Post-Translational Modifications of Proteins of Malaria Parasites during the Life Cycle
Abstract
1. Introduction
2. Protein PTMs in Plasmodium Parasite Growth and Development
2.1. Sporozoites in the Host
2.2. Liver Forms (from Sporozoites to Exoerythrocytic Forms, EEFs)
2.3. Asexual Forms in the Host (Erythrocytic Stage)
2.3.1. Studies of Multiply PTMs
2.3.2. Phosphorylation and Dephosphorylation
2.3.3. Acetylation and Methylation
2.3.4. Protein Cleavage and Processing
2.3.5. Nitrosylation
2.3.6. Glycosylation
2.3.7. Glutathionylation
2.3.8. Lipidation
2.3.9. Lipoxidation
2.3.10. Autophagy, Ubiquitination
2.3.11. Biotinylation
2.3.12. Hemozoin-Related PTMs
2.4. Sexual Forms in the Host (Gametocytes)
2.5. Sexual Forms in the Vector and Sporozoite Formation
3. Antimalarials and Protein Modifications
3.1. Phosphorylation
3.2. Proteolysis
3.3. Acetylation, Methylation
3.4. Nitrosylation
3.5. Lipid Modifications
3.6. Alkylation, Glycosylation, Lipoxidation
3.7. Ubiquitination and Others
4. Discussion
5. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Malaria Report 2023, Updated 30 November 2023. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2023 (accessed on 23 April 2024).
- Poespoprodjo, J.R.; Douglas, N.M.; Ansong, D.; Kho, S.; Anstey, N.M. Malaria. Lancet 2023, 402, 2328–2345. [Google Scholar] [CrossRef] [PubMed]
- Malaria Report by CDC (The Centers for Disease Control and Prevention). Updated in 2024. Available online: https://www.cdc.gov/dpdx/malaria/index.html (accessed on 21 April 2024).
- Sato, S. Plasmodium—A brief introduction to the parasites causing human malaria and their basic biology. J. Physiol. Anthropol. 2021, 40, 1, Erratum in J. Physiol. Anthropol. 2021, 40, 3. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pattaradilokrat, S.; Wu, J.; Xu, F.; Su, X.Z. The origins, isolation, and biological characterization of rodent malaria parasites. Parasitol. Int. 2022, 91, 102636. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Srivastava, A.; Philip, N.; Hughes, K.R.; Georgiou, K.; MacRae, J.I.; Barrett, M.P.; Creek, D.J.; McConville, M.J.; Waters, A.P. Stage-Specific Changes in Plasmodium Metabolism Required for Differentiation and Adaptation to Different Host and Vector Environments. PLoS Pathog. 2016, 12, e1006094. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chung, D.W.; Ponts, N.; Cervantes, S.; Le Roch, K.G. Post-translational modifications in Plasmodium: More than you think! Mol. Biochem. Parasitol. 2009, 168, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Doerig, C.; Rayner, J.C.; Scherf, A.; Tobin, A.B. Post-translational protein modifications in malaria parasites. Nat. Rev. Microbiol. 2015, 13, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Rashidi, S.; Tuteja, R.; Mansouri, R.; Ali-Hassanzadeh, M.; Shafiei, R.; Ghani, E.; Karimazar, M.; Nguewa, P.; Manzano-Román, R. The main post-translational modifications and related regulatory pathways in the malaria parasite Plasmodium falciparum: An update. J. Proteom. 2021, 245, 104279. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.M.; McRobert, L.; Grainger, M.; Sicard, A.; Dluzewski, A.R.; Hopp, C.S.; Holder, A.A.; Baker, D.A. The malaria parasite cyclic GMP-dependent protein kinase plays a central role in blood-stage schizogony. Eukaryot. Cell 2010, 9, 37–45. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lasonder, E.; Green, J.L.; Camarda, G.; Talabani, H.; Holder, A.A.; Langsley, G.; Alano, P. The Plasmodium falciparum schizont phosphoproteome reveals extensive phosphatidylinositol and cAMP-protein kinase A signaling. J. Proteome Res. 2012, 11, 5323–5337. [Google Scholar] [CrossRef] [PubMed]
- Guttery, D.S.; Poulin, B.; Ramaprasad, A.; Wall, R.J.; Ferguson, D.J.; Brady, D.; Patzewitz, E.M.; Whipple, S.; Straschil, U.; Wright, M.H.; et al. Genome-wide functional analysis of Plasmodium protein phosphatases reveals key regulators of parasite development and differentiation. Cell Host Microbe 2014, 16, 128–140. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ong, H.W.; Adderley, J.; Tobin, A.B.; Drewry, D.H.; Doerig, C. Parasite and host kinases as targets for antimalarials. Expert Opin. Ther. Targets 2023, 27, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Borba, J.V.V.B.; Silva, A.C.E.; do Nascimento, M.N.; Ferreira, L.T.; Rimoldi, A.; Starling, L.; Ramos, P.I.P.; Costa, F.T.M.; Andrade, C.H. Update and elucidation of Plasmodium kinomes: Prioritization of kinases as potential drug targets for malaria. Comput. Struct. Biotechnol. J. 2022, 20, 3708–3717. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Doerig, C.; Tobin, A.B. Parasite protein kinases: At home and abroad. Cell Host Microbe 2010, 8, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Ripp, J.; Smyrnakou, X.; Neuhoff, M.T.; Hentzschel, F.; Frischknecht, F. Phosphorylation of myosin A regulates gliding motility and is essential for Plasmodium transmission. EMBO Rep. 2022, 23, e54857. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Le Roch, K.G.; Zhou, Y.; Blair, P.L.; Grainger, M.; Moch, J.K.; Haynes, J.D.; De La Vega, P.; Holder, A.A.; Batalov, S.; Carucci, D.J.; et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 2003, 301, 1503–1508. [Google Scholar] [CrossRef] [PubMed]
- Coppi, A.; Tewari, R.; Bishop, J.R.; Bennett, B.L.; Lawrence, R.; Esko, J.D.; Billker, O.; Sinnis, P. Heparan sulfate proteoglycans provide a signal to Plasmodium sporozoites to stop migrating and productively invade host cells. Cell Host Microbe 2007, 2, 316–327. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, P.B.; Ding, S.; Zanghì, G.; Soulard, V.; DiMaggio, P.A.; Fuchter, M.J.; Mecheri, S.; Mazier, D.; Scherf, A.; Malmquist, N.A. Plasmodium falciparum PfSET7: Enzymatic characterization and cellular localization of a novel protein methyltransferase in sporozoite, liver and erythrocytic stage parasites. Sci. Rep. 2016, 6, 21802. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Doud, M.B.; Koksal, A.C.; Mi, L.Z.; Song, G.; Lu, C.; Springer, T.A. Unexpected fold in the circumsporozoite protein target of malaria vaccines. Proc. Natl. Acad. Sci. USA 2012, 109, 7817–7822. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Swearingen, K.E.; Lindner, S.E.; Flannery, E.L.; Vaughan, A.M.; Morrison, R.D.; Patrapuvich, R.; Koepfli, C.; Muller, I.; Jex, A.; Moritz, R.L.; et al. Proteogenomic analysis of the total and surface-exposed proteomes of Plasmodium vivax salivary gland sporozoites. PLoS Negl. Trop. Dis. 2017, 11, e0005791. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Swearingen, K.E.; Lindner, S.E.; Shi, L.; Shears, M.J.; Harupa, A.; Hopp, C.S.; Vaughan, A.M.; Springer, T.A.; Moritz, R.L.; Kappe, S.H.; et al. Interrogating the Plasmodium Sporozoite Surface: Identification of Surface-Exposed Proteins and Demonstration of Glycosylation on CSP and TRAP by Mass Spectrometry-Based Proteomics. PLoS Pathog. 2016, 12, e1005606. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lopaticki, S.; Yang, A.S.P.; John, A.; Scott, N.E.; Lingford, J.P.; O’Neill, M.T.; Erickson, S.M.; McKenzie, N.C.; Jennison, C.; Whitehead, L.W.; et al. Protein O-fucosylation in Plasmodium falciparum ensures efficient infection of mosquito and vertebrate hosts. Nat. Commun. 2017, 8, 561. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sanz, S.; Aquilini, E.; Tweedell, R.E.; Verma, G.; Hamerly, T.; Hritzo, B.; Tripathi, A.; Machado, M.; Churcher, T.S.; Rodrigues, J.A.; et al. Protein O-Fucosyltransferase 2 Is Not Essential for Plasmodium berghei Development. Front. Cell. Infect. Microbiol. 2019, 9, 238. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, M.; Fennell, C.; Ranford-Cartwright, L.; Sakthivel, R.; Gueirard, P.; Meister, S.; Caspi, A.; Doerig, C.; Nussenzweig, R.S.; Tuteja, R.; et al. The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J. Exp. Med. 2010, 207, 1465–1474. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, M.; Mishra, S.; Sakthivel, R.; Fontoura, B.M.; Nussenzweig, V. UIS2: A Unique Phosphatase Required for the Development of Plasmodium Liver Stages. PLoS Pathog. 2016, 12, e1005370. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Turque, O.; Tsao, T.; Li, T.; Zhang, M. Translational Repression in Malaria Sporozoites. Microb. Cell 2016, 3, 227–229. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fennell, C.; Babbitt, S.; Russo, I.; Wilkes, J.; Ranford-Cartwright, L.; Goldberg, D.E.; Doerig, C. PfeIK1, a eukaryotic initiation factor 2alpha kinase of the human malaria parasite Plasmodium falciparum, regulates stress-response to amino-acid starvation. Malar. J. 2009, 8, 99. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Aly, A.S.; Matuschewski, K. A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J. Exp. Med. 2005, 202, 225–230. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Silvie, O.; Franetich, J.F.; Rénia, L.; Mazier, D. Malaria sporozoite: Migrating for a living. Trends Mol. Med. 2004, 10, 97–100; Discussion 100-1. [Google Scholar] [CrossRef] [PubMed]
- Uddin, N.; Hoessli, D.C.; Butt, A.; Kaleem, A.; Iqbal, Z.; Afzal, I.; Muhammad, H.; Zamani, Z.; Shakoori, A.R. O-GlcNAc modification of the anti-malarial vaccine candidate PfAMA1: In silico-defined structural changes and potential to generate a better vaccine. Mol. Biol. Rep. 2012, 39, 4663–4672. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Noda, N. Autophagy-regulating protease Atg4: Structure, function, regulation and inhibition. J. Antibiot. 2018, 71, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Varshney, A.; Mishra, S. Regulation of Atg8 membrane deconjugation by cysteine proteases in the malaria parasite Plasmodium berghei. Cell. Mol. Life Sci. 2023, 80, 344. [Google Scholar] [CrossRef] [PubMed]
- Dalhuisen, T.; Plenderleith, L.J.; Ursani, I.; Philip, N.; Hahn, B.H.; Sharp, P.M. Unusually Divergent Ubiquitin Genes and Proteins in Plasmodium Species. Genome Biol. Evol. 2023, 15, evad137. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Imhoff, R.D.; Rosenthal, M.R.; Ashraf, K.; Bhanot, P.; Ng, C.L.; Flaherty, D.P. Identification of covalent fragment inhibitors for Plasmodium falciparum UCHL3 with anti-malarial efficacy. Bioorg. Med. Chem. Lett. 2023, 94, 129458. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hopp, C.S.; Balaban, A.E.; Bushell, E.S.; Billker, O.; Rayner, J.C.; Sinnis, P. Palmitoyl transferases have critical roles in the development of mosquito and liver stages of Plasmodium. Cell. Microbiol. 2016, 18, 1625–1641. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ghartey-Kwansah, G.; Yin, Q.; Li, Z.; Gumpper, K.; Sun, Y.; Yang, R.; Wang, D.; Jones, O.; Zhou, X.; Wang, L.; et al. Calcium-dependent Protein Kinases in Malaria Parasite Development and Infection. Cell Transplant. 2020, 29, 963689719884888. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tewari, R.; Straschil, U.; Bateman, A.; Böhme, U.; Cherevach, I.; Gong, P.; Pain, A.; Billker, O. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe 2010, 8, 377–387. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sebastian, S.; Brochet, M.; Collins, M.O.; Schwach, F.; Jones, M.L.; Goulding, D.; Rayner, J.C.; Choudhary, J.S.; Billker, O. A Plasmodium calcium-dependent protein kinase controls zygote development and transmission by translationally activating repressed mRNAs. Cell Host Microbe 2012, 12, 9–19. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Coppi, A.; Pinzon-Ortiz, C.; Hutter, C.; Sinnis, P. The Plasmodium circumsporozoite protein is proteolytically processed during cell invasion. J. Exp. Med. 2005, 201, 27–33. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Geens, R.; Stanisich, J.; Beyens, O.; D’Hondt, S.; Thiberge, J.M.; Ryckebosch, A.; De Groot, A.; Magez, S.; Vertommen, D.; Amino, R.; et al. Biophysical characterization of the Plasmodium falciparum circumsporozoite protein’s N-terminal domain. Protein Sci. 2024, 33, e4852. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Voss, C.; Ehrenman, K.; Mlambo, G.; Mishra, S.; Kumar, K.A.; Sacci, J.B., Jr.; Sinnis, P.; Coppens, I. Overexpression of Plasmodium berghei ATG8 by Liver Forms Leads to Cumulative Defects in Organelle Dynamics and to Generation of Noninfectious Merozoites. mBio 2016, 7, e00682-16. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Goerdeler, F.; Seeberger, P.H.; Moscovitz, O. Unveiling the Sugary Secrets of Plasmodium Parasites. Front. Microbiol. 2021, 12, 712538. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Srivastava, P.N.; Paul, P.; Mishra, S. Protein O-Fucosyltransferase Is Required for the Efficient Invasion of Hepatocytes by Plasmodium berghei Sporozoites. ACS Infect. Dis. 2024; epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Govindasamy, K.; Khan, R.; Snyder, M.; Lou, H.J.; Du, P.; Kudyba, H.M.; Muralidharan, V.; Turk, B.E.; Bhanot, P. Plasmodium falciparum Cyclic GMP-Dependent Protein Kinase Interacts with a Subunit of the Parasite Proteasome. Infect. Immunol. 2018, 87, e00523-18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Raphemot, R.; Eubanks, A.L.; Toro-Moreno, M.; Geiger, R.A.; Hughes, P.F.; Lu, K.Y.; Haystead, T.A.J.; Derbyshire, E.R. Plasmodium PK9 Inhibitors Promote Growth of Liver-Stage Parasites. Cell. Chem. Biol. 2019, 26, 411–419.e7. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jaijyan, D.K.; Verma, P.K.; Singh, A.P. A novel FIKK kinase regulates the development of mosquito and liver stages of the malaria. Sci. Rep. 2016, 6, 39285. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kaushansky, A.; Douglass, A.N.; Arang, N.; Vigdorovich, V.; Dambrauskas, N.; Kain, H.S.; Austin, L.S.; Sather, D.N.; Kappe, S.H. Malaria parasites target the hepatocyte receptor EphA2 for successful host infection. Science 2015, 350, 1089–1092. [Google Scholar] [CrossRef] [PubMed]
