Knockdown of Esr1 from DRD1-Rich Brain Regions Affects Adipose Tissue Metabolism: Potential Crosstalk between Nucleus Accumbens and Adipose Tissue
Abstract
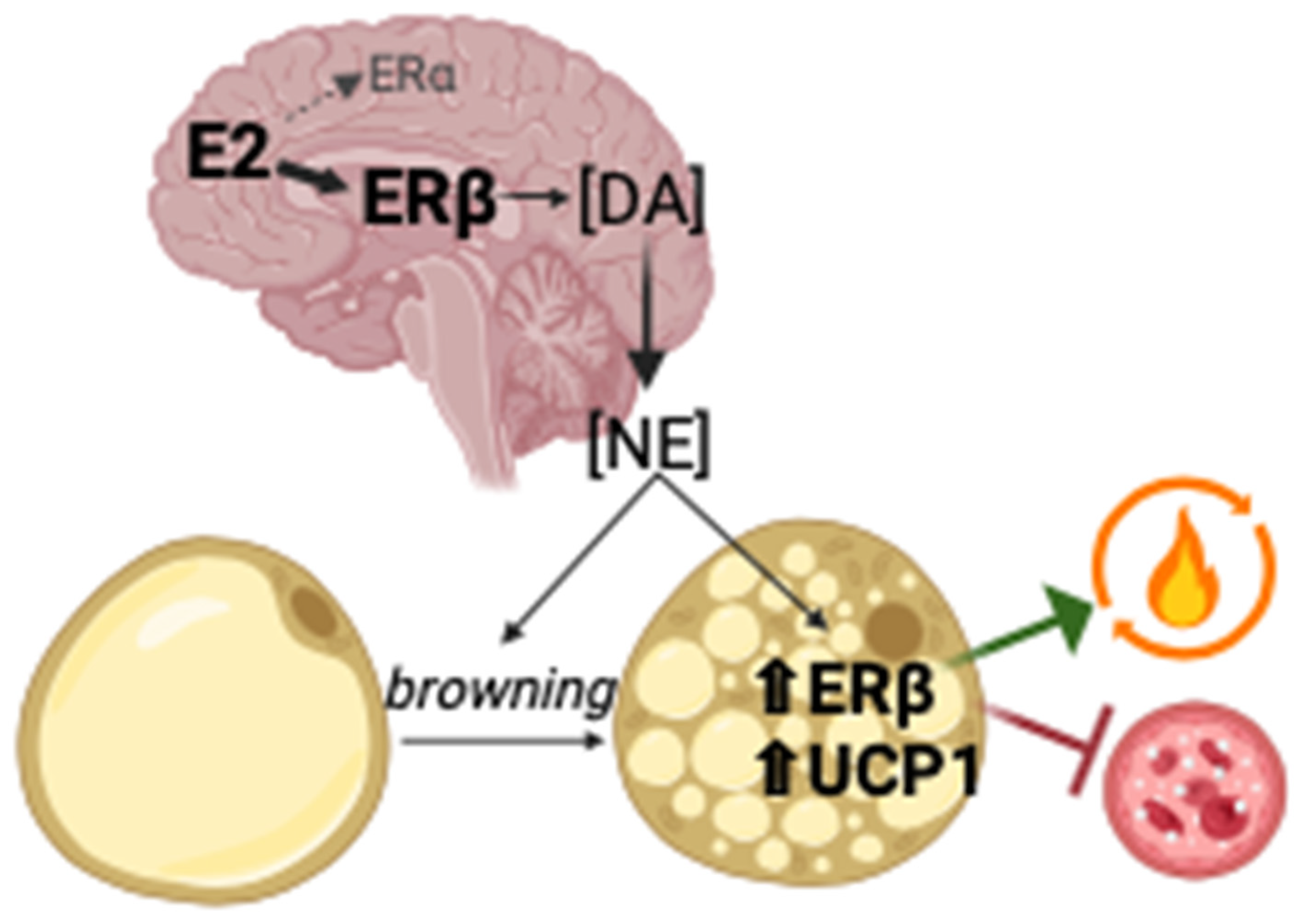
:1. Introduction
2. Results
2.1. Animal Model Confirmation
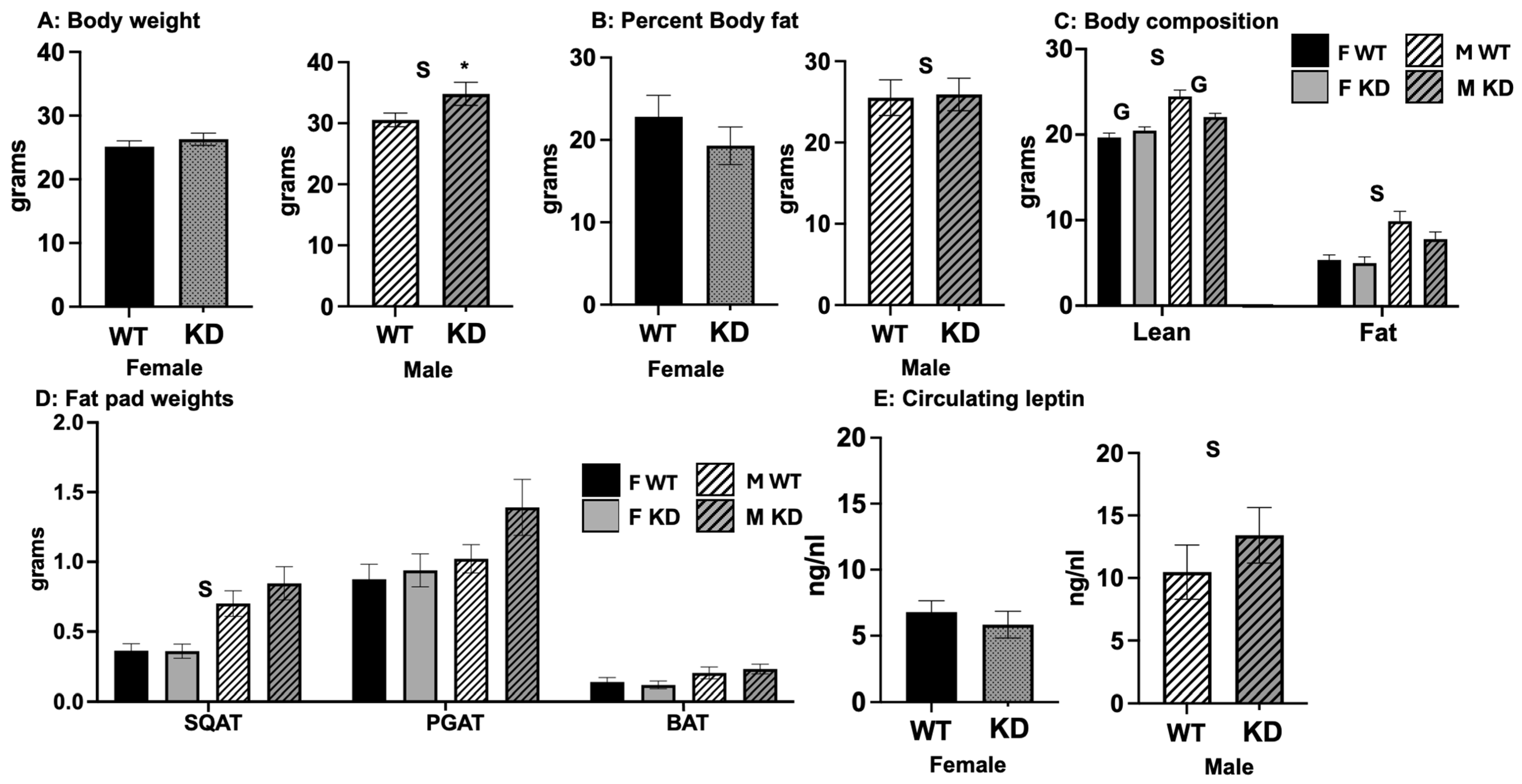
2.2. Effects of DRD1-Specific Esr1 Deletion on Body Weight and Body Composition
2.3. Effects of DRD1-Specific Esr1 Deletion on Energy Intake and Expenditure
2.4. Effects of DRD1-Specific Esr1 Deletion on Brown Adipose Tissue Phenotype
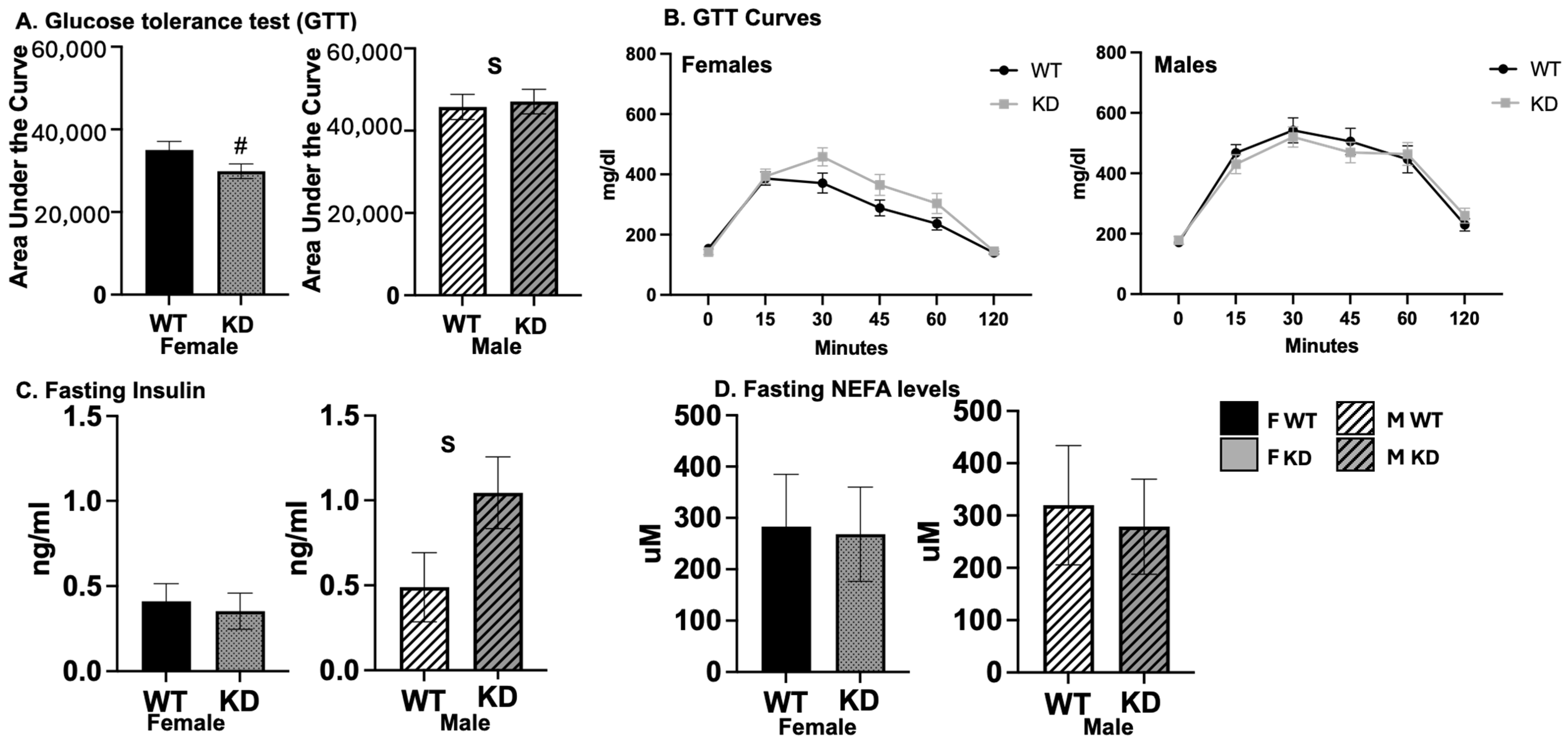
2.5. Effects of DRD1-Specific Esr1 Deletion on Insulin Sensitivity and Glucose Tolerance
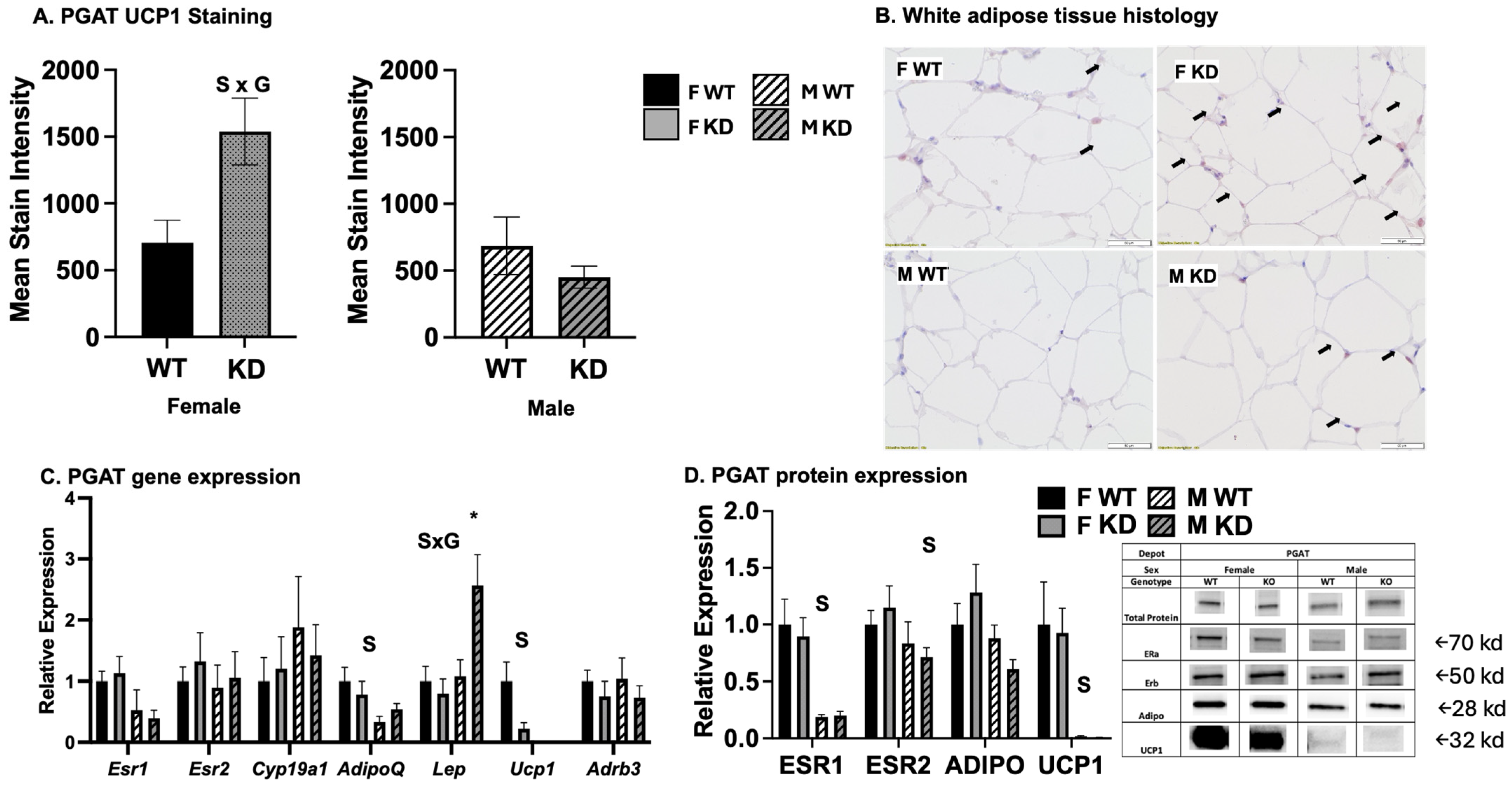
2.6. Effects of DRD1-Specific Esr1 Deletion on White Adipose Tissue Browning
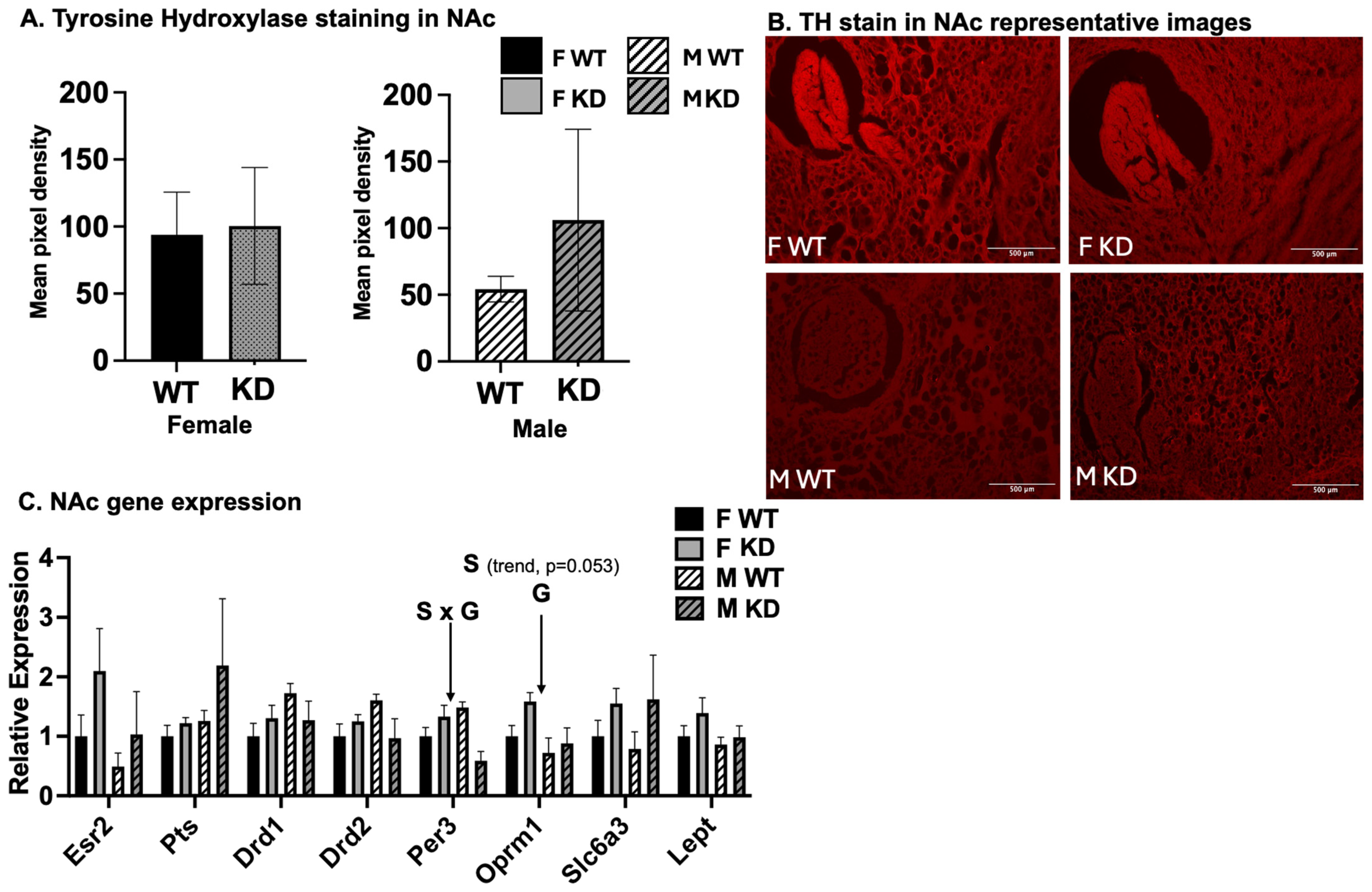
2.7. Effects of Sex and D1ERKO Genotype on Nucleus Accumbens (NAc) Gene Expression and Tyrosine Hydroxylase Staining
2.8. Correlations among Nucleus Accumbens Gene Expression, Adipose Tissue Phenotype, Activity Levels, and Metabolism
3. Discussion
Study Limitations
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, Y.-M.; Kanaley, J.A.; Padilla, J.; Zidon, T.; Welly, R.J.; Will, M.J.; Britton, S.L.; Koch, L.G.; Ruegsegger, G.N.; Booth, F.W.; et al. Effects of intrinsic aerobic capacity and ovariectomy on voluntary wheel running and nucleus accumbens dopamine receptor gene expression. Physiol. Behav. 2016, 164 Pt A, 383–389. [Google Scholar] [CrossRef]
- Kim, D.-I.; Park, Y.-M. Effects of Menopause on Physical Activity and Dopamine Signaling in Women. Iran. J. Public Health 2022, 51, 2372–2374. [Google Scholar] [CrossRef] [PubMed]
- Bracht, J.R.; Vieira-Potter, V.J.; Santos, R.D.S.; Öz, O.K.; Palmer, B.F.; Clegg, D.J. The role of estrogens in the adipose tissue milieu. Ann. N. Y. Acad. Sci. 2020, 1461, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Marsh, M.L.; Oliveira, M.N.; Vieira-Potter, V.J. Adipocyte Metabolism and Health after the Menopause: The Role of Exercise. Nutrients 2023, 15, 444. [Google Scholar] [CrossRef] [PubMed]
- Clegg, D.; Hevener, A.L.; Moreau, K.L.; Morselli, E.; Criollo, A.; Van Pelt, R.E.; Vieira-Potter, V.J. Sex Hormones and Cardiometabolic Health: Role of Estrogen and Estrogen Receptors. Endocrinology 2017, 158, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Barros, R.P.A.; Gustafsson, J.Å. Estrogen receptors and the metabolic network. Cell Metab. 2011, 14, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Heine, P.A.; Taylor, J.A.; Iwamoto, G.A.; Lubahn, D.B.; Cooke, P.S. Increased adipose tissue in male and female estrogen receptor-α knockout mice. Proc. Natl. Acad. Sci. USA 2000, 97, 12729–12734. [Google Scholar] [CrossRef] [PubMed]
- Savva, C.; Korach-André, M. Estrogen Receptor beta (ERβ) Regulation of Lipid Homeostasis—Does Sex Matter? Metabolites 2020, 10, 116. [Google Scholar] [CrossRef] [PubMed]
