [CrossRef] [PubMed]
- Fischl, A.M.; Heron, P.M.; Stromberg, A.J.; McClintock, T.S. Activity-dependent genes in mouse olfactory sensory neurons. Chem. Senses 2014, 39, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Oztokatli, H.; Hornberg, M.; Berghard, A.; Bohm, S. Retinoic acid receptor and CNGA2 channel signaling are part of a regulatory feedback loop controlling axonal convergence and survival of olfactory sensory neurons. FASEB J. 2012, 26, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.K.; Kulaga, H.M.; Reed, R.R. Odor-evoked gene regulation and visualization in olfactory receptor neurons. Mol. Cell. Neurosci. 2010, 43, 353–362. [Google Scholar] [CrossRef]
- Wang, Q.; Titlow, W.B.; McClintock, D.A.; Stromberg, A.J.; McClintock, T.S. Activity-Dependent Gene Expression in the Mammalian Olfactory Epithelium. Chem. Senses 2017, 42, 611–624. [Google Scholar] [CrossRef]
- Ibarra-Soria, X.; Nakahara, T.S.; Lilue, J.; Jiang, Y.; Trimmer, C.; Souza, M.A.; Netto, P.H.; Ikegami, K.; Murphy, N.R.; Kusma, M.; et al. Variation in olfactory neuron repertoires is genetically controlled and environmentally modulated. eLife 2017, 6, e21476. [Google Scholar] [CrossRef] [PubMed]
- D’Hulst, C.; Mina, R.B.; Gershon, Z.; Jamet, S.; Cerullo, A.; Tomoiaga, D.; Bai, L.; Belluscio, L.; Rogers, M.E.; Sirotin, Y.; et al. MouSensor: A Versatile Genetic Platform to Create Super Sniffer Mice for Studying Human Odor Coding. Cell Rep. 2016, 16, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- Livermore, A.; Hummel, T. The influence of training on chemosensory event-related potentials and interactions between the olfactory and trigeminal systems. Chem. Senses 2004, 29, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Hossain, K.; Smith, M.; Santoro, S.W. In mice, discrete odors can selectively promote the neurogenesis of sensory neuron subtypes that they stimulate. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Sarnat, H.B.; Yu, W. Maturation and Dysgenesis of the Human Olfactory Bulb. Brain Pathol. 2016, 26, 301–318. [Google Scholar] [CrossRef] [PubMed]
- Todrank, J.; Heth, G.; Restrepo, D. Effects of in utero odorant exposure on neuroanatomical development of the olfactory bulb and odour preferences. Proc. Biol. Sci. 2011, 278, 1949–1955. [Google Scholar] [CrossRef] [PubMed]
- Prenzel, N.; Zwick, E.; Daub, H.; Leserer, M.; Abraham, R.; Wallasch, C.; Ullrich, A. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature 1999, 402, 884–888. [Google Scholar] [CrossRef] [PubMed]
- Fischer, O.M.; Hart, S.; Gschwind, A.; Ullrich, A. EGFR signal transactivation in cancer cells. Biochem. Soc. Trans. 2003, 31 Pt 6, 1203–1208. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.; Fischer, O.M.; Prenzel, N.; Zwick-Wallasch, E.; Schneider, M.; Hennighausen, L.; Ullrich, A. GPCR-induced migration of breast carcinoma cells depends on both EGFR signal transactivation and EGFR-independent pathways. Biol. Chem. 2005, 386, 845–855. [Google Scholar] [CrossRef]
- Watt, W.C.; Storm, D.R. Odorants stimulate the ERK/mitogen-activated protein kinase pathway and activate cAMP-response element-mediated transcription in olfactory sensory neurons. J. Biol. Chem. 2001, 276, 2047–2052. [Google Scholar] [CrossRef]
- Orecchioni, M.; Kobiyama, K.; Winkels, H.; Ghosheh, Y.; McArdle, S.; Mikulski, Z.; Kiosses, W.B.; Fan, Z.; Wen, L.; Jung, Y.; et al. Olfactory receptor 2 in vascular macrophages drives atherosclerosis by NLRP3-dependent IL-1 production. Science 2022, 375, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Vadevoo, S.M.P.; Gunassekaran, G.R.; Lee, C.; Lee, N.; Lee, J.; Chae, S.; Park, J.Y.; Koo, J.; Lee, B. The macrophage odorant receptor Olfr78 mediates the lactate-induced M2 phenotype of tumor-associated macrophages. Proc. Natl. Acad. Sci. USA 2021, 118, e2102434118. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitsch, A.I.; Maretzky, T.; Weskamp, G.; Haxaire, C.; Tueshaus, J.; Lichtenthaler, S.F.; Monette, S.; Blobel, C.P. Analysis of the function of ADAM17 in iRhom2 curly-bare and tylosis with esophageal cancer mutant mice. J. Cell Sci. 2023, 136, jcs260910. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Smedley, D.; Haider, S.; Ballester, B.; Holland, R.; London, D.; Thorisson, G.; Kasprzyk, A. BioMart—Biological queries made easy. BMC Genom. 2009, 10, 22. [Google Scholar] [CrossRef]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzopardi, S.A.; Lu, H.-Y.; Monette, S.; Rabinowitsch, A.I.; Salmon, J.E.; Matsunami, H.; Blobel, C.P. Role of iRhom2 in Olfaction: Implications for Odorant Receptor Regulation and Activity-Dependent Adaptation. Int. J. Mol. Sci. 2024, 25, 6079. https://doi.org/10.3390/ijms25116079
Azzopardi SA, Lu H-Y, Monette S, Rabinowitsch AI, Salmon JE, Matsunami H, Blobel CP. Role of iRhom2 in Olfaction: Implications for Odorant Receptor Regulation and Activity-Dependent Adaptation. International Journal of Molecular Sciences. 2024; 25(11):6079. https://doi.org/10.3390/ijms25116079
Chicago/Turabian StyleAzzopardi, Stephanie A., Hsiu-Yi Lu, Sebastien Monette, Ariana I. Rabinowitsch, Jane E. Salmon, Hiroaki Matsunami, and Carl P. Blobel. 2024. "Role of iRhom2 in Olfaction: Implications for Odorant Receptor Regulation and Activity-Dependent Adaptation" International Journal of Molecular Sciences 25, no. 11: 6079. https://doi.org/10.3390/ijms25116079
APA StyleAzzopardi, S. A., Lu, H.-Y., Monette, S., Rabinowitsch, A. I., Salmon, J. E., Matsunami, H., & Blobel, C. P. (2024). Role of iRhom2 in Olfaction: Implications for Odorant Receptor Regulation and Activity-Dependent Adaptation. International Journal of Molecular Sciences, 25(11), 6079. https://doi.org/10.3390/ijms25116079