- Akhouri, R.R.; Sharma, A.; Malhotra, P.; Sharma, A. Role of Plasmodium falciparum thrombospondin-related anonymous protein in host-cell interactions. Malar. J. 2008, 7, 63. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tawk, L.; Lacroix, C.; Gueirard, P.; Kent, R.; Gorgette, O.; Thiberge, S.; Mercereau-Puijalon, O.; Ménard, R.; Barale, J.C. A key role for Plasmodium subtilisin-like SUB1 protease in egress of malaria parasites from host hepatocytes. J. Biol. Chem. 2013, 288, 33336–33346. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dellibovi-Ragheb, T.A.; Jhun, H.; Goodman, C.D.; Walters, M.S.; Ragheb, D.R.T.; Matthews, K.A.; Rajaram, K.; Mishra, S.; McFadden, G.I.; Sinnis, P.; et al. Host biotin is required for liver stage development in malaria parasites. Proc. Natl. Acad. Sci. USA 2018, 115, E2604–E2613. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mikolajczak, S.A.; Sacci, J.B., Jr.; De La Vega, P.; Camargo, N.; VanBuskirk, K.; Krzych, U.; Cao, J.; Jacobs-Lorena, M.; Cowman, A.F.; Kappe, S.H. Disruption of the Plasmodium falciparum liver-stage antigen-1 locus causes a differentiation defect in late liver-stage parasites. Cell. Microbiol. 2011, 13, 1250–1260. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nicoll, W.S.; Sacci, J.B.; Rodolfo, C.; Di Giacomo, G.; Piacentini, M.; Holland, Z.J.; Doerig, C.; Hollingdale, M.R.; Lanar, D.E. Plasmodium falciparum liver stage antigen-1 is cross-linked by tissue transglutaminase. Malar. J. 2011, 10, 14. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Koussis, K.; Goulielmaki, E.; Chalari, A.; Withers-Martinez, C.; Siden-Kiamos, I.; Matuschewski, K.; Loukeris, T.G. Targeted Deletion of a Plasmodium Site-2 Protease Impairs Life Cycle Progression in the Mammalian Host. PLoS ONE 2017, 12, e0170260. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sharma, P.K.; Kalia, I.; Kaushik, V.; Brünnert, D.; Quadiri, A.; Kashif, M.; Chahar, K.R.; Agrawal, A.; Singh, A.P.; Goyal, P. STK35L1 regulates host cell cycle-related genes and is essential for Plasmodium infection during the liver stage of malaria. Exp. Cell Res. 2021, 406, 112764. [Google Scholar] [CrossRef] [PubMed]
- Kluck, G.E.G.; Wendt, C.H.C.; Imperio, G.E.D.; Araujo, M.F.C.; Atella, T.C.; da Rocha, I.; Miranda, K.R.; Atella, G.C. Plasmodium Infection Induces Dyslipidemia and a Hepatic Lipogenic State in the Host through the Inhibition of the AMPK-ACC Pathway. Sci. Rep. 2019, 9, 14695. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Prado, M.; Eickel, N.; De Niz, M.; Heitmann, A.; Agop-Nersesian, C.; Wacker, R.; Schmuckli-Maurer, J.; Caldelari, R.; Janse, C.J.; Khan, S.M.; et al. Long-term live imaging reveals cytosolic immune responses of host hepatocytes against Plasmodium infection and parasite escape mechanisms. Autophagy 2015, 11, 1561–1579. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zheng, H.; Lu, X.; Li, K.; Zhu, F.; Zhao, C.; Liu, T.; Ding, Y.; Fu, Y.; Zhang, K.; Zhou, T.; et al. ATG Ubiquitination Is Required for Circumsporozoite Protein to Subvert Host Innate Immunity Against Rodent Malaria Liver Stage. Front. Immunol. 2022, 13, 815936. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Agop-Nersesian, C.; De Niz, M.; Niklaus, L.; Prado, M.; Eickel, N.; Heussler, V.T. Shedding of host autophagic proteins from the parasitophorous vacuolar membrane of Plasmodium berghei. Sci. Rep. 2017, 7, 2191. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Maruthi, M.; Singh, D.; Reddy, S.R.; Mastan, B.S.; Mishra, S.; Kumar, K.A. Modulation of host cell SUMOylation facilitates efficient development of Plasmodium berghei and Toxoplasma gondii. Cell. Microbiol. 2017, 19, 7. [Google Scholar] [CrossRef] [PubMed]
- Martín Moyano, P.; Němec, V.; Paruch, K. Cdc-Like Kinases (CLKs): Biology, Chemical Probes, and Therapeutic Potential. Int. J. Mol. Sci. 2020, 21, 7549. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gray, K.A.; Gresty, K.J.; Chen, N.; Zhang, V.; Gutteridge, C.E.; Peatey, C.L.; Chavchich, M.; Waters, N.C.; Cheng, Q. Correlation between Cyclin Dependent Kinases and Artemisinin-Induced Dormancy in Plasmodium falciparum In Vitro. PLoS ONE 2016, 11, e0157906. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Arisue, N.; Palacpac, N.M.Q.; Tougan, T.; Horii, T. Characteristic features of the SERA multigene family in the malaria parasite. Parasit. Vectors 2020, 13, 170. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, J.; Jiang, N.; Sang, X.; Yang, N.; Feng, Y.; Chen, R.; Wang, X.; Chen, Q. Protein Modification Characteristics of the Malaria Parasite Plasmodium falciparum and the Infected Erythrocytes. Mol. Cell. Proteom. 2021, 20, 100001. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Painter, H.J.; Chung, N.C.; Sebastian, A.; Albert, I.; Storey, J.D.; Llinás, M. Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development. Nat. Commun. 2018, 9, 2656, Erratum in: Nat. Commun. 2022, 13, 1497. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wilkes, J.M.; Doerig, C. The protein-phosphatome of the human malaria parasite Plasmodium falciparum. BMC Genom. 2008, 9, 412. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wilde, M.L.; Triglia, T.; Marapana, D.; Thompson, J.K.; Kouzmitchev, A.A.; Bullen, H.E.; Gilson, P.R.; Cowman, A.F.; Tonkin, C.J. Protein Kinase A Is Essential for Invasion of Plasmodium falciparum into Human Erythrocytes. mBio 2019, 10, e01972-19. [Google Scholar] [CrossRef] [PubMed]
- Merckx, A.; Nivez, M.P.; Bouyer, G.; Alano, P.; Langsley, G.; Deitsch, K.; Thomas, S.; Doerig, C.; Egée, S. Plasmodium falciparum regulatory subunit of cAMP-dependent PKA and anion channel conductance. PLoS Pathog. 2008, 4, e19. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wurtz, N.; Chapus, C.; Desplans, J.; Parzy, D. cAMP-dependent protein kinase from Plasmodium falciparum: An update. Parasitology 2011, 138, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Dorin, D.; Semblat, J.P.; Poullet, P.; Alano, P.; Goldring, J.P.; Whittle, C.; Patterson, S.; Chakrabarti, D.; Doerig, C. PfPK7, an atypical MEK-related protein kinase, reflects the absence of classical three-component MAPK pathways in the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 2005, 55, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Koyama, F.C.; Ribeiro, R.Y.; Garcia, J.L.; Azevedo, M.F.; Chakrabarti, D.; Garcia, C.R. Ubiquitin proteasome system and the atypical kinase PfPK7 are involved in melatonin signaling in Plasmodium falciparum. J. Pineal. Res. 2012, 53, 147–153. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pease, B.N.; Huttlin, E.L.; Jedrychowski, M.P.; Dorin-Semblat, D.; Sebastiani, D.; Segarra, D.T.; Roberts, B.F.; Chakrabarti, R.; Doerig, C.; Gygi, S.P.; et al. Characterization of Plasmodium falciparum Atypical Kinase PfPK7- Dependent Phosphoproteome. J. Proteome Res. 2018, 17, 2112–2123. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, A.S.; Agarwal, M.; Mehra, P.; Gupta, A.; Gupta, N.; Doerig, C.D.; Dhar, S.K. Regulation of Plasmodium falciparum Origin Recognition Complex subunit 1 (PfORC1) function through phosphorylation mediated by CDK-like kinase PK5. Mol. Microbiol. 2015, 98, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Mishra, S.; Sakthivel, R.; Rojas, M.; Ranjan, R.; Sullivan, W.J., Jr.; Fontoura, B.M.; Ménard, R.; Dever, T.E.; Nussenzweig, V. PK4, a eukaryotic initiation factor 2α(eIF2α) kinase, is essential for the development of the erythrocytic cycle of Plasmodium. Proc. Natl. Acad. Sci. USA 2012, 109, 3956–3961. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wernimont, A.K.; Artz, J.D.; Finerty, P., Jr.; Lin, Y.H.; Amani, M.; Allali-Hassani, A.; Senisterra, G.; Vedadi, M.; Tempel, W.; Mackenzie, F.; et al. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat. Struct. Mol. Biol. 2010, 17, 596–601. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hopp, C.S.; Bowyer, P.W.; Baker, D.A. The role of cGMP signalling in regulating life cycle progression of Plasmodium. Microbes Infect. 2012, 14, 831–837. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Baker, D.A.; Stewart, L.B.; Large, J.M.; Bowyer, P.W.; Ansell, K.H.; Jiménez-Díaz, M.B.; El Bakkouri, M.; Birchall, K.; Dechering, K.J.; Bouloc, N.S.; et al. A potent series targeting the malarial cGMP-dependent protein kinase clears infection and blocks transmission. Nat. Commun. 2017, 8, 430. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Droucheau, E.; Primot, A.; Thomas, V.; Mattei, D.; Knockaert, M.; Richardson, C.; Sallicandro, P.; Alano, P.; Jafarshad, A.; Baratte, B.; et al. Plasmodium falciparum glycogen synthase kinase-3: Molecular model, expression, intracellular localisation and selective inhibitors. Biochim. Biophys. Acta 2004, 1697, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Masch, A.; Nasereddin, A.; Alder, A.; Bird, M.J.; Schweda, S.I.; Preu, L.; Doerig, C.; Dzikowski, R.; Gilberger, T.W.; Kunick, C. Structure-activity relationships in a series of antiplasmodial thieno [2,3-b]pyridines. Malar. J. 2019, 18, 89. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Moolman, C.; van der Sluis, R.; Beteck, R.M.; Legoabe, L.J. Exploration of benzofuran-based compounds as potent and selective Plasmodium falciparum glycogen synthase kinase-3 (PfGSK-3) inhibitors. Bioorg. Chem. 2021, 112, 104839. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.C.; Ahmed, A.; Gilberger, T.W.; Sharma, P. Regulation of Plasmodium falciparum glideosome associated protein 45 (PfGAP45) phosphorylation. PLoS ONE 2012, 7, e35855. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Green, J.L.; Rees-Channer, R.R.; Howell, S.A.; Martin, S.R.; Knuepfer, E.; Taylor, H.M.; Grainger, M.; Holder, A.A. The motor complex of Plasmodium falciparum: Phosphorylation by a calcium-dependent protein kinase. J. Biol. Chem. 2008, 283, 30980–30989. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hitz, E.; Wiedemar, N.; Passecker, A.; Graça, B.A.S.; Scheurer, C.; Wittlin, S.; Brancucci, N.M.B.; Vakonakis, I.; Mäser, P.; Voss, T.S. The 3-phosphoinositide-dependent protein kinase 1 is an essential upstream activator of protein kinase A in malaria parasites. PLoS Biol. 2021, 19, e3001483. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chandra, B.R.; Olivieri, A.; Silvestrini, F.; Alano, P.; Sharma, A. Biochemical characterization of the two nucleosome assembly proteins from Plasmodium falciparum. Mol. Biochem. Parasitol. 2005, 142, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Barik, S.; Taylor, R.E.; Chakrabarti, D. Identification, cloning, and mutational analysis of the casein kinase 1 cDNA of the malaria parasite, Plasmodium falciparum. Stage-specific expression of the gene. J. Biol. Chem. 1997, 272, 26132–26138. [Google Scholar] [CrossRef] [PubMed]
- Dastidar, E.G.; Dayer, G.; Holland, Z.M.; Dorin-Semblat, D.; Claes, A.; Chêne, A.; Sharma, A.; Hamelin, R.; Moniatte, M.; Lopez-Rubio, J.J.; et al. Involvement of Plasmodium falciparum protein kinase CK2 in the chromatin assembly pathway. BMC Biol. 2012, 10, 5. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dehury, B.; Behera, S.K.; Mahapatra, N. Structural dynamics of Casein Kinase I (CKI) from malarial parasite Plasmodium falciparum (Isolate 3D7): Insights from theoretical modelling and molecular simulations. J. Mol. Graph. Model. 2017, 71, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Hitz, E.; Grüninger, O.; Passecker, A.; Wyss, M.; Scheurer, C.; Wittlin, S.; Beck, H.P.; Brancucci, N.M.B.; Voss, T.S. The catalytic subunit of Plasmodium falciparum casein kinase 2 is essential for gametocytogenesis. Commun. Biol. 2021, 4, 336. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Treeck, M.; Sanders, J.L.; Elias, J.E.; Boothroyd, J.C. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe 2011, 10, 410–419. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Watzlowik, M.T.; Das, S.; Meissner, M.; Längst, G. Peculiarities of Plasmodium falciparum Gene Regulation and Chromatin Structure. Int. J. Mol. Sci. 2021, 22, 5168. [Google Scholar] [CrossRef] [PubMed]