- Aldiss, P.; Betts, J.; Sale, C.; Pope, M.; Budge, H.; Symonds, M.E. Exercise-induced ‘browning’ of adipose tissues. Metabolism 2018, 81, 63–70. [Google Scholar] [CrossRef]
- Caron, A.; Lee, S.; Elmquist, J.K.; Gautron, L. Leptin and brain-adipose crosstalks. Nat. Rev. Neurosci. 2018, 19, 153–165. [Google Scholar] [CrossRef]
- Basso, J.C.; Morrell, J.I. The medial prefrontal cortex and nucleus accumbens mediate the motivation for voluntary wheel running in the rat. Behav. Neurosci. 2015, 129, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Almey, A.; Milner, T.A.; Brake, W.G. Estrogen receptors in the central nervous system and their implication for dopamine-dependent cognition in females. Horm. Behav. 2015, 74, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Thompson, T.L.; Moore, C.C.; Smith, B. Estrogen priming modulates autoreceptor-mediated potentiation of dopamine uptake. Eur. J. Pharmacol. 2000, 401, 357–363. [Google Scholar] [CrossRef]
- Dluzen, D.E. Neuroprotective effects of estrogen upon the nigrostriatal dopaminergic system. J. Neurocytol. 2000, 29, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Shay, D.A.; Welly, R.J.; Givan, S.A.; Bivens, N.; Kanaley, J.; Marshall, B.L.; Lubahn, D.B.; Rosenfeld, C.S.; Vieira-Potter, V.J. Changes in nucleus accumbens gene expression accompany sex-specific suppression of spontaneous physical activity in aromatase knockout mice. Horm. Behav. 2020, 121, 104719. [Google Scholar] [CrossRef] [PubMed]
- Korner, G.; Scherer, T.; Adamsen, D.; Rebuffat, A.; Crabtree, M.; Rassi, A.; Scavelli, R.; Homma, D.; Ledermann, B.; Konrad, D.; et al. Mildly compromised tetrahydrobiopterin cofactor biosynthesis due to Pts variants leads to unusual body fat distribution and abdominal obesity in mice. J. Inherit. Metab. Dis. 2016, 39, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Townsend, L.K.; Wright, D.C. Looking on the “brite” side exercise-induced browning of white adipose tissue. Pflugers Arch. 2019, 471, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Clookey, S.L.; Welly, R.J.; Shay, D.; Woodford, M.L.; Fritsche, K.L.; Rector, R.S.; Padilla, J.; Lubahn, D.B.; Vieira-Potter, V.J. Beta 3 Adrenergic Receptor Activation Rescues Metabolic Dysfunction in Female Estrogen Receptor Alpha-Null Mice. Front. Physiol. 2019, 10, 9. [Google Scholar] [CrossRef]
- Aldiss, P.; Dellschaft, N.; Sacks, H.; Budge, H.; Symonds, M.E. Beyond obesity—Thermogenic adipocytes and cardiometabolic health. Horm. Mol. Biol. Clin. Investig. 2017, 31, 20170007. [Google Scholar] [CrossRef]
- Scheele, C.; Wolfrum, C. Brown Adipose Crosstalk in Tissue Plasticity and Human Metabolism. Endocr. Rev. 2020, 41, 53–65. [Google Scholar] [CrossRef]
- Queathem, E.D.; Welly, R.J.; Clart, L.M.; Rowles, C.C.; Timmons, H.; Fitzgerald, M.; Eichen, P.A.; Lubahn, D.B.; Vieira-Potter, V.J. White Adipose Tissue Depots Respond to Chronic Beta-3 Adrenergic Receptor Activation in a Sexually Dimorphic and Depot Divergent Manner. Cells 2021, 10, 3453. [Google Scholar] [CrossRef]
- Fernández-Peña, C.; Reimúndez, A.; Viana, F.; Arce, V.M.; Señarís, R. Sex differences in thermoregulation in mammals: Implications for energy homeostasis. Front. Endocrinol. 2023, 14, 1093376. [Google Scholar] [CrossRef]
- Vieira-Potter, V.J.; Padilla, J.; Park, Y.-M.; Welly, R.J.; Scroggins, R.J.; Britton, S.L.; Koch, L.G.; Jenkins, N.T.; Crissey, J.M.; Zidon, T.; et al. Female rats selectively bred for high intrinsic aerobic fitness are protected from ovariectomy-associated metabolic dysfunction. Am. J. Physiol. Integr. Comp. Physiol. 2015, 308, R530–R542. [Google Scholar] [CrossRef]
- Zidon, T.M.; Padilla, J.; Fritsche, K.L.; Welly, R.J.; McCabe, L.T.; Stricklin, O.E.; Frank, A.; Park, Y.; Clegg, D.J.; Lubahn, D.B.; et al. Effects of ERbeta and ERalpha on OVX-induced changes in adiposity and insulin resistance. J. Endocrinol. 2020, 245, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Diaz, J.C.; Dunaway, K.; Zuniga, C.; Sheil, E.; Sadeghian, K.; Auger, A.P.; Baldo, B.A. Delayed estrogen actions diminish food consumption without changing food approach, motor activity, or hypothalamic activation elicited by corticostriatal µ-opioid signaling. Neuropsychopharmacology 2023, 48, 1952–1962. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Moore, T.M.; Drew, B.G.; Ribas, V.; Wanagat, J.; Civelek, M.; Segawa, M.; Wolf, D.M.; Norheim, F.; Seldin, M.M.; et al. Estrogen receptor α controls metabolism in white and brown adipocytes by regulating Polg1 and mitochondrial remodeling. Sci. Transl. Med. 2020, 12, eaax8096. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, V.C.T.; Jiang, Y.; He, Y.; Xu, P. Hypothalamic Estrogen Signaling and Adipose Tissue Metabolism in Energy Homeostasis. Front. Endocrinol. 2022, 13, 898139. [Google Scholar] [CrossRef] [PubMed]
- de Morentin, P.B.M.; González-García, I.; Martins, L.; Lage, R.; Fernández-Mallo, D.; Martínez-Sánchez, N.; Ruíz-Pino, F.; Liu, J.; Morgan, D.A.; Pinilla, L.; et al. Estradiol regulates brown adipose tissue thermogenesis via hypothalamic AMPK. Cell Metab. 2014, 20, 41–53. [Google Scholar]
- Miao, Y.F.; Su, W.; Dai, Y.B.; Wu, W.F.; Huang, B.; Barros, R.P.; Nguyen, H.; Maneix, L.; Guan, Y.F.; Warner, M.; et al. An ERbeta agonist induces browning of subcutaneous abdominal fat pad in obese female mice. Sci. Rep. 2016, 6, 38579. [Google Scholar] [CrossRef]
- Clart, L.M.; Welly, R.J.; Queathem, E.D.; Rector, R.S.; Padilla, J.; Baines, C.P.; Kanaley, J.A.; Lubahn, D.B.; Vieira-Potter, V.J. Role of ERbeta in adipocyte metabolic response to wheel running following ovariectomy. J. Endocrinol. 2021, 249, 223–237. [Google Scholar] [CrossRef]