- Hora, R.; Bridges, D.J.; Craig, A.; Sharma, A. Erythrocytic casein kinase II regulates cytoadherence of Plasmodium falciparum-infected red blood cells. J. Biol. Chem. 2009, 284, 6260–6269. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Doerig, C.; Endicott, J.; Chakrabarti, D. Cyclin-dependent kinase homologues of Plasmodium falciparum. Int. J. Parasitol. 2002, 32, 1575–1585. [Google Scholar] [CrossRef] [PubMed]
- Bracchi-Ricard, V.; Barik, S.; Delvecchio, C.; Doerig, C.; Chakrabarti, R.; Chakrabarti, D. PfPK6, a novel cyclin-dependent kinase/mitogen-activated protein kinase-related protein kinase from Plasmodium falciparum. Biochem. J. 2000, 347 Pt 1, 255–263. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Le Roch, K.; Sestier, C.; Dorin, D.; Waters, N.; Kappes, B.; Chakrabarti, D.; Meijer, L.; Doerig, C. Activation of a Plasmodium falciparum cdc2-related kinase by heterologous p25 and cyclin H. Functional characterization of a P. falciparum cyclin homologue. J. Biol. Chem. 2000, 275, 8952–8958. [Google Scholar] [CrossRef] [PubMed]
- Merckx, A.; Le Roch, K.; Nivez, M.P.; Dorin, D.; Alano, P.; Gutierrez, G.J.; Nebreda, A.R.; Goldring, D.; Whittle, C.; Patterson, S.; et al. Identification and initial characterization of three novel cyclin-related proteins of the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 2003, 278, 39839–39850. [Google Scholar] [CrossRef] [PubMed]
- Machado, M.; Klaus, S.; Klaschka, D.; Guizetti, J.; Ganter, M. Plasmodium falciparum CRK4 links early mitotic events to the onset of S-phase during schizogony. mBio 2023, 14, e0077923. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Geyer, J.A.; Keenan, S.M.; Woodard, C.L.; Thompson, P.A.; Gerena, L.; Nichols, D.A.; Gutteridge, C.E.; Waters, N.C. Selective inhibition of Pfmrk, a Plasmodium falciparum CDK.; by antimalarial 1,3-diaryl-2-propenones. Bioorg. Med. Chem. Lett. 2009, 19, 1982–1985. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, T.G.; Doerig, C.; Reininger, L. Nima- and Aurora-related kinases of malaria parasites. Biochim. Biophys. Acta 2013, 1834, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Dorin-Semblat, D.; Quashie, N.; Halbert, J.; Sicard, A.; Doerig, C.; Peat, E.; Ranford-Cartwright, L.; Doerig, C. Functional characterization of both MAP kinases of the human malaria parasite Plasmodium falciparum by reverse genetics. Mol. Microbiol. 2007, 65, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Rangarajan, R.; Bei, A.K.; Jethwaney, D.; Maldonado, P.; Dorin, D.; Sultan, A.A.; Doerig, C. A mitogen-activated protein kinase regulates male gametogenesis and transmission of the malaria parasite Plasmodium berghei. EMBO Rep. 2005, 6, 464–469. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hitz, E.; Balestra, A.C.; Brochet, M.; Voss, T.S. PfMAP-2 is essential for male gametogenesis in the malaria parasite Plasmodium falciparum. Sci. Rep. 2020, 10, 11930. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Siddiqui, G.; Proellochs, N.I.; Cooke, B.M. Identification of essential exported Plasmodium falciparum protein kinases in malaria-infected red blood cells. Br. J. Haematol. 2020, 188, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Anil Kumar, D.; Shrivastava, D.; Sahasrabuddhe, A.A.; Habib, S.; Trivedi, V. Plasmodium falciparum FIKK9.1 is a monomeric serine-threonine protein kinase with features to exploit as a drug target. Chem. Biol. Drug Des. 2021, 97, 962–977. [Google Scholar] [CrossRef] [PubMed]
- Ekka, R.; Gupta, A.; Bhatnagar, S.; Malhotra, P.; Sharma, P. Phosphorylation of Rhoptry Protein RhopH3 Is Critical for Host Cell Invasion by the Malaria Parasite. mBio 2020, 11, e00166-20. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dixit, A.; Singh, P.K.; Sharma, G.P.; Malhotra, P.; Sharma, P. PfSRPK1, a novel splicing-related kinase from Plasmodium falciparum. J. Biol. Chem. 2010, 285, 38315–38323. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pandey, R.; Mohmmed, A.; Pierrot, C.; Khalife, J.; Malhotra, P.; Gupta, D. Genome wide in silico analysis of Plasmodium falciparum phosphatome. BMC Genom. 2014, 15, 1024. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumar, R.; Adams, B.; Oldenburg, A.; Musiyenko, A.; Barik, S. Characterisation and expression of a PP1 serine/threonine protein phosphatase (PfPP1) from the malaria parasite, Plasmodium falciparum: Demonstration of its essential role using RNA interference. Malar. J. 2002, 1, 5. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ali, F.; Wali, H.; Jan, S.; Zia, A.; Aslam, M.; Ahmad, I.; Afridi, S.G.; Shams, S.; Khan, A. Analysing the essential proteins set of Plasmodium falciparum PF3D7 for novel drug targets identification against malaria. Malar. J. 2021, 20, 335. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Philip, N.; Waters, A.P. Conditional Degradation of Plasmodium Calcineurin Reveals Functions in Parasite Colonization of both Host and Vector. Cell Host Microbe 2015, 18, 122–131. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumar, R.; Musiyenko, A.; Barik, S. Plasmodium falciparum calcineurin and its association with heat shock protein 90: Mechanisms for the antimalarial activity of cyclosporin A and synergism with geldanamycin. Mol. Biochem. Parasitol. 2005, 141, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, F.; Jalallou, N.; Ebrahimi, M. Analysis of Apical Membrane Antigen (AMA)-1 characteristics using bioinformatics tools in order to vaccine design against Plasmodium vivax. Infect. Genet. Evol. 2019, 71, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Lawrence, M.; Jeffers, V.; Zhao, F.; Parker, D.; Ge, Y.; Sullivan, W.J., Jr.; Cui, L. Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during Plasmodium falciparum intraerythrocytic development. Mol. Microbiol. 2013, 89, 660–675. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Trelle, M.B.; Salcedo-Amaya, A.M.; Cohen, A.M.; Stunnenberg, H.G.; Jensen, O.N. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J. Proteome Res. 2009, 8, 3439–3450. [Google Scholar] [CrossRef] [PubMed]
- Connacher, J.; von Grüning, H.; Birkholtz, L. Histone Modification Landscapes as a Roadmap for Malaria Parasite Development. Front. Cell Dev. Biol. 2022, 10, 848797. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Coleman, B.I.; Skillman, K.M.; Jiang, R.H.Y.; Childs, L.M.; Altenhofen, L.M.; Ganter, M.; Leung, Y.; Goldowitz, I.; Kafsack, B.F.C.; Marti, M.; et al. A Plasmodium falciparum histone deacetylase regulates antigenic variation and gametocyte conversion. Cell Host Microbe 2014, 16, 177–186. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mancio-Silva, L.; Lopez-Rubio, J.J.; Claes, A.; Scherf, A. Sir2a regulates rDNA transcription and multiplication rate in the human malaria parasite Plasmodium falciparum. Nat. Commun. 2013, 4, 1530. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kanyal, A.; Rawat, M.; Gurung, P.; Choubey, D.; Anamika, K.; Karmodiya, K. Genome-wide survey and phylogenetic analysis of histone acetyltransferases and histone deacetylases of Plasmodium falciparum. FEBS J. 2018, 285, 1767–1782. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rubio, J.J.; Mancio-Silva, L.; Scherf, A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 2009, 5, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Wang, C.; Lucky, A.B.; Liang, X.; Min, H.; Adapa, S.R.; Jiang, R.; Kim, K.; Cui, L. A unique GCN5 histone acetyltransferase complex controls erythrocyte invasion and virulence in the malaria parasite Plasmodium falciparum. PLoS Pathog. 2021, 17, e1009351. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shekhar, S.; Bhowmick, K.; Dhar, S.K. Role of PfMYST in DNA replication in Plasmodium falciparum. Exp. Parasitol. 2022, 242, 108396. [Google Scholar] [CrossRef] [PubMed]
- Zeeshan, M.; Kaur, I.; Joy, J.; Saini, E.; Paul, G.; Kaushik, A.; Dabral, S.; Mohmmed, A.; Gupta, D.; Malhotra, P. Proteomic Identification and Analysis of Arginine-Methylated Proteins of Plasmodium falciparum at Asexual Blood Stages. J. Proteome Res. 2017, 16, 368–383. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Miao, J.; Cui, L.; Cui, L. Characterization of PRMT1 from Plasmodium falciparum. Biochem. J. 2009, 421, 107–118. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Volz, J.C.; Bártfai, R.; Petter, M.; Langer, C.; Josling, G.A.; Tsuboi, T.; Schwach, F.; Baum, J.; Rayner, J.C.; Stunnenberg, H.G.; et al. PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe 2012, 11, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Mu, J.; Zhang, Q.; Ni, T.; Srinivasan, P.; Rayavara, K.; Yang, W.; Turner, L.; Lavstsen, T.; Theander, T.G.; et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 2013, 499, 223–227. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cao, X.; Wen, Y.; Li, Y.; Ma, X.; Jing, Q.; Jiang, L.; Wei, G. PfSET2 Is Involved in Genome Organization of Var Gene Family in Plasmodium falciparum. Microbiol. Spectr. 2023, 11, e0389122. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kaur, I.; Zeeshan, M.; Saini, E.; Kaushik, A.; Mohmmed, A.; Gupta, D.; Malhotra, P. Widespread occurrence of lysine methylation in Plasmodium falciparum proteins at asexual blood stages. Sci. Rep. 2016, 6, 35432. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hossain, M.; Sharma, S.; Korde, R.; Kanodia, S.; Chugh, M.; Rawat, K.; Malhotra, P. Organization of Plasmodium falciparum spliceosomal core complex and role of arginine methylation in its assembly. Malar. J. 2013, 12, 333. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cui, L.; Fan, Q.; Cui, L.; Miao, J. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int. J. Parasitol. 2008, 38, 1083–1097. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Matthews, K.A.; Senagbe, K.M.; Nötzel, C.; Gonzales, C.A.; Tong, X.; Rijo-Ferreira, F.; Bhanu, N.V.; Miguel-Blanco, C.; Lafuente-Monasterio, M.J.; Garcia, B.A.; et al. Disruption of the Plasmodium falciparum Life Cycle through Transcriptional Reprogramming by Inhibitors of Jumonji Demethylases. ACS Infect. Dis. 2020, 6, 1058–1075. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rosenthal, P.J. Cysteine proteases of malaria parasites. Int. J. Parasitol. 2004, 34, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, P.J. Falcipain cysteine proteases of malaria parasites: An update. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140362. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.C.; Dixit, R. Structure-function of falcipains: Malarial cysteine proteases. J. Trop. Med. 2012, 2012, 345195. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pei, Y.; Miller, J.L.; Lindner, S.E.; Vaughan, A.M.; Torii, M.; Kappe, S.H.I. Plasmodium yoelii inhibitor of cysteine proteases is exported to exomembrane structures and interacts with yoelipain-2 during asexual blood-stage development. Cell. Microbiol. 2013, 15, 1508–1526. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Patra, J.; Rana, D.; Arora, S.; Pal, M.; Mahindroo, N. Falcipains: Biochemistry, target validation and structure-activity relationship studies of inhibitors as antimalarials. Eur. J. Med. Chem. 2023, 252, 115299. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, C.; Tan, M.S.Y.; de Vries, L.E.; Russo, I.; Sanchez, M.I.; Goldberg, D.E.; Deu, E. Plasmodium falciparum dipeptidyl aminopeptidase 3 activity is important for efficient erythrocyte invasion by the malaria parasite. PLoS Pathog. 2018, 14, e1007031. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gomes, M.M.; Budu, A.; Ventura, P.D.; Bagnaresi, P.; Cotrin, S.S.; Cunha, R.L.; Carmona, A.K.; Juliano, L.; Gazarini, M.L. Specific calpain activity evaluation in Plasmodium parasites. Anal. Biochem. 2015, 468, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Nasamu, A.S.; Rubiano, K.C.; Goldberg, D.E. Activation of the Plasmodium Egress Effector Subtilisin-Like Protease 1 Is Mediated by Plasmepsin X Destruction of the Prodomain. mBio 2023, 14, e0067323. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lemberg, M.K.; Freeman, M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007, 17, 1634–1646. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vera, I.M.; Beatty, W.L.; Sinnis, P.; Kim, K. Plasmodium protease ROM1 is important for proper formation of the parasitophorous vacuole. PLoS Pathog. 2011, 7, e1002197. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Baker, R.P.; Wijetilaka, R.; Urban, S. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathog. 2006, 2, e113. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sleebs, B.E.; Lopaticki, S.; Marapana, D.S.; O’Neill, M.T.; Rajasekaran, P.; Gazdik, M.; Günther, S.; Whitehead, L.W.; Lowes, K.N.; Barfod, L.; et al. Inhibition of Plasmepsin V activity demonstrates its essential role in protein export, PfEMP1 display, and survival of malaria parasites. PLoS Biol. 2014, 12, e1001897. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nasamu, A.S.; Polino, A.J.; Istvan, E.S.; Goldberg, D.E. Malaria parasite plasmepsins: More than just plain old degradative pepsins. J. Biol. Chem. 2020, 295, 8425–8441. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bhaumik, P.; Xiao, H.; Parr, C.L.; Kiso, Y.; Gustchina, A.; Yada, R.Y.; Wlodawer, A. Crystal structures of the histo-aspartic protease (HAP) from Plasmodium falciparum. J. Mol. Biol. 2009, 388, 520–540. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Murata, C.E.; Goldberg, D.E. Plasmodium falciparum falcilysin: A metalloprotease with dual specificity. J. Biol. Chem. 2003, 278, 38022–38028. [Google Scholar] [CrossRef] [PubMed]