- Zheng, R.; Yang, L.; Sikorski, M.A.; Enns, L.C.; Czyzyk, T.A.; Ladiges, W.C.; McKnight, G.S. Deficiency of the RIIbeta subunit of PKA affects locomotor activity and energy homeostasis in distinct neuronal populations. Proc. Natl. Acad. Sci. USA 2013, 110, E1631–E1640. [Google Scholar] [CrossRef] [PubMed]
- Grigsby, K.B.; Kovarik, C.M.; Mao, X.; Booth, F.W. Medial preoptic estrogen receptor-beta blunts the estrogen receptor-alpha mediated increases in wheel-running behavior of female rats. Behav. Brain Res. 2020, 379, 112341. [Google Scholar] [CrossRef]
- Zhao, L.; Woody, S.K.; Chhibber, A. Estrogen receptor beta in Alzheimer’s disease: From mechanisms to therapeutics. Ageing Res. Rev. 2015, 24 Pt B, 178–190. [Google Scholar] [CrossRef]
- Ponnusamy, S.; Tran, Q.T.; Harvey, I.; Smallwood, H.S.; Thiyagarajan, T.; Banerjee, S.; Johnson, D.L.; Dalton, J.T.; Sullivan, R.D.; Miller, D.D.; et al. Pharmacologic activation of estrogen receptor beta increases mitochondrial function, energy expenditure, and brown adipose tissue. FASEB J. 2017, 31, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Cho, Y.; Lee, J.; Jeong, H.; Lee, Y.; Kim, S.I.; Kim, C.H.; Lee, H.W.; Nam, K.T. Sexually dimorphic leanness and hypermobility in p16(Ink4a)/CDKN2A-deficient mice coincides with phenotypic changes in the cerebellum. Sci. Rep. 2019, 9, 11167. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Santos, R.d.S.; Gao, S.; Ávalos, Y.; Criollo, A.; Palmer, B.F.; Clegg, D.J. Impact of estrogens and estrogen receptor-α in brain lipid metabolism. Am. J. Physiol. Metab. 2018, 315, E7–E14. [Google Scholar] [CrossRef] [PubMed]
- Faltraco, F.; Palm, D.; Uzoni, A.; Borchert, L.; Simon, F.; Tucha, O.; Thome, J. Dopamine adjusts the circadian gene expression of Per2 and Per3 in human dermal fibroblasts from ADHD patients. J. Neural Transm. 2021, 128, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, A.; Costa, M.J.; Rivero-Gutiérrez, B.; Ji, L.; Morgan, S.L.; Feldman, B.J. The Circadian Clock Regulates Adipogenesis by a Per3 Crosstalk Pathway to Klf15. Cell Rep. 2017, 21, 2367–2375. [Google Scholar] [CrossRef]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef]


| Lean mass (rel BW) | PGAT (g) | BAT (g) | GTT AUC | SPA (daily) | REE (relBW) | INSULIN [ng/mL] | LEPTIN [ng/mL] | NEFA [μM] | TH stain | NAc Oprm1 | NAc Per3 | NAc Pts | NAc Slc6a3 | NAc Esr2 | BAT Ucp1 | BAT Adrb3 | BAT Esr1 | BAT Esr2 | BAT Adipoq | BAT Lept | PGAT Ucp1 | PGAT Adrb3 | PGAT Esr2 | PGAT Adipoq | PGAT Lept | BAT UCP1 stain | PGAT UCP1 stain | PGAT ESR1 | PGAT ESR2 | PGAT UCP1 | PGAT ADIPOQ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lean mass (rel BW) | 1 | −0.823 ** | −0.437 ** | −0.600 ** | 0.053 | 0.375 ** | −0.644 ** | −0.829 ** | −0.1 | 0.355 | 0.431 * | −0.01 | 0.041 | 0.383 * | 0.354 | −0.144 | 0.132 | 0.101 | 0.203 | 0.138 | 0.157 | −0.01 | 0.189 | 0.231 | 0.154 | −0.528 ** | 0.31 | 0.38 | 0.451 * | 0.547 ** | 0.356 * | 0.176 |
| PGAT (g) | −0.823 ** | 1 | 0.458 ** | 0.514 ** | 0.016 | −0.302 ** | 0.680 ** | 0.854 ** | 0.124 | −0.19 | −0.208 | 0.051 | −0.234 | −0.228 | −0.124 | 0.091 | −0.183 | −0.144 | −0.199 | −0.148 | −0.207 | −0.046 | −0.196 | −0.258 | −0.161 | 0.652 ** | −0.204 | −0.382 | −0.285 | −0.400 * | −0.191 | −0.31 |
| BAT (g) | −0.437 ** | 0.458 ** | 1 | 0.433 ** | 0.013 | −0.257 * | 0.601 ** | 0.570 ** | 0.025 | −0.415 | −0.254 | −0.163 | −0.437 * | −0.17 | 0.139 | 0.023 | −0.198 | −0.235 | −0.213 | −0.21 | −0.232 | −0.153 | −0.232 | −0.264 | −0.236 | 0.471 ** | −0.246 | −0.423 | −0.435 * | −0.356 | −0.421 * | −0.343 |
| GTT AUC | −0.600 ** | 0.514 ** | 0.433 ** | 1 | −0.258 | −0.399 ** | 0.322 * | 0.472 ** | −0.014 | −0.181 | −0.730 ** | −0.425 | −0.091 | −0.594 * | −0.666 * | 0.098 | −0.066 | −0.102 | −0.246 | −0.01 | −0.207 | −0.051 | 0.146 | −0.052 | −0.084 | 0.543 ** | −0.318 | −0.697 ** | −0.385 * | −0.388 * | −0.376 * | −0.331 |
| SPA (daily) | 0.053 | 0.016 | 0.013 | −0.258 | 1 | 0.314 ** | 0.141 | 0.072 | −0.031 | 0.227 | 0.131 | 0.323 | −0.103 | 0.085 | −0.046 | 0.097 | 0.066 | 0.065 | 0.166 | 0.151 | 0.325 * | −0.024 | −0.215 | −0.233 | −0.175 | −0.251 | 0.16 | 0.122 | 0.467 ** | 0.448 * | 0.314 | 0.3 |
| REE (relBW) | 0.375 ** | −0.302 ** | −0.257 * | −0.399 ** | 0.314 ** | 1 | −0.272 * | −0.410 ** | 0 | 0.498 * | 0.254 | 0.141 | −0.125 | 0.262 | 0.057 | 0.069 | 0.141 | 0.202 | 0.244 | 0.189 | 0.197 | 0.144 | 0.024 | −0.077 | −0.044 | −0.375 ** | 0.236 | 0.543 * | 0.324 | 0.351 | 0.379 * | 0.28 |
| INSULIN [ng/mL] | −0.644 ** | 0.680 ** | 0.601 ** | 0.322 * | 0.141 | −0.272 * | 1 | 0.806 ** | 0.237 | −0.267 | −0.286 | −0.215 | −0.249 | −0.22 | −0.235 | −0.062 | −0.284 * | −0.226 | −0.204 | −0.238 | −0.315 * | −0.087 | −0.243 | −0.249 | −0.279 * | 0.439 ** | 0.022 | −0.332 | −0.333 | −0.35 | −0.236 | −0.376 * |
| LEPTIN [ng/mL] | −0.829 ** | 0.854 ** | 0.570 ** | 0.472 ** | 0.072 | −0.410 ** | 0.806 ** | 1 | 0.19 | −0.336 | −0.331 | −0.009 | −0.066 | −0.303 | −0.305 | −0.047 | −0.264 | −0.256 | −0.275 * | −0.264 | −0.317 * | −0.099 | −0.266 | −0.319 * | −0.218 | 0.521 ** | −0.118 | −0.540 * | −0.396 * | −0.377 * | −0.343 | −0.301 |
| NEFA [μM] | −0.1 | 0.124 | 0.025 | −0.014 | −0.031 | 0 | 0.237 | 0.19 | 1 | −0.564 * | −0.212 | 0.14 | 0.366 | −0.153 | −0.301 | −0.320 * | −0.372 ** | −0.340 * | −0.372 ** | −0.293 * | −0.480 ** | −0.226 | 0.035 | −0.067 | −0.205 | 0.036 | 0.133 | −0.228 | −0.1 | 0.006 | −0.203 | −0.304 |
| TH stain | 0.355 | −0.19 | −0.415 | −0.181 | 0.227 | 0.498 * | −0.267 | −0.336 | −0.564 * | 1 | -- | -- | -- | -- | -- | 0.713 ** | 0.456 | 0.641 ** | 0.545 * | 0.709 ** | 0.482 | −0.198 | 0.007 | 0.286 | −0.243 | −0.13 | 0.052 | 0.703 | 0.002 | −0.16 | 0.218 | 0.004 |