- Drinkwater, N.; Sivaraman, K.K.; Bamert, R.S.; Rut, W.; Mohamed, K.; Vinh, N.B.; Scammells, P.J.; Drag, M.; McGowan, S. Structure and substrate fingerprint of aminopeptidase P from Plasmodium falciparum. Biochem. J. 2016, 473, 3189–3204. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, H.; Oh, S.S.; Chishti, A.H. A Presenilin-like protease associated with Plasmodium falciparum micronemes is involved in erythrocyte invasion. Mol. Biochem. Parasitol. 2008, 158, 22–31. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lauterbach, S.B.; Coetzer, T.L. The M18 aspartyl aminopeptidase of Plasmodium falciparum binds to human erythrocyte spectrin in vitro. Malar. J. 2008, 7, 161. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Akman-Anderson, L.; Olivier, M.; Luckhart, S. Induction of nitric oxide synthase and activation of signaling proteins in Anopheles mosquitoes by the malaria pigment, hemozoin. Infect. Immunol. 2007, 75, 4012–4019. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumar, A.; Singh, K.P.; Bali, P.; Anwar, S.; Kaul, A.; Singh, O.P.; Gupta, B.K.; Kumari, N.; Noor Alam, M.; Raziuddin, M.; et al. iNOS polymorphism modulates iNOS/NO expression via impaired antioxidant and ROS content in P. vivax and P. falciparum infection. Redox Biol. 2018, 15, 192–206. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Torgler, R.; Bongfen, S.E.; Romero, J.C.; Tardivel, A.; Thome, M.; Corradin, G. Sporozoite-mediated hepatocyte wounding limits Plasmodium parasite development via MyD88-mediated NF-kappa B activation and inducible NO synthase expression. J. Immunol. 2008, 180, 3990–3999. [Google Scholar] [CrossRef] [PubMed]
- Kordes, M.; Ormond, L.; Rausch, S.; Matuschewski, K.; Hafalla, J.C.R. TLR9 signalling inhibits Plasmodium liver infection by macrophage activation. Eur. J. Immunol. 2022, 52, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Skorokhod, O.A.; Schwarzer, E.; Ceretto, M.; Arese, P. Malarial pigment haemozoin, IFN-gamma, TNF-alpha, IL-1beta and LPS do not stimulate expression of inducible nitric oxide synthase and production of nitric oxide in immuno-purified human monocytes. Malar. J. 2007, 6, 73. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, L.; Delahunty, C.; Prieto, J.H.; Rahlfs, S.; Jortzik, E.; Yates, J.R., 3rd; Becker, K. Protein S-nitrosylation in Plasmodium falciparum. Antioxid. Redox Signal. 2014, 20, 2923–2935. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kimura, E.A.; Couto, A.S.; Peres, V.J.; Casal, O.L.; Katzin, A.M. N-linked glycoproteins are related to schizogony of the intraerythrocytic stage in Plasmodium falciparum. J. Biol. Chem. 1996, 271, 14452–14461. [Google Scholar] [CrossRef] [PubMed]
- Gowda, D.C.; Miller, L.H. Glycosylation in malaria parasites: What do we know? Trends. Parasitol. 2024, 40, 131–146. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dieckmann-Schuppert, A.; Bause, E.; Schwarz, R.T. Glycosylation reactions in Plasmodium falciparum, Toxoplasma gondii, and Trypanosoma brucei brucei probed by the use of synthetic peptides. Biochim. Biophys. Acta 1994, 1199, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Bandini, G.; Albuquerque-Wendt, A.; Hegermann, J.; Samuelson, J.; Routier, F.H. Protein O- and C-Glycosylation pathways in Toxoplasma gondii and Plasmodium falciparum. Parasitology 2019, 146, 1755–1766. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tajik, S.; Sadeghi, S.; Iravani, A.; Khalili, M.; Arjmand, M.; Din, N.U.; Vahabi, F.; Feiz-Haddad, H.; Lame-Rad, B.; Naddaf, S.R.; et al. Characterization of Glycoproteins of Native 19kDa C-Terminal Merozoite Surface Protein-1 from Native Antigen of Plasmodium falciparum. J. Arthropod. Borne Dis. 2019, 13, 324–333. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kehr, S.; Jortzik, E.; Delahunty, C.; Yates, J.R., 3rd; Rahlfs, S.; Becker, K. Protein S-glutathionylation in malaria parasites. Antioxid. Redox Signal. 2011, 15, 2855–2865. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schlott, A.C.; Knuepfer, E.; Green, J.L.; Hobson, P.; Borg, A.J.; Morales-Sanfrutos, J.; Perrin, A.J.; Maclachlan, C.; Collinson, L.M.; Snijders, A.P.; et al. Inhibition of protein N-myristoylation blocks Plasmodium falciparum intraerythrocytic development, egress and invasion. PLoS Biol. 2021, 19, e3001408. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Counihan, N.A.; Chernih, H.C.; de Koning-Ward, T.F. Post-translational lipid modifications in Plasmodium parasites. Curr. Opin. Microbiol. 2022, 69, 102196. [Google Scholar] [CrossRef] [PubMed]
- Rees-Channer, R.R.; Martin, S.R.; Green, J.L.; Bowyer, P.W.; Grainger, M.; Molloy, J.E.; Holder, A.A. Dual acylation of the 45 kDa gliding-associated protein (GAP45) in Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 2006, 149, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Collins, M.O.; Goulding, D.; Choudhary, J.S.; Rayner, J.C. Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe 2012, 12, 246–258. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cabrera, A.; Herrmann, S.; Warszta, D.; Santos, J.M.; John Peter, A.T.; Kono, M.; Debrouver, S.; Jacobs, T.; Spielmann, T.; Ungermann, C.; et al. Dissection of minimal sequence requirements for rhoptry membrane targeting in the malaria parasite. Traffic 2012, 13, 1335–1350. [Google Scholar] [CrossRef] [PubMed]
- Geiger, M.; Brown, C.; Wichers, J.S.; Strauss, J.; Lill, A.; Thuenauer, R.; Liffner, B.; Wilcke, L.; Lemcke, S.; Heincke, D.; et al. Structural Insights Into PfARO and Characterization of its Interaction With PfAIP. J. Mol. Biol. 2020, 432, 878–896. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Hertrich, N.; Perrin, A.J.; Withers-Martinez, C.; Collins, C.R.; Jones, M.L.; Watermeyer, J.M.; Fobes, E.T.; Martin, S.R.; Saibil, H.R.; et al. Processing of Plasmodium falciparum Merozoite Surface Protein MSP1 Activates a Spectrin-Binding Function Enabling Parasite Egress from RBCs. Cell Host Microbe 2015, 18, 433–444. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Egan, A.F.; Morris, J.; Barnish, G.; Allen, S.; Greenwood, B.M.; Kaslow, D.C.; Holder, A.A.; Riley, E.M. Clinical immunity to Plasmodium falciparum malaria is associated with serum antibodies to the 19-kDa C-terminal fragment of the merozoite surface antigen, PfMSP-1. J. Infect. Dis. 1996, 173, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.P.; Moliner Cubel, S.; Nyboer, B.; Jankowska-Döllken, M.; Schaeffer-Reiss, C.; Ayoub, D.; Planelles, G.; Lanzer, M. Phosphomimetic substitution at Ser-33 of the chloroquine resistance transporter PfCRT reconstitutes drug responses in Plasmodium falciparum. J. Biol. Chem. 2019, 294, 12766–12778. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hodson, N.; Invergo, B.; Rayner, J.C.; Choudhary, J.S. Palmitoylation and palmitoyl-transferases in Plasmodium parasites. Biochem. Soc. Trans. 2015, 43, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Frénal, K.; Tay, C.L.; Mueller, C.; Bushell, E.S.; Jia, Y.; Graindorge, A.; Billker, O.; Rayner, J.C.; Soldati-Favre, D. Global analysis of apicomplexan protein S-acyl transferases reveals an enzyme essential for invasion. Traffic 2013, 14, 895–911. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qian, P.; Wang, X.; Zhong, C.Q.; Wang, J.; Cai, M.; Nguitragool, W.; Li, J.; Cui, H.; Yuan, J. Inner membrane complex proteomics reveals a palmitoylation regulation critical for intraerythrocytic development of malaria parasite. eLife 2022, 11, e77447. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Suazo, K.F.; Schaber, C.; Palsuledesai, C.C.; Odom John, A.R.; Distefano, M.D. Global proteomic analysis of prenylated proteins in Plasmodium falciparum using an alkyne-modified isoprenoid analogue. Sci. Rep. 2016, 6, 38615. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jung, D.; Bachmann, H.S. Regulation of protein prenylation. Biomed. Pharmacother. 2023, 164, 114915. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, D.; Azam, T.; DelVecchio, C.; Qiu, L.; Park, Y.I.; Allen, C.M. Protein prenyl transferase activities of Plasmodium falciparum. Mol. Biochem. Parasitol. 1998, 94, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.H.; Clough, B.; Rackham, M.D.; Rangachari, K.; Brannigan, J.A.; Grainger, M.; Moss, D.K.; Bottrill, A.R.; Heal, W.P.; Broncel, M.; et al. Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat. Chem. 2014, 6, 112–121. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Howe, R.; Kelly, M.; Jimah, J.; Hodge, D.; Odom, A.R. Isoprenoid biosynthesis inhibition disrupts Rab5 localization and food vacuolar integrity in Plasmodium falciparum. Eukaryot. Cell 2013, 12, 215–223. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ayong, L.; DaSilva, T.; Mauser, J.; Allen, C.M.; Chakrabarti, D. Evidence for prenylation-dependent targeting of a Ykt6 SNARE in Plasmodium falciparum. Mol. Biochem. Parasitol. 2011, 175, 162–168. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mayer, D.C.G. Protein Sorting in Plasmodium falciparum. Life 2021, 11, 937. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gisselberg, J.E.; Zhang, L.; Elias, J.E.; Yeh, E. The Prenylated Proteome of Plasmodium falciparum Reveals Pathogen-specific Prenylation Activity and Drug Mechanism-of-action. Mol. Cell. Proteom. 2017, 16 (Suppl. S1), S54–S64. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, D.; Da Silva, T.; Barger, J.; Paquette, S.; Patel, H.; Patterson, S.; Allen, C.M. Protein farnesyltransferase and protein prenylation in Plasmodium falciparum. J. Biol. Chem. 2002, 277, 42066–42073. [Google Scholar] [CrossRef] [PubMed]
- Rzepczyk, C.M.; Saul, A.J.; Ferrante, A. Polyamine oxidase-mediated intraerythrocytic killing of Plasmodium falciparum: Evidence against the role of reactive oxygen metabolites. Infect. Immunol. 1984, 43, 238–244. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pabón, A.; Carmona, J.; Burgos, L.C.; Blair, S. Oxidative stress in patients with non-complicated malaria. Clin. Biochem. 2003, 36, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, E.; Arese, P.; Skorokhod, O.A. Role of the lipoperoxidation product 4-hydroxynonenal in the pathogenesis of severe malaria anemia and malaria immunodepression. Oxid. Med. Cell. Longev. 2015, 2015, 638416. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mueangson, O.; Mahittikorn, A.; Anabire, N.G.; Mala, W.; Kotepui, M. Increased Blood Concentrations of Malondialdehyde in Plasmodium Infection: A Systematic Review and Meta-Analysis. Antioxidants 2023, 12, 1502. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Muller, S. Redox and antioxidant systems of the malaria parasite Plasmodium falciparum. Mol. Microbiol. 2004, 53, 1291–1305. [Google Scholar] [CrossRef] [PubMed]
- Bozdech, Z.; Ginsburg, H. Antioxidant defense in Plasmodium falciparum—Data mining of the transcriptome. Malar. J. 2004, 3, 23. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- Hain, A.U.; Bosch, J. Autophagy in Plasmodium, a multifunctional pathway? Comput. Struct. Biotechnol. J. 2013, 8, e201308002. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ponder, E.L.; Bogyo, M. Ubiquitin-like modifiers and their deconjugating enzymes in medically important parasitic protozoa. Eukaryot. Cell 2007, 6, 1943–1952. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chung, D.W.; Ponts, N.; Prudhomme, J.; Rodrigues, E.M.; Le Roch, K.G. Characterization of the ubiquitylating components of the human malaria parasite’s protein degradation pathway. PLoS ONE 2012, 7, e43477. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sumam de Oliveira, D.; Kronenberger, T.; Palmisano, G.; Wrenger, C.; de Souza, E.E. Targeting SUMOylation in Plasmodium as a Potential Target for Malaria Therapy. Front. Cell. Infect. Microbiol. 2021, 11, 685866. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mata-Cantero, L.; Azkargorta, M.; Aillet, F.; Xolalpa, W.; LaFuente, M.J.; Elortza, F.; Carvalho, A.S.; Martin-Plaza, J.; Matthiesen, R.; Rodriguez, M.S. New insights into host-parasite ubiquitin proteome dynamics in P. falciparum infected red blood cells using a TUBEs-MS approach. J. Proteom. 2016, 139, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Ponts, N.; Saraf, A.; Chung, D.W.; Harris, A.; Prudhomme, J.; Washburn, M.P.; Florens, L.; Le Roch, K.G. Unraveling the ubiquitome of the human malaria parasite. J. Biol. Chem. 2011, 286, 40320–40330. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Karpiyevich, M.; Adjalley, S.; Mol, M.; Ascher, D.B.; Mason, B.; van der Heden van Noort, G.J.; Laman, H.; Ovaa, H.; Lee, M.C.S.; Artavanis-Tsakonas, K. Nedd8 hydrolysis by UCH proteases in Plasmodium parasites. PLoS Pathog. 2019, 15, e1008086. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Philip, N.; Haystead, T.A. Characterization of a UBC13 kinase in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2007, 104, 7845–7850. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Narwal, S.K.; Nayak, B.; Mehra, P.; Mishra, S. Protein kinase 9 is not required for completion of the Plasmodium berghei life cycle. Microbiol. Res. 2022, 260, 127051. [Google Scholar] [CrossRef] [PubMed]
- Pisciotta, J.M.; Scholl, P.F.; Shuman, J.L.; Shualev, V.; Sullivan, D.J. Quantitative characterization of hemozoin in Plasmodium berghei and vivax. Int. J. Parasitol. Drugs Drug Resist. 2017, 7, 110–119. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schwarzer, E.; Turrini, F.; Arese, P. A luminescence method for the quantitative determination of phagocytosis of erythrocytes, of malaria-parasitized erythrocytes and of malarial pigment. Br. J. Haematol. 1994, 88, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Deroost, K.; Lays, N.; Noppen, S.; Martens, E.; Opdenakker, G.; Van den Steen, P.E. Improved methods for haemozoin quantification in tissues yield organ-and parasite-specific information in malaria-infected mice. Malar. J. 2012, 11, 166. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thamarath, S.S.; Xiong, A.; Lin, P.H.; Preiser, P.R.; Han, J. Enhancing the sensitivity of micro magnetic resonance relaxometry detection of low parasitemia Plasmodium falciparum in human blood. Sci. Rep. 2019, 9, 2555. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Di Gregorio, E.; Ferrauto, G.; Schwarzer, E.; Gianolio, E.; Valente, E.; Ulliers, D.; Aime, S.; Skorokhod, O. Relaxometric studies of erythrocyte suspensions infected by Plasmodium falciparum: A tool for staging infection and testing anti-malarial drugs. Magn. Reason. Med. 2020, 84, 3366–3378. [Google Scholar] [CrossRef] [PubMed]
- Karl, S.; Gutiérrez, L.; House, M.J.; Davis, T.M.; St Pierre, T.G. Nuclear magnetic resonance: A tool for malaria diagnosis? Am. J. Trop. Med. Hyg. 2011, 85, 815–817. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Poli, G.; Schaur, R.J.; Siems, W.G.; Leonarduzzi, G. 4-Hydroxynonenal: A membrane lipid oxidation product of medicinal interest. Med. Res. Rev. 2008, 28, 569–631. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K.; Stadtman, E.R. Modification of histidine residues in proteins by reaction with 4-hydroxynonenal. Proc. Natl. Acad. Sci. USA 1992, 89, 4544–4548. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schaur, R.J. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol. Asp. Med. 2003, 24, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Viedma-Poyatos, Á.; González-Jiménez, P.; Langlois, O.; Company-Marín, I.; Spickett, C.M.; Pérez-Sala, D. Protein Lipoxidation: Basic Concepts and Emerging Roles. Antioxidants 2021, 10, 295. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Buffinton, G.D.; Hunt, N.H.; Cowden, W.B.; Clark, I.A. Detection of short-chain carbonyl products of lipid peroxidation from malaria-parasite (Plasmodium vinckei)-infected red blood cells exposed to oxidative stress. Biochem. J. 1988, 249, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Van Den Ham, K.; Shio, M.T.; Kassa, F.A.; Fougeray, S. Malarial pigment hemozoin and the innate inflammatory response. Front. Immunol. 2014, 5, 25. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Uyoga, S.; Skorokhod, O.A.; Opiyo, M.; Orori, E.N.; Williams, T.N.; Arese, P.; Schwarzer, E. Transfer of 4-hydroxynonenal from parasitized to non-parasitized erythrocytes in rosettes. Proposed role in severe malaria anemia. Br. J. Haematol. 2012, 157, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, R.; Marrocco, T.; Skorokhod, O.A.; Barbosa, A.; Nhabomba, A.; Manaca, M.N.; Guinovart, C.; Quintó, L.; Arese, P.; Alonso, P.L.; et al. Blood oxidative stress markers and Plasmodium falciparum malaria in non-immune African children. Br. J. Haematol. 2014, 164, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Skorokhod, O.A.; Khoo, S.K.; Aguilar, R.; Wiertsema, S.; Nhabomba, A.J.; Marrocco, T.; McNamara-Smith, M.; Manaca, M.N.; Barbosa, A.; et al. Plasma advanced oxidative protein products are associated with anti-oxidative stress pathway genes and malaria in a longitudinal cohort. Malar. J. 2014, 13, 134. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vennerstrom, J.L.; Eaton, J.W. Oxidants, oxidant drugs, and malaria. J. Med. Chem. 1988, 31, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.P.; Chitnis, C.E. Lipid peroxidation and its repair in malaria parasites. Trends. Parasitol. 2023, 39, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.F.; Campo, B.; Alexander, S.P.H.; Arendse, L.B.; Cheng, X.; Davenport, A.P.; Faccenda, E.; Fidock, D.A.; Godinez-Macias, K.P.; Harding, S.D.; et al. Advances in malaria pharmacology and the online guide to MALARIA PHARMACOLOGY: IUPHAR review 38. Br. J. Pharmacol. 2023, 180, 1899–1929. [Google Scholar] [CrossRef] [PubMed]
- Barrera, V.; Skorokhod, O.A.; Baci, D.; Gremo, G.; Arese, P.; Schwarzer, E. Host fibrinogen stably bound to hemozoin rapidly activates monocytes via TLR-4 and CD11b/CD18-integrin: A new paradigm of hemozoin action. Blood 2011, 117, 5674–5682. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, E.; Turrini, F.; Ulliers, D.; Giribaldi, G.; Ginsburg, H.; Arese, P. Impairment of macrophage functions after ingestion of Plasmodium falciparum-infected erythrocytes or isolated malarial pigment. J. Exp. Med. 1992, 176, 1033–1041. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Metzger, W.G.; Mordmüller, B.G.; Kremsner, P.G. Malaria pigment in leucocytes. Trans. R. Soc. Trop. Med. Hyg. 1995, 89, 637–638. [Google Scholar] [CrossRef] [PubMed]
- Gallo, V.; Skorokhod, O.A.; Schwarzer, E.; Arese, P. Simultaneous determination of phagocytosis of Plasmodium falciparum-parasitized and non-parasitized red blood cells by flow cytometry. Malar. J. 2012, 11, 428. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Skorokhod, O.A.; Alessio, M.; Mordmüller, B.; Arese, P.; Schwarzer, E. Hemozoin (malarial pigment) inhibits differentiation and maturation of human monocyte-derived dendritic cells: A peroxisome proliferator-activated receptor-gamma-mediated effect. J. Immunol. 2004, 173, 4066–4074. [Google Scholar] [CrossRef] [PubMed]
- Urban, B.C.; Todryk, S. Malaria pigment paralyzes dendritic cells. J. Biol. 2006, 5, 4. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- Hänscheid, T.; Egan, T.J.; Grobusch, M.P. Haemozoin: From melatonin pigment to drug target, diagnostic tool, and immune modulator. Lancet Infect. Dis. 2007, 7, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, E.; Skorokhod, O.A.; Barrera, V.; Arese, P. Hemozoin and the human monocyte—A brief review of their interactions. Parassitologia 2008, 50, 143–145. [Google Scholar] [PubMed]
- Skorokhod, O.A.; Barrera, V.; Heller, R.; Carta, F.; Turrini, F.; Arese, P.; Schwarzer, E. Malarial pigment hemozoin impairs chemotactic motility and transendothelial migration of monocytes via 4-hydroxynonenal. Free Radic. Biol. Med. 2014, 75, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Skorokhod, O.; Barrera, V.; Mandili, G.; Costanza, F.; Valente, E.; Ulliers, D.; Schwarzer, E. Malaria Pigment Hemozoin Impairs GM-CSF Receptor Expression and Function by 4-Hydroxynonenal. Antioxidants 2021, 10, 1259. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Skorokhod, O.; Triglione, V.; Barrera, V.; Di Nardo, G.; Valente, E.; Ulliers, D.; Schwarzer, E.; Gilardi, G. Posttranslational Modification of Human Cytochrome CYP4F11 by 4-Hydroxynonenal Impairs ω-Hydroxylation in Malaria Pigment Hemozoin-Fed Monocytes: The Role in Malaria Immunosuppression. Int. J. Mol. Sci. 2023, 24, 10232. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cambos, M.; Bazinet, S.; Abed, E.; Sanchez-Dardon, J.; Bernard, C.; Moreau, R.; Olivier, M.; Scorza, T. The IL-12p70/IL-10 interplay is differentially regulated by free heme and hemozoin in murine bone-marrow-derived macrophages. Int. J. Parasitol. 2010, 40, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Bujila, I.; Schwarzer, E.; Skorokhod, O.; Weidner, J.M.; Troye-Blomberg, M.; Östlund Farrants, A.K. Malaria-derived hemozoin exerts early modulatory effects on the phenotype and maturation of human dendritic cells. Cell. Microbiol. 2016, 18, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Lasaviciute, G.; Tariq, K.; Sugathan, A.; Quin, J.E.; Bujila, I.; Skorokhod, O.; Troye-Blomberg, M.; Sverremark-Ekstrom, E.; Farrants, A.-K.O. Malaria-derived hemozoin alters chromatin remodelling and skews dendritic cell responses to subsequent bacterial infections. bioRxiv 2024. [Google Scholar] [CrossRef]
- Skorokhod, O.A.; Caione, L.; Marrocco, T.; Migliardi, G.; Barrera, V.; Arese, P.; Piacibello, W.; Schwarzer, E. Inhibition of erythropoiesis in malaria anemia: Role of hemozoin and hemozoin-generated 4-hydroxynonenal. Blood 2010, 116, 4328–4337. [Google Scholar] [CrossRef] [PubMed]
- Dumarchey, A.; Lavazec, C.; Verdier, F. Erythropoiesis and Malaria, a Multifaceted Interplay. Int. J. Mol. Sci. 2022, 23, 12762. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, E.; Müller, O.; Arese, P.; Siems, W.G.; Grune, T. Increased levels of 4-hydroxynonenal in human monocytes fed with malarial pigment hemozoin. A possible clue for hemozoin toxicity. FEBS Lett. 1996, 388, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, F.; Lee, P.C.; Abdul Wahab, H.; Mustaffa, K.M.F.; Leow, C.H.; Azhar, R.; Lai, N.S. Plasmodium falciparum protein kinase as a potential therapeutic target for antimalarial drugs development. Trop. Biomed. 2020, 37, 822–841. [Google Scholar] [CrossRef] [PubMed]
- Alder, A.; Wilcke, L.; Pietsch, E.; von Thien, H.; Pazicky, S.; Löw, C.; Mesen-Ramirez, P.; Bachmann, A.; Burda, P.C.; Kunick, C.; et al. Functional inactivation of Plasmodium falciparum glycogen synthase kinase GSK3 modulates erythrocyte invasion and blocks gametocyte maturation. J. Biol. Chem. 2022, 298, 102360. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Blomqvist, K.; Helmel, M.; Wang, C.; Absalon, S.; Labunska, T.; Rudlaff, R.M.; Adapa, S.; Jiang, R.; Steen, H.; Dvorin, J.D. Influence of Plasmodium falciparum Calcium-Dependent Protein Kinase 5 (PfCDPK5) on the Late Schizont Stage Phosphoproteome. mSphere 2020, 5, e00921-19. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jeninga, M.D.; Quinn, J.E.; Petter, M. ApiAP2 Transcription Factors in Apicomplexan Parasites. Pathogens 2019, 8, 47. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Reyser, T.; Paloque, L.; Augereau, J.M.; Di Stefano, L.; Benoit-Vical, F. Epigenetic regulation as a therapeutic target in the malaria parasite Plasmodium falciparum. Malar. J. 2024, 23, 44. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- López-Gutiérrez, B.; Cova, M.; Izquierdo, L.A. Plasmodium falciparum C-mannosyltransferase is dispensable for parasite asexual blood stage development. Parasitology 2019, 146, 1767–1772. [Google Scholar] [CrossRef] [PubMed]