| NAc Oprm1 | 0.431 * | −0.208 | −0.254 | −0.730 ** | 0.131 | 0.254 | −0.286 | −0.331 | −0.212 | -- | 1 | 0.537 ** | 0.525 ** | 0.560 ** | 0.657 ** | −0.102 | −0.073 | −0.072 | −0.064 | −0.16 | −0.153 | 0.419 | −0.105 | −0.394 | −0.151 | −0.724 ** | -- | -- | 0.315 | 0.694 | 0.447 | 0.857 |
| NAc Per3 | −0.01 | 0.051 | −0.163 | −0.425 | 0.323 | 0.141 | −0.215 | −0.009 | 0.14 | -- | 0.537 ** | 1 | 0.547 ** | 0.231 | 0.346 | 0.498 | 0.489 | 0.494 | 0.514 | 0.444 | 0.41 | −0.026 | −0.132 | 0.054 | −0.106 | −0.458 | -- | -- | 0.133 | 0.961 | 0.268 | -- |
| NAc Pts | 0.041 | −0.234 | −0.437 * | −0.091 | −0.103 | −0.125 | −0.249 | −0.066 | 0.366 | -- | 0.525 ** | 0.547 ** | 1 | 0.333 | 0.292 | −0.538 | −0.468 | −0.438 | −0.471 | −0.654 * | −0.660 * | 0.225 | 0.281 | 0.119 | 0.058 | 0.168 | -- | -- | 0.574 | 0.217 | 0.792 | 0.463 |
| NAc Slc6a3 | 0.383 * | −0.228 | −0.17 | −0.594 * | 0.085 | 0.262 | −0.22 | −0.303 | −0.153 | -- | 0.560 ** | 0.231 | 0.333 | 1 | 0.431 * | −0.221 | −0.19 | −0.206 | −0.204 | −0.217 | −0.183 | 0.109 | −0.228 | −0.321 | −0.3 | −0.443 | -- | -- | 0.838 | −0.853 | 0.779 | −0.688 |
| NAc Esr2 | 0.354 | −0.124 | 0.139 | −0.666 * | −0.046 | 0.057 | −0.235 | −0.305 | −0.301 | -- | 0.657 ** | 0.346 | 0.292 | 0.431 * | 1 | −0.2 | −0.195 | −0.183 | −0.193 | −0.19 | −0.197 | 0.129 | −0.327 | −0.331 | −0.482 | −0.476 | -- | -- | 0.902 | −0.295 | 0.980 * | −0.037 |
| BAT Ucp1 | −0.144 | 0.091 | 0.023 | 0.098 | 0.097 | 0.069 | −0.062 | −0.047 | −0.320 * | 0.713 ** | −0.102 | 0.498 | −0.538 | −0.221 | −0.2 | 1 | 0.786 ** | 0.755 ** | 0.536 ** | 0.721 ** | 0.812 ** | 0.059 | 0.179 | 0.336 * | 0.255 | 0.229 | −0.211 | −0.183 | −0.262 | −0.162 | −0.156 | 0.15 |
| BAT Adrb3 | 0.132 | −0.183 | −0.198 | −0.066 | 0.066 | 0.141 | −0.284 * | −0.264 | −0.372 ** | 0.456 | −0.073 | 0.489 | −0.468 | −0.19 | −0.195 | 0.786 ** | 1 | 0.875 ** | 0.612 ** | 0.740 ** | 0.866 ** | 0.053 | 0.219 | 0.242 | 0.306 * | 0.021 | −0.137 | 0.127 | −0.098 | 0.14 | 0.028 | 0.197 |
| BAT Esr1 | 0.101 | −0.144 | −0.235 | −0.102 | 0.065 | 0.202 | −0.226 | −0.256 | −0.340 * | 0.641 ** | −0.072 | 0.494 | −0.438 | −0.206 | −0.183 | 0.755 ** | 0.875 ** | 1 | 0.662 ** | 0.770 ** | 0.772 ** | 0.096 | 0.225 | 0.321 * | 0.394 ** | 0.053 | −0.122 | 0.214 | 0.019 | 0.165 | 0.165 | 0.414 * |
| BAT Esr2 | 0.203 | −0.199 | −0.213 | −0.246 | 0.166 | 0.244 | −0.204 | −0.275 * | −0.372 ** | 0.545 * | −0.064 | 0.514 | −0.471 | −0.204 | −0.193 | 0.536 ** | 0.612 ** | 0.662 ** | 1 | 0.489 ** | 0.830 ** | −0.077 | −0.007 | 0.112 | 0.114 | −0.005 | −0.112 | 0.057 | 0.063 | 0.201 | 0.314 | 0.274 |
| BAT Adipoq | 0.138 | −0.148 | −0.21 | −0.01 | 0.151 | 0.189 | −0.238 | −0.264 | −0.293 * | 0.709 ** | −0.16 | 0.444 | −0.654 * | −0.217 | −0.19 | 0.721 ** | 0.740 ** | 0.770 ** | 0.489 ** | 1 | 0.600 ** | −0.094 | 0.147 | 0.267 | 0.103 | −0.06 | −0.07 | 0.293 | −0.076 | 0.078 | 0.126 | −0.035 |