- Kupferschmid, M.; Aquino-Gil, M.O.; Shams-Eldin, H.; Schmidt, J.; Yamakawa, N.; Krzewinski, F.; Schwarz, R.T.; Lefebvre, T. Identification of O-GlcNAcylated proteins in Plasmodium falciparum. Malar. J. 2017, 16, 485. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Howell, S.A.; Well, I.; Fleck, S.L.; Kettleborough, C.; Collins, C.R.; Blackman, M.J. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J. Biol. Chem. 2003, 278, 23890–23898. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.R.; Hackett, F.; Howell, S.A.; Snijders, A.P.; Russell, M.R.; Collinson, L.M.; Blackman, M.J. The malaria parasite sheddase SUB2 governs host red blood cell membrane sealing at invasion. eLife 2020, 9, e61121. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumar, S.; Gargaro, O.R.; Kappe, S.H.I. Plasmodium falciparum CRK5 Is Critical for Male Gametogenesis and Infection of the Mosquito. mBio 2022, 13, e0222722. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Renn, J.P.; Doritchamou, J.Y.A.; Tentokam, B.C.N.; Morrison, R.D.; Cowles, M.V.; Burkhardt, M.; Ma, R.; Mahamar, A.; Attaher, O.; Diarra, B.S.; et al. Allelic variants of full-length VAR2CSA.; the placental malaria vaccine candidate, differ in antigenicity and receptor binding affinity. Commun. Biol. 2021, 4, 1309, Erratum in Commun. Biol. 2022, 5, 404. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kennedy, M.C.; Wang, J.; Zhang, Y.; Miles, A.P.; Chitsaz, F.; Saul, A.; Long, C.A.; Miller, L.H.; Stowers, A.W. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): Production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect. Immunol. 2002, 70, 6948–6960. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mathews, E.S.; Jezewski, A.J.; Odom John, A.R. Protein Prenylation and Hsp40 in Thermotolerance of Plasmodium falciparum Malaria Parasites. mBio 2021, 12, e0076021. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Josling, G.A.; Williamson, K.C.; Llinás, M. Regulation of Sexual Commitment and Gametocytogenesis in Malaria Parasites. Annu. Rev. Microbiol. 2018, 72, 501–519. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tibúrcio, M.; Silvestrini, F.; Bertuccini, L.; Sander, A.F.; Turner, L.; Lavstsen, T.; Alano, P. Early gametocytes of the malaria parasite Plasmodium falciparum specifically remodel the adhesive properties of infected erythrocyte surface. Cell. Microbiol. 2013, 15, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Lucky, A.B.; Brashear, A.M.; Li, X.; Cui, L.; Miao, J. Distinct Histone Post-translational Modifications during Plasmodium falciparum Gametocyte Development. J. Proteome Res. 2022, 21, 1857–1867. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jamjoom, G.A. Evidence for a role of hemozoin in metabolism and gametocytogenesis. MalariaWorld J. 2017, 8, 10. [Google Scholar] [PubMed] [PubMed Central]
- Orjih, A.U. Hemozoin accumulation in Garnham bodies of Plasmodium falciparum gametocytes. Parasitol. Res. 2012, 111, 2353–2359. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.M.; Franke-Fayard, B.; Mair, G.R.; Lasonder, E.; Janse, C.J.; Mann, M.; Waters, A.P. Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 2005, 121, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Lasonder, E.; Rijpma, S.R.; van Schaijk, B.C.; Hoeijmakers, W.A.; Kensche, P.R.; Gresnigt, M.S.; Italiaander, A.; Vos, M.W.; Woestenenk, R.; Bousema, T.; et al. Integrated transcriptomic and proteomic analyses of P. falciparum gametocytes: Molecular insight into sex-specific processes and translational repression. Nucleic Acids Res. 2016, 44, 6087–6101. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Santolamazza, F.; Avellino, P.; Siciliano, G.; Yao, F.A.; Lombardo, F.; Ouédraogo, J.B.; Modiano, D.; Alano, P.; Mangano, V.D. Detection of Plasmodium falciparum male and female gametocytes and determination of parasite sex ratio in human endemic populations by novel, cheap and robust RTqPCR assays. Malar. J. 2017, 16, 468. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Balestra, A.C.; Zeeshan, M.; Rea, E.; Pasquarello, C.; Brusini, L.; Mourier, T.; Subudhi, A.K.; Klages, N.; Arboit, P.; Pandey, R.; et al. A divergent cyclin/cyclin-dependent kinase complex controls the atypical replication of a malaria parasite during gametogony and transmission. eLife 2020, 9, e56474. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Camarda, G.; Bertuccini, L.; Singh, S.K.; Salzano, A.M.; Lanfrancotti, A.; Olivieri, A.; Scaloni, A.; Sharma, A.; Alano, P. Regulated oligomerisation and molecular interactions of the early gametocyte protein Pfg27 in Plasmodium falciparum sexual differentiation. Int. J. Parasitol. 2010, 40, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Sannella, A.R.; Olivieri, A.; Bertuccini, L.; Ferrè, F.; Severini, C.; Pace, T.; Alano, P. Specific tagging of the egress-related osmiophilic bodies in the gametocytes of Plasmodium falciparum. Malar. J. 2012, 11, 88. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Suárez-Cortés, P.; Sharma, V.; Bertuccini, L.; Costa, G.; Bannerman, N.L.; Sannella, A.R.; Williamson, K.; Klemba, M.; Levashina, E.A.; Lasonder, E.; et al. Comparative Proteomics and Functional Analysis Reveal a Role of Plasmodium falciparum Osmiophilic Bodies in Malaria Parasite Transmission. Mol. Cell. Proteom. 2016, 15, 3243–3255. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Silvestrini, F.; Lasonder, E.; Olivieri, A.; Camarda, G.; van Schaijk, B.; Sanchez, M.; Younis Younis, S.; Sauerwein, R.; Alano, P. Protein export marks the early phase of gametocytogenesis of the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteom. 2010, 9, 1437–1448. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grasso, F.; Fratini, F.; Albanese, T.G.; Mochi, S.; Ciardo, M.; Pace, T.; Ponzi, M.; Pizzi, E.; Olivieri, A. Identification and preliminary characterization of Plasmodium falciparum proteins secreted upon gamete formation. Sci. Rep. 2022, 12, 9592. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jennison, C.; Lucantoni, L.; O’Neill, M.T.; McConville, R.; Erickson, S.M.; Cowman, A.F.; Sleebs, B.E.; Avery, V.M.; Boddey, J.A. Inhibition of Plasmepsin V Activity Blocks Plasmodium falciparum Gametocytogenesis and Transmission to Mosquitoes. Cell Rep. 2019, 29, 3796–3806.e4. [Google Scholar] [CrossRef] [PubMed]
- Abugri, J.; Ayariga, J.; Sunwiale, S.S.; Wezena, C.A.; Gyamfi, J.A.; Adu-Frimpong, M.; Agongo, G.; Dongdem, J.T.; Abugri, D.; Dinko, B. Targeting the Plasmodium falciparum proteome and organelles for potential antimalarial drug candidates. Heliyon 2022, 8, e10390. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ngwa, C.J.; Kiesow, M.J.; Orchard, L.M.; Farrukh, A.; Llinás, M.; Pradel, G. The G9a Histone Methyltransferase Inhibitor BIX-01294 Modulates Gene Expression during Plasmodium falciparum Gametocyte Development and Transmission. Int. J. Mol. Sci. 2019, 20, 5087. [Google Scholar] [CrossRef] [PubMed]
- Josling, G.A.; Russell, T.J.; Venezia, J.; Orchard, L.; van Biljon, R.; Painter, H.J.; Llinás, M. Dissecting the role of PfAP2-G in malaria gametocytogenesis. Nat. Commun. 2020, 11, 1503. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tay, C.L.; Jones, M.L.; Hodson, N.; Theron, M.; Choudhary, J.S.; Rayner, J.C. Study of Plasmodium falciparum DHHC palmitoyl transferases identifies a role for PfDHHC9 in gametocytogenesis. Cell. Microbiol. 2016, 18, 1596–1610. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Johnson, N.; Philip, N. Beyond phosphorylation: Putative roles of post-translational modifications in Plasmodium sexual stages. Mol. Biochem. Parasitol. 2021, 245, 111406. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, P.P.; Jiang, X.; Zhu, L.; Zhou, D.; Hong, M.; He, L.; Chen, L.; Yao, S.; Zhao, Y.; Chen, G.; et al. A G-Protein-Coupled Receptor Modulates Gametogenesis via PKG-Mediated Signaling Cascade in Plasmodium berghei. Microbiol. Spectr. 2022, 10, e0015022. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ruberto, A.A.; Bourke, C.; Vantaux, A.; Maher, S.P.; Jex, A.; Witkowski, B.; Snounou, G.; Mueller, I. Single-cell RNA sequencing of Plasmodium vivax sporozoites reveals stage- and species-specific transcriptomic signatures. PLoS Negl. Trop. Dis. 2022, 16, e0010633. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ouologuem, D.T.; Dara, A.; Kone, A.; Ouattara, A.; Djimde, A.A. Plasmodium falciparum Development from Gametocyte to Oocyst: Insight from Functional Studies. Microorganisms 2023, 11, 1966. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Invergo, B.M.; Brochet, M.; Yu, L.; Choudhary, J.; Beltrao, P.; Billker, O. Sub-minute Phosphoregulation of Cell Cycle Systems during Plasmodium Gamete Formation. Cell Rep. 2017, 21, 2017–2029. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Garcia, C.H.S.; Depoix, D.; Queiroz, R.M.L.; Souza, J.M.F.; Fontes, W.; de Sousa, M.V.; Santos, M.D.M.; Carvalho, P.C.; Grellier, P.; Charneau, S. Dynamic molecular events associated to Plasmodium berghei gametogenesis through proteomic approach. J. Proteom. 2018, 180, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Morales, A.; González-López, L.; Cázares-Raga, F.E.; Cortés-Martínez, L.; Torres-Monzón, J.A.; Gallegos-Pérez, J.L.; Rodríguez, M.H.; James, A.A.; Hernández-Hernández Fde, L. Protein phosphorylation during Plasmodium berghei gametogenesis. Exp. Parasitol. 2015, 156, 49–60. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hall, N.; Karras, M.; Raine, J.D.; Carlton, J.M.; Kooij, T.W.; Berriman, M.; Florens, L.; Janssen, C.S.; Pain, A.; Christophides, G.K.; et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 2005, 307, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Lasonder, E.; Janse, C.J.; van Gemert, G.J.; Mair, G.R.; Vermunt, A.M.; Douradinha, B.G.; van Noort, V.; Huynen, M.A.; Luty, A.J.; Kroeze, H.; et al. Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog. 2008, 4, e1000195. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Preira, C.M.F.; Pizzi, E.; Fratini, F.; Grasso, F.; Boccolini, D.; Mochi, S.; Favia, G.; Piselli, E.; Damiani, C.; Siden-Kiamos, I.; et al. A time point proteomic analysis reveals protein dynamics of Plasmodium oocysts. Mol. Cell. Proteom. 2024, 23, 100736. [Google Scholar] [CrossRef] [PubMed]
- Armistead, J.S.; Jennison, C.; O’Neill, M.T.; Lopaticki, S.; Liehl, P.; Hanson, K.K.; Annoura, T.; Rajasekaran, P.; Erickson, S.M.; Tonkin, C.J.; et al. Plasmodium falciparum subtilisin-like ookinete protein SOPT plays an important and conserved role during ookinete infection of the Anopheles stephensi midgut. Mol. Microbiol. 2018, 109, 458–473. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.S.; Thompson, J.; Kafatos, F.C.; Barillas-Mury, C. Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: The time bomb theory of ookinete invasion of mosquitoes. EMBO J. 2000, 19, 6030–6040, Erratum in EMBO J. 2001, 20, 1483. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zeeshan, M.; Rea, E.; Abel, S.; Vukušić, K.; Markus, R.; Brady, D.; Eze, A.; Rashpa, R.; Balestra, A.C.; Bottrill, A.R.; et al. Plasmodium ARK2 and EB1 drive unconventional spindle dynamics, during chromosome segregation in sexual transmission stages. Nat. Commun. 2023, 14, 5652. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guttery, D.S.; Poulin, B.; Ferguson, D.J.; Szoor, B.; Wickstead, B.; Carroll, P.L.; Ramakrishnan, C.; Brady, D.; Patzewitz, E.M.; Straschil, U.; et al. A unique protein phosphatase with kelch-like domains (PPKL) in Plasmodium modulates ookinete differentiation, motility and invasion. PLoS Pathog. 2012, 8, e1002948. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.P.; Chin, W.H.; Zhu, L.; Mok, S.; Luah, Y.H.; Lim, E.H.; Bozdech, Z. Dynamic epigenetic regulation of gene expression during the life cycle of malaria parasite Plasmodium falciparum. PLoS Pathog. 2013, 9, e1003170. [Google Scholar] [CrossRef] [PubMed]
- von Gruning, H.; Coradin, M.; Mendoza, M.R.; Reader, J.; Sidoli, S.; Garcia, B.A.; Birkholtz, L.M. A Dynamic and Combinatorial Histone Code Drives Malaria Parasite Asexual and Sexual Development. Mol. Cell. Proteom. 2022, 21, 100199. [Google Scholar] [CrossRef] [PubMed]