| BAT Lept | 0.157 | −0.207 | −0.232 | −0.207 | 0.325 * | 0.197 | −0.315 * | −0.317 * | −0.480 ** | 0.482 | −0.153 | 0.41 | −0.660 * | −0.183 | −0.197 | 0.812 ** | 0.866 ** | 0.772 ** | 0.830 ** | 0.600 ** | 1 | −0.191 | −0.135 | 0.05 | 0.087 | −0.166 | −0.112 | 0.08 | −0.017 | 0.113 | 0.122 | 0.426 * |
| PGAT Ucp1 | −0.01 | −0.046 | −0.153 | −0.051 | −0.024 | 0.144 | −0.087 | −0.099 | −0.226 | −0.198 | 0.419 | −0.026 | 0.225 | 0.109 | 0.129 | 0.059 | 0.053 | 0.096 | −0.077 | −0.094 | −0.191 | 1 | 0.269 | 0.09 | 0.341 * | 0.116 | −0.076 | −0.119 | 0.307 | −0.055 | 0.682 ** | −0.157 |
| PGAT Adrb3 | 0.189 | −0.196 | −0.232 | 0.146 | −0.215 | 0.024 | −0.243 | −0.266 | 0.035 | 0.007 | −0.105 | −0.132 | 0.281 | −0.228 | −0.327 | 0.179 | 0.219 | 0.225 | −0.007 | 0.147 | −0.135 | 0.269 | 1 | 0.636 ** | 0.557 ** | 0.324 * | 0.193 | −0.297 | −0.05 | 0.101 | −0.012 | −0.073 |
| PGAT Esr2 | 0.231 | −0.258 | −0.264 | −0.052 | −0.233 | −0.077 | −0.249 | −0.319 * | −0.067 | 0.286 | −0.394 | 0.054 | 0.119 | −0.321 | −0.331 | 0.336 * | 0.242 | 0.321 * | 0.112 | 0.267 | 0.05 | 0.09 | 0.636 ** | 1 | 0.604 ** | 0.186 | 0.009 | −0.164 | 0.049 | −0.026 | −0.059 | 0.226 |
| PGAT Adipoq | 0.154 | −0.161 | −0.236 | −0.084 | −0.175 | −0.044 | −0.279 * | −0.218 | −0.205 | −0.243 | −0.151 | −0.106 | 0.058 | −0.3 | −0.482 | 0.255 | 0.306 * | 0.394 ** | 0.114 | 0.103 | 0.087 | 0.341 * | 0.557 ** | 0.604 ** | 1 | 0.255 | 0.041 | −0.373 | 0.134 | −0.107 | 0.148 | 0.410 * |
| PGAT Lept | −0.528 ** | 0.652 ** | 0.471 ** | 0.543 ** | −0.251 | −0.375 ** | 0.439 ** | 0.521 ** | 0.036 | −0.13 | −0.724 ** | −0.458 | 0.168 | −0.443 | −0.476 | 0.229 | 0.021 | 0.053 | −0.005 | −0.06 | −0.166 | 0.116 | 0.324 * | 0.186 | 0.255 | 1 | −0.13 | −0.648 ** | −0.244 | −0.316 | −0.105 | −0.305 |
| BAT UCP1 stain | 0.31 | −0.204 | −0.246 | −0.318 | 0.16 | 0.236 | 0.022 | −0.118 | 0.133 | 0.052 | -- | -- | -- | -- | -- | −0.211 | −0.137 | −0.122 | −0.112 | −0.07 | −0.112 | −0.076 | 0.193 | 0.009 | 0.041 | −0.13 | 1 | 0.234 | 0.283 | 0.09 | 0.308 | 0.265 |
| PGAT UCP1 stain | 0.38 | −0.382 | −0.423 | −0.697 ** | 0.122 | 0.543 * | −0.332 | −0.540 * | −0.228 | 0.703 | -- | -- | -- | -- | -- | −0.183 | 0.127 | 0.214 | 0.057 | 0.293 | 0.08 | −0.119 | −0.297 | −0.164 | −0.373 | −0.648 ** | 0.234 | 1 | 0.199 | 0.027 | 0.024 | 0.197 |
| PGAT ESR1 | 0.451 * | −0.285 | −0.435 * | −0.385 * | 0.467 ** | 0.324 | −0.333 | −0.396 * | −0.1 | 0.002 | 0.315 | 0.133 | 0.574 | 0.838 | 0.902 | −0.262 | −0.098 | 0.019 | 0.063 | −0.076 | −0.017 | 0.307 | −0.05 | 0.049 | 0.134 | −0.244 | 0.283 | 0.199 | 1 | 0.668 ** | 0.569 ** | 0.2 |
| PGAT ESR2 | 0.547 ** | −0.400 * | −0.356 | −0.388 * | 0.448 * | 0.351 | −0.35 | −0.377 * | 0.006 | −0.16 | 0.694 | 0.961 | 0.217 | −0.853 | −0.295 | −0.162 | 0.14 | 0.165 | 0.201 | 0.078 | 0.113 | −0.055 | 0.101 | −0.026 | −0.107 | −0.316 | 0.09 | 0.027 | 0.668 ** | 1 | 0.324 | 0.15 |