- Rashpa, R.; Klages, N.; Schvartz, D.; Pasquarello, C.; Brochet, M. The Skp1-Cullin1-FBXO1 complex is a pleiotropic regulator required for the formation of gametes and motile forms in Plasmodium berghei. Nat. Commun. 2023, 14, 1312. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wetzel, J.; Herrmann, S.; Swapna, L.S.; Prusty, D.; John Peter, A.T.; Kono, M.; Saini, S.; Nellimarla, S.; Wong, T.W.; Wilcke, L.; et al. The role of palmitoylation for protein recruitment to the inner membrane complex of the malaria parasite. J. Biol. Chem. 2015, 290, 1712–1728. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.M.; Duarte, N.; Kehrer, J.; Ramesar, J.; Avramut, M.C.; Koster, A.J.; Dessens, J.T.; Frischknecht, F.; Chevalley-Maurel, S.; Janse, C.J.; et al. Maternally supplied S-acyl-transferase is required for crystalloid organelle formation and transmission of the malaria parasite. Proc. Natl. Acad. Sci. USA 2016, 113, 7183–7188. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, X.; Qian, P.; Cui, H.; Yao, L.; Yuan, J. A protein palmitoylation cascade regulates microtubule cytoskeleton integrity in Plasmodium. EMBO J. 2020, 39, e104168, Erratum in EMBO J. 2021, 40, e109070. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kolli, S.K.; Molina-Cruz, A.; Araki, T.; Geurten, F.J.A.; Ramesar, J.; Chevalley-Maurel, S.; Kroeze, H.J.; Bezemer, S.; de Korne, C.; Withers, R.; et al. Malaria parasite evades mosquito immunity by glutaminyl cyclase-mediated posttranslational protein modification. Proc. Natl. Acad. Sci. USA 2022, 119, e2209729119. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pinheiro-Silva, R.; Borges, L.; Coelho, L.P.; Cabezas-Cruz, A.; Valdés, J.J.; do Rosário, V.; de la Fuente, J.; Domingos, A. Gene expression changes in the salivary glands of Anopheles coluzzii elicited by Plasmodium berghei infection. Parasit. Vectors 2015, 8, 485. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Klaus, S.; Binder, P.; Kim, J.; Machado, M.; Funaya, C.; Schaaf, V.; Klaschka, D.; Kudulyte, A.; Cyrklaff, M.; Laketa, V.; et al. Asynchronous nuclear cycles in multinucleated Plasmodium falciparum facilitate rapid proliferation. Sci. Adv. 2022, 8, eabj5362. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kaushik, M.; Nehra, A.; Gill, S.S.; Gill, R. Unraveling CAF-1 family in Plasmodium falciparum: Comparative genome-wide identification and phylogenetic analysis among eukaryotes, expression profiling and protein-protein interaction studies. 3 Biotech 2020, 10, 143. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fang, H.; Klages, N.; Baechler, B.; Hillner, E.; Yu, L.; Pardo, M.; Choudhary, J.; Brochet, M. Multiple short windows of calcium-dependent protein kinase 4 activity coordinate distinct cell cycle events during Plasmodium gametogenesis. eLife 2017, 6, e26524. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mair, G.R.; Lasonder, E.; Garver, L.S.; Franke-Fayard, B.M.; Carret, C.K.; Wiegant, J.C.; Dirks, R.W.; Dimopoulos, G.; Janse, C.J.; Waters, A.P. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010, 6, e1000767. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Singh, M.; Suryanshu; Kanika; Singh, G.; Dubey, A.; Chaitanya, R.K. Plasmodium’s journey through the, A.nopheles mosquito: A comprehensive review. Biochimie 2021, 181, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Klug, D.; Blandin, S.A. Activation of complement-like antiparasitic responses in Anopheles mosquitoes. Curr. Opin. Microbiol. 2023, 72, 102280. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Molina-Cruz, A.; Brzostowski, J.; Mu, J.; Miller, L.H. Plasmodium falciparum Calcium-Dependent Protein Kinase 2 Is Critical for Male Gametocyte Exflagellation but Not Essential for Asexual Proliferation. mBio 2017, 8, e01656-17. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lopaticki, S.; McConville, R.; John, A.; Geoghegan, N.; Mohamed, S.D.; Verzier, L.; Steel, R.W.J.; Evelyn, C.; O’Neill, M.T.; Soler, N.M.; et al. Tryptophan C-mannosylation is critical for Plasmodium falciparum transmission. Nat. Commun. 2022, 13, 4400. [Google Scholar] [CrossRef]
- Yadav, D.K.; Kumar, S.; Teli, M.K.; Yadav, R.; Chaudhary, S. Molecular Targets for Malarial Chemotherapy: A Review. Curr. Top. Med. Chem. 2019, 19, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Alves, F.M.; Bellei, J.C.B.; Barbosa, C.S.; Duarte, C.L.; Fonseca, A.L.D.; Pinto, A.C.S.; Raimundo, F.O.; Carpinter, B.A.; Lemos, A.S.O.; Coimbra, E.S.; et al. Rational-Based Discovery of Novel β-Carboline Derivatives as Potential Antimalarials: From In Silico Identification of Novel Targets to Inhibition of Experimental Cerebral Malaria. Pathogens 2022, 11, 1529. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mogwera, K.S.P.; Chibale, K.; Arendse, L.B. Developing kinase inhibitors for malaria: An opportunity or liability? Trends. Parasitol. 2023, 39, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Adderley, J.; Doerig, C. Comparative analysis of the kinomes of Plasmodium falciparum, Plasmodium vivax and their host Homo sapiens. BMC Genom. 2022, 23, 237. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Munro, B.A.; McMorran, B.J. Antimalarial Drug Strategies to Target Plasmodium Gametocytes. Parasitologia 2022, 2, 101–124. [Google Scholar] [CrossRef]
- Baker, D.A.; Matralis, A.N.; Osborne, S.A.; Large, J.M.; Penzo, M. Targeting the Malaria Parasite cGMP-Dependent Protein Kinase to Develop New Drugs. Front. Microbiol. 2020, 11, 602803. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vanaerschot, M.; Murithi, J.M.; Pasaje, C.F.A.; Ghidelli-Disse, S.; Dwomoh, L.; Bird, M.; Spottiswoode, N.; Mittal, N.; Arendse, L.B.; Owen, E.S.; et al. Inhibition of Resistance-Refractory P. falciparum Kinase PKG Delivers Prophylactic, Blood Stage, and Transmission-Blocking Antiplasmodial Activity. Cell. Chem. Biol. 2020, 27, 806–816.e8. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Diaz, C.A.; Allocco, J.; Powles, M.A.; Yeung, L.; Donald, R.G.; Anderson, J.W.; Liberator, P.A. Characterization of Plasmodium falciparum cGMP-dependent protein kinase (PfPKG): Antiparasitic activity of a PKG inhibitor. Mol. Biochem. Parasitol. 2006, 146, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Sahu, N.K.; Kohli, D.V. Structural insight for imidazopyridazines as malarial kinase PfPK7 inhibitors using QSAR techniques. Med. Chem. 2012, 8, 636–648. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Dinér, P.; Dorin-Semblat, D.; Doerig, C.; Grøtli, M. Synthesis of 3-(1,2,3-triazol-1-yl)- and 3-(1,2,3-triazol-4-yl)-substituted pyrazolo [3,4-d]pyrimidin-4-amines via click chemistry: Potential inhibitors of the Plasmodium falciparum PfPK7 protein kinase. Org. Biomol. Chem. 2009, 7, 3421–3429. [Google Scholar] [CrossRef] [PubMed]
- Bouloc, N.; Large, J.M.; Smiljanic, E.; Whalley, D.; Ansell, K.H.; Edlin, C.D.; Bryans, J.S. Synthesis and in vitro evaluation of imidazopyridazines as novel inhibitors of the malarial kinase PfPK7. Bioorg. Med. Chem. Lett. 2008, 18, 5294–5298. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Batovska, D.I.; Medhi, B.; Radotra, B.D.; Bhalla, A.; Markova, N.; Sehgal, R. In vitro anti-malarial efficacy of chalcones: Cytotoxicity profile, mechanism of action and their effect on erythrocytes. Malar. J. 2019, 18, 421. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ali, A.H.; Sudi, S.; Basir, R.; Embi, N.; Sidek, H.M. The Antimalarial Effect of Curcumin Is Mediated by the Inhibition of Glycogen Synthase Kinase-3β. J. Med. Food 2017, 20, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Arendse, L.B.; Wyllie, S.; Chibale, K.; Gilbert, I.H. Plasmodium Kinases as Potential Drug Targets for Malaria: Challenges and Opportunities. ACS Infect. Dis. 2021, 7, 518–534. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mustière, R.; Vanelle, P.; Primas, N. Plasmodial Kinase Inhibitors Targeting Malaria: Recent Developments. Molecules 2020, 25, 5949. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pantaleo, A.; Ferru, E.; Pau, M.C.; Khadjavi, A.; Mandili, G.; Mattè, A.; Spano, A.; De Franceschi, L.; Pippia, P.; Turrini, F. Band 3 Erythrocyte Membrane Protein Acts as Redox Stress Sensor Leading to Its Phosphorylation by p (72) Syk. Oxid. Med. Cell. Longev. 2016, 2016, 6051093. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pantaleo, A.; Kesely, K.R.; Pau, M.C.; Tsamesidis, I.; Schwarzer, E.; Skorokhod, O.A.; Chien, H.D.; Ponzi, M.; Bertuccini, L.; Low, P.S.; et al. Syk inhibitors interfere with erythrocyte membrane modification during P falciparum growth and suppress parasite egress. Blood 2017, 130, 1031–1040. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tsamesidis, I.; Reybier, K.; Marchetti, G.; Pau, M.C.; Virdis, P.; Fozza, C.; Nepveu, F.; Low, P.S.; Turrini, F.M.; Pantaleo, A. Syk Kinase Inhibitors Synergize with Artemisinins by Enhancing Oxidative Stress in Plasmodium falciparum-Parasitized Erythrocytes. Antioxidants 2020, 9, 753. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Baragaña, B.; Hallyburton, I.; Lee, M.C.; Norcross, N.R.; Grimaldi, R.; Otto, T.D.; Proto, W.R.; Blagborough, A.M.; Meister, S.; Wirjanata, G.; et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 2015, 522, 315–320, Erratum in Nature 2016, 537, 122. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khandelwal, A.; Arez, F.; Alves, P.M.; Badolo, L.; Brito, C.; Fischli, C.; Fontinha, D.; Oeuvray, C.; Prudêncio, M.; Rottmann, M.; et al. Translation of liver stage activity of M5717, a Plasmodium elongation factor 2 inhibitor: From bench to bedside. Malar. J. 2022, 21, 151. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McCarthy, J.S.; Yalkinoglu, Ö.; Odedra, A.; Webster, R.; Oeuvray, C.; Tappert, A.; Bezuidenhout, D.; Giddins, M.J.; Dhingra, S.K.; Fidock, D.A.; et al. Safety, pharmacokinetics, and antimalarial activity of the novel plasmodium eukaryotic translation elongation factor 2 inhibitor M5717: A first-in-human, randomised, placebo-controlled, double-blind, single ascending dose study and volunteer infection study. Lancet Infect. Dis. 2021, 21, 1713–1724. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Deu, E. Proteases as antimalarial targets: Strategies for genetic, chemical, and therapeutic validation. FEBS J. 2017, 284, 2604–2628. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ng, C.L.; Fidock, D.A.; Bogyo, M. Protein Degradation Systems as Antimalarial Therapeutic Targets. Trends. Parasitol. 2017, 33, 731–743. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Madhav, H.; Patel, T.S.; Rizvi, Z.; Reddy, G.S.; Rahman, A.; Rahman, M.A.; Ahmedi, S.; Fatima, S.; Saxena, K.; Manzoor, N.; et al. Development of diphenylmethylpiperazine hybrids of chloroquinoline and triazolopyrimidine using Petasis reaction as new cysteine proteases inhibitors for malaria therapeutics. Eur. J. Med. Chem. 2023, 258, 115564. [Google Scholar] [CrossRef] [PubMed]
- Skorokhod, O.; Valente, E.; Mandili, G.; Ulliers, D.; Schwarzer, E. Micromolar Dihydroartemisinin Concentrations Elicit Lipoperoxidation in Plasmodium falciparum-Infected Erythrocytes. Antioxidants 2023, 12, 1468. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.; Chen, H.; Bahamontes-Rosa, N.; Kun, J.F.; Traore, B.; Crompton, P.D.; Chishti, A.H. Plasmodium falciparum signal peptide peptidase is a promising drug target against blood stage malaria. Biochem. Biophys. Res. Commun. 2009, 380, 454–459. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Parvanova, I.; Epiphanio, S.; Fauq, A.; Golde, T.E.; Prudêncio, M.; Mota, M.M. A Small Molecule Inhibitor of Signal Peptide Peptidase Inhibits Plasmodium Development in the Liver and Decreases Malaria Severity. PLoS ONE 2009, 4, e5078. [Google Scholar] [CrossRef] [PubMed]
- Harbut, M.B.; Patel, B.A.; Yeung, B.K.; McNamara, C.W.; Bright, A.T.; Ballard, J.; Supek, F.; Golde, T.E.; Winzeler, E.A.; Diagana, T.T.; et al. Targeting the ERAD pathway via inhibition of signal peptide peptidase for antiparasitic therapeutic design. Proc. Natl. Acad. Sci. USA 2012, 109, 21486–21491. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pino, P.; Caldelari, R.; Mukherjee, B.; Vahokoski, J.; Klages, N.; Maco, B.; Collins, C.R.; Blackman, M.J.; Kursula, I.; Heussler, V.; et al. A multistage antimalarial targets the plasmepsins IX and X essential for invasion and egress. Science 2017, 358, 522–528. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lisauskaitė, M.; Nixon, G.L.; Woodley, C.M.; Berry, N.G.; Coninckx, A.; Qie, L.C.; Leung, S.C.; Taramelli, D.; Basilico, N.; Parapini, S.; et al. Design, synthesis and modelling of photoreactive chemical probes for investigating target engagement of plasmepsin IX and X in Plasmodium falciparum. RSC Chem. Biol. 2023, 5, 19–29. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McConville, M.; Fernández, J.; Angulo-Barturen, Í.; Bahamontes-Rosa, N.; Ballell-Pages, L.; Castañeda, P.; de Cózar, C.; Crespo, B.; Guijarro, L.; Jiménez-Díaz, M.B.; et al. Carbamoyl Triazoles, Known Serine Protease Inhibitors, Are a Potent New Class of Antimalarials. J. Med. Chem. 2015, 58, 6448–6455. [Google Scholar] [CrossRef] [PubMed]