| PGAT UCP1 | 0.356 * | −0.191 | −0.421 * | −0.376 * | 0.314 | 0.379 * | −0.236 | −0.343 | −0.203 | 0.218 | 0.447 | 0.268 | 0.792 | 0.779 | 0.980 * | −0.156 | 0.028 | 0.165 | 0.314 | 0.126 | 0.122 | 0.682 ** | −0.012 | −0.059 | 0.148 | −0.105 | 0.308 | 0.024 | 0.569 ** | 0.324 | 1 | −0.009 |
| PGAT ADIPOQ | 0.176 | −0.31 | −0.343 | −0.331 | 0.3 | 0.28 | −0.376 * | −0.301 | −0.304 | 0.004 | 0.857 | -- | 0.463 | −0.688 | −0.037 | 0.15 | 0.197 | 0.414 * | 0.274 | −0.035 | 0.426 * | −0.157 | −0.073 | 0.226 | 0.410 * | −0.305 | 0.265 | 0.197 | 0.2 | 0.15 | −0.009 | 1 |
| Gene ID | Company | Forward | Reverse | Product Size |
|---|---|---|---|---|
| 18s | IDT (Coralville, IA) | TCAAGAACGAAAGTCGGAGG | GGACATCTAAGGGCATCAC | 488 |
| PTS | Sigma (Aizu, Japan) | CGATGAAGAGAACTTAAGAGTG | TGTAACAGGATCAATCTCTCC | 106 |
| Per3 | Sigma | GAGAGTATGTCATTCTGGATTC | TCATTTAATGGACTCGTTCG | 109 |
| DRD1 | IDT | TCTCCCAGATCGGGCATTT | GGGCCTCTTCCTGGTCAATC | 131 |
| DRD2 | Sigma | TTGTTCTTGGTGTGTTCATC | TATAGATGATGGGGTTCACG | 150 |
| OPRM1 | Sigma | ACTCATGTTGAAAAACCCTG | CTGTGTTCAGATGACATTCAC | 125 |
| Slc6a3 | Sigma | CCAATAACTGCTATAGAGATGC | GTAGATGATGAAGATCAACCC | 167 |
| Esr1 | Sigma | CAAAGGTAAATGTGTGGAAGG | GTGTACACTCCGGAATTAAG | 141 |
| Esr2 | Sigma | CTCAACTCCAGTATGTACCC | CATGAGAAAGAAGCATCAGG | 178 |
| Cyp19a1 | Sigma | AACATCATTCTGAACATCGG | AGGGAACATTCTTCTCAAAG | 94 |
| Adipoq | IDT | GCACTGGCAAGTTCTACTGCAA | GTAGGTGAAGAGAACGGCCTTGT | |
| Leptin | IDT | CCTATTGATGGGTCTGCCCA | TGAGCGCTACCTGCATAGAC | |
| Ucp1 | IDT | CACGGGGACCTACAATGCTT | ACAGTAAATGGCAGGGGACG | |
| Adrb3 | IDT | AGGACCTGACCCTGTCATCC | TCTAAGCCTTTCATGCCCACA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shay, D.; Welly, R.; Mao, J.; Kinkade, J.; Brown, J.K.; Rosenfeld, C.S.; Vieira-Potter, V.J. Knockdown of Esr1 from DRD1-Rich Brain Regions Affects Adipose Tissue Metabolism: Potential Crosstalk between Nucleus Accumbens and Adipose Tissue. Int. J. Mol. Sci. 2024, 25, 6130. https://doi.org/10.3390/ijms25116130
Shay D, Welly R, Mao J, Kinkade J, Brown JK, Rosenfeld CS, Vieira-Potter VJ. Knockdown of Esr1 from DRD1-Rich Brain Regions Affects Adipose Tissue Metabolism: Potential Crosstalk between Nucleus Accumbens and Adipose Tissue. International Journal of Molecular Sciences. 2024; 25(11):6130. https://doi.org/10.3390/ijms25116130
Chicago/Turabian StyleShay, Dusti, Rebecca Welly, Jiude Mao, Jessica Kinkade, Joshua K. Brown, Cheryl S. Rosenfeld, and Victoria J. Vieira-Potter. 2024. "Knockdown of Esr1 from DRD1-Rich Brain Regions Affects Adipose Tissue Metabolism: Potential Crosstalk between Nucleus Accumbens and Adipose Tissue" International Journal of Molecular Sciences 25, no. 11: 6130. https://doi.org/10.3390/ijms25116130