- Nizi, E.; Sferrazza, A.; Fabbrini, D.; Nardi, V.; Andreini, M.; Graziani, R.; Gennari, N.; Bresciani, A.; Paonessa, G.; Harper, S. Peptidomimetic nitrile inhibitors of malarial protease falcipain-2 with high selectivity against human cathepsins. Bioorg. Med. Chem. Lett. 2018, 28, 1540–1544. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Wollenberg, K.; Sellers, M.; Zainabadi, K.; Galinsky, K.; Moss, E.; Nguitragool, W.; Neafsey, D.; Desai, S.A. An epigenetic antimalarial resistance mechanism involving parasite genes linked to nutrient uptake. J. Biol. Chem. 2013, 288, 19429–19440. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mira-Martínez, S.; Rovira-Graells, N.; Crowley, V.M.; Altenhofen, L.M.; Llinás, M.; Cortés, A. Epigenetic switches in clag3 genes mediate blasticidin S resistance in malaria parasites. Cell. Microbiol. 2013, 15, 1913–1923. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rawat, M.; Kanyal, A.; Sahasrabudhe, A.; Vembar, S.S.; Lopez-Rubio, J.J.; Karmodiya, K. Histone acetyltransferase PfGCN5 regulates stress responsive and artemisinin resistance related genes in Plasmodium falciparum. Sci. Rep. 2021, 11, 852. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lucky, A.B.; Wang, C.; Shakri, A.R.; Kalamuddin, M.; Chim-Ong, A.; Li, X.; Miao, J. Plasmodium falciparum GCN5 plays a key role in regulating artemisinin resistance-related stress responses. Antimicrob. Agents Chemother. 2023, 67, e0057723. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumar, A.; Bhowmick, K.; Vikramdeo, K.S.; Mondal, N.; Subbarao, N.; Dhar, S.K. Designing novel inhibitors against histone acetyltransferase (HAT: GCN5) of Plasmodium falciparum. Eur. J. Med. Chem. 2017, 138, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Sen, U.; Nayak, A.; Khurana, J.; Sharma, D.; Gupta, A. Inhibition of PfMYST Histone Acetyltransferase Activity Blocks Plasmodium falciparum Growth and Survival. Antimicrob. Agents Chemother. 2020, 65, e00953-20. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sabnis, R.W. Novel Histone Acetyltransferase (HAT) Inhibitors for Treating Diseases. ACS Med. Chem. Lett. 2021, 12, 1198–1199. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Andrews, K.T.; Tran, T.N.; Lucke, A.J.; Kahnberg, P.; Le, G.T.; Boyle, G.M.; Gardiner, D.L.; Skinner-Adams, T.S.; Fairlie, D.P. Potent antimalarial activity of histone deacetylase inhibitor analogues. Antimicrob. Agents Chemother. 2008, 52, 1454–1461. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Andrews, K.T.; Gupta, A.P.; Tran, T.N.; Fairlie, D.P.; Gobert, G.N.; Bozdech, Z. Comparative gene expression profiling of P. falciparum malaria parasites exposed to three different histone deacetylase inhibitors. PLoS ONE 2012, 7, e31847. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jublot, D.; Cavaillès, P.; Kamche, S.; Francisco, D.; Fontinha, D.; Prudêncio, M.; Guichou, J.-F.; Labesse, G.; Sereno, D.; Loeuillet, C. A Histone Deacetylase (HDAC) Inhibitor with Pleiotropic In Vitro Anti-Toxoplasma and Anti-Plasmodium Activities Controls Acute and Chronic Toxoplasma Infection in Mice. Int. J. Mol. Sci. 2022, 23, 3254. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, T.K.; Nayak, R.R.; Ganeshpurkar, A.; Tiwari, P.; Kumar, D. Opportunities and Difficulties in the Repurposing of HDAC Inhibitors as Antiparasitic Agents. Drugs Drug Candidates 2024, 3, 70–101. [Google Scholar] [CrossRef]
- von Bredow, L.; Schäfer, T.M.; Hogenkamp, J.; Tretbar, M.; Stopper, D.; Kraft, F.B.; Schliehe-Diecks, J.; Schöler, A.; Borkhardt, A.; Bhatia, S.; et al. Synthesis, Antiplasmodial, and Antileukemia Activity of Dihydroartemisinin–HDAC Inhibitor Hybrids as Multitarget Drugs. Pharmaceuticals 2022, 15, 333. [Google Scholar] [CrossRef] [PubMed]
- Malmquist, N.A.; Sundriyal, S.; Caron, J.; Chen, P.; Witkowski, B.; Menard, D.; Suwanarusk, R.; Renia, L.; Nosten, F.; Jiménez-Díaz, M.B.; et al. Histone methyltransferase inhibitors are orally bioavailable, fast-acting molecules with activity against different species causing malaria in humans. Antimicrob. Agents Chemother. 2015, 59, 950–959. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gomes, A.R.Q.; Cunha, N.; Varela, E.L.P.; Brígido, H.P.C.; Vale, V.V.; Dolabela, M.F.; De Carvalho, E.P.; Percário, S. Oxidative Stress in Malaria: Potential Benefits of Antioxidant Therapy. Int. J. Mol. Sci. 2022, 23, 5949. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zheng, Z.; Liu, H.; Wang, X.; Zhang, Y.; Qu, S.; Yang, Y.; Deng, S.; Chen, L.; Zhu, X.; Li, Y. Artesunate and Tetramethylpyrazine Exert Effects on Experimental Cerebral Malaria in a Mechanism of Protein S-Nitrosylation. ACS Infect. Dis. 2021, 7, 2836–2849. [Google Scholar] [CrossRef] [PubMed]
- Ben Mamoun, C.; Prigge, S.T.; Vial, H. Targeting the Lipid Metabolic Pathways for the Treatment of Malaria. Drug Dev. Res. 2010, 71, 44–55. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schlott, A.C.; Holder, A.A.; Tate, E.W. N-Myristoylation as a Drug Target in Malaria: Exploring the Role of N-Myristoyltransferase Substrates in the Inhibitor Mode of Action. ACS Infect. Dis. 2018, 4, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Schlott, A.C.; Mayclin, S.; Reers, A.R.; Coburn-Flynn, O.; Bell, A.S.; Green, J.; Knuepfer, E.; Charter, D.; Bonnert, R.; Campo, B.; et al. Structure-Guided Identification of Resistance Breaking Antimalarial N-Myristoyltransferase Inhibitors. Cell. Chem. Biol. 2019, 26, 991–1000.e7. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, J.; Tan, Y.Z.; Wicht, K.J.; Erramilli, S.K.; Dhingra, S.K.; Okombo, J.; Vendome, J.; Hagenah, L.M.; Giacometti, S.I.; Warren, A.L.; et al. Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature 2019, 576, 315–320. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wurtz, N.; Fall, B.; Pascual, A.; Fall, M.; Baret, E.; Camara, C.; Nakoulima, A.; Diatta, B.; Fall, K.B.; Mbaye, P.S.; et al. Role of Pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob. Agents Chemother. 2014, 58, 7032–7040. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Knak, T.; Abdullaziz, M.A.; Höfmann, S.; Alves Avelar, L.A.; Klein, S.; Martin, M.; Fischer, M.; Tanaka, N.; Kurz, T. Over 40 Years of Fosmidomycin Drug Research: A Comprehensive Review and Future Opportunities. Pharmaceuticals 2022, 15, 1553. [Google Scholar] [CrossRef]
- Bofill Verdaguer, I.; Sussmann, R.A.C.; Santiago, V.F.; Palmisano, G.; Moura, G.C.; Mesquita, J.T.; Yamaguchi, L.F.; Kato, M.J.; Katzin, A.M.; Crispim, M. Isoprenoid alcohols utilization by malaria parasites. Front. Chem. 2022, 10, 1035548. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meshnick, S.R. Artemisinin antimalarials: Mechanisms of action and resistance. Med. Trop. (Mars) 1998, 58 (Suppl. S3), 13–17. [Google Scholar] [PubMed]
- Tilley, L.; Straimer, J.; Gnädig, N.F.; Ralph, S.A.; Fidock, D.A. Artemisinin Action and Resistance in Plasmodium falciparum. Trends. Parasitol. 2016, 32, 682–696. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jourdan, J.; Walz, A.; Matile, H.; Schmidt, A.; Wu, J.; Wang, X.; Dong, Y.; Vennerstrom, J.L.; Schmidt, R.S.; Wittlin, S.; et al. Stochastic Protein Alkylation by Antimalarial Peroxides. ACS Infect. Dis. 2019, 5, 2067–2075. [Google Scholar] [CrossRef] [PubMed]
- Embo-Ibouanga, A.W.; Nguyen, M.; Paloque, L.; Coustets, M.; Joly, J.P.; Augereau, J.M.; Vanthuyne, N.; Bikanga, R.; Coquin, N.; Robert, A.; et al. Hybrid Peptide-Alkoxyamine Drugs: A Strategy for the Development of a New Family of Antiplasmodial Drugs. Molecules 2024, 29, 1397. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Keita, A.; Franetich, J.F.; Carraz, M.; Valentin, L.; Bordessoules, M.; Baron, L.; Bigeard, P.; Dupuy, F.; Geay, V.; Tefit, M.; et al. Potent Antiplasmodial Derivatives of Dextromethorphan Reveal the Ent-Morphinan Pharmacophore of Tazopsine-Type Alkaloids. Pharmaceutics 2022, 14, 372. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Skorokhod, O.A.; Davalos-Schafler, D.; Gallo, V.; Valente, E.; Ulliers, D.; Notarpietro, A.; Mandili, G.; Novelli, F.; Persico, M.; Taglialatela-Scafati, O.; et al. Oxidative stress-mediated antimalarial activity of plakortin, a natural endoperoxide from the tropical sponge Plakortis simplex. Free Radic. Biol. Med. 2015, 89, 624–637. [Google Scholar] [CrossRef] [PubMed]
- Ismail, H.M.; Barton, V.; Phanchana, M.; Charoensutthivarakul, S.; Wong, M.H.; Hemingway, J.; Biagini, G.A.; O’Neill, P.M.; Ward, S.A. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc. Natl. Acad. Sci. USA 2016, 113, 2080–2085. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Talman, A.M.; Clain, J.; Duval, R.; Ménard, R.; Ariey, F. Artemisinin Bioactivity and Resistance in Malaria Parasites. Trends. Parasitol. 2019, 35, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Eckstein-Ludwig, U.; Webb, R.J.; Van Goethem, I.D.; East, J.M.; Lee, A.G.; Kimura, M.; O’Neill, P.M.; Bray, P.G.; Ward, S.A.; Krishna, S. Artemisinins target the SERCA of Plasmodium falciparum. Nature 2003, 424, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Woodley, C.M.; Amado, P.S.M.; Cristiano, M.L.S.; O’Neill, P.M. Artemisinin inspired synthetic endoperoxide drug candidates: Design, synthesis, and mechanism of action studies. Med. Res. Rev. 2021, 41, 3062–3095. [Google Scholar] [CrossRef] [PubMed]
- Noel, S.; Sharma, S.; Shankar, R.; Rath, S.K. Identification of differentially expressed genes after acute exposure to bulaquine (CDRI 80/53) in mice liver. Basic Clin. Pharmacol. Toxicol. 2008, 103, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Siciliano, G.; Di Paolo, V.; Rotili, D.; Migale, R.; Pedini, F.; Casella, M.; Camerini, S.; Dalzoppo, D.; Henderson, R.; Huijs, T.; et al. The Nitrobenzoxadiazole Derivative NBDHEX Behaves as Plasmodium falciparum Gametocyte Selective Inhibitor with Malaria Parasite Transmission Blocking Activity. Pharmaceuticals 2022, 15, 168. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Florens, L.; Washburn, M.P.; Raine, J.D.; Anthony, R.M.; Grainger, M.; Haynes, J.D.; Moch, J.K.; Muster, N.; Sacci, J.B.; Tabb, D.L.; et al. A proteomic view of the Plasmodium falciparum life cycle. Nature 2002, 419, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Florens, L.; Carucci, D.J.; Yates, J.R., 3rd. Proteomics in malaria. J. Proteome Res. 2004, 3, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Venkadesan, S.; Mohanty, D. Pf-Phospho: A machine learning-based phosphorylation sites prediction tool for Plasmodium proteins. Brief. Bioinform. 2022, 23, bbac249. [Google Scholar] [CrossRef] [PubMed]
- Gunalan, K.; Rowley, E.H.; Miller, L.H. A Way Forward for Culturing Plasmodium vivax. Trends. Parasitol. 2020, 36, 512–519. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Menkin-Smith, L.; Winders, W.T. Plasmodium vivax Malaria. In StatPearls [Internet]; Updated 17 July 2023; StatPearls Publishing: Treasure Island, FL, USA, January 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK538333/ (accessed on 3 May 2024).
- Molina-Franky, J.; Reyes, C.; Picón Jaimes, Y.A.; Kalkum, M.; Patarroyo, M.A. The Black Box of Cellular and Molecular Events of Plasmodium vivax Merozoite Invasion into Reticulocytes. Int. J. Mol. Sci. 2022, 23, 14528. [Google Scholar] [CrossRef] [PubMed]
- Divya, M.; Prabhu, S.R.; Satyamoorthy, K.; Saadi, A.V. Therapeutics through glycobiology: An approach for targeted elimination of malaria. Biologia 2023, 78, 1807–1811. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tran, P.N.; Brown, S.H.; Rug, M.; Ridgway, M.C.; Mitchell, T.W.; Maier, A.G. Changes in lipid composition during sexual development of the malaria parasite Plasmodium falciparum. Malar. J. 2016, 15, 73. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tokumasu, F.; Hayakawa, E.H.; Fukumoto, J.; Tokuoka, S.M.; Miyazaki, S. Creative interior design by Plasmodium falciparum: Lipid metabolism and the parasite’s secret chamber. Parasitol. Int. 2021, 83, 102369. [Google Scholar] [CrossRef] [PubMed]
- Spickett, C.M. Formation of Oxidatively Modified Lipids as the Basis for a Cellular Epilipidome. Front. Endocrinol. 2020, 11, 602771. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Adigun, R.A.; Malan, F.P.; Balogun, M.O.; October, N. Rational Optimization of Dihydropyrimidinone-Quinoline Hybrids as Plasmodium falciparum Glutathione Reductase Inhibitors. ChemMedChem 2022, 17, e202200034. [Google Scholar] [CrossRef] [PubMed]
- Shafi, S.; Gupta, S.; Jain, R.; Shoaib, R.; Munjal, A.; Maurya, P.; Kumar, P.; Kalam Najmi, A.; Singh, S. Tackling the emerging Artemisinin-resistant malaria parasite by modulation of defensive oxido-reductive mechanism via nitrofurantoin repurposing. Biochem. Pharmacol. 2023, 215, 115756. [Google Scholar] [CrossRef] [PubMed]
- Berneburg, I.; Peddibhotla, S.; Heimsch, K.C.; Haeussler, K.; Maloney, P.; Gosalia, P.; Preuss, J.; Rahbari, M.; Skorokhod, O.; Valente, E.; et al. An optimized dihydrodibenzothiazepine lead compound (SBI-0797750) as a potent and selective inhibitor of Plasmodium falciparum and P. vivax glucose 6-phosphate dehydrogenase 6-phosphogluconolactonase. Antimicrob. Agents Chemother. 2022, 66, e0210921. [Google Scholar] [CrossRef] [PubMed]
- Skorokhod, O.; Vostokova, E.; Gilardi, G. The role of P450 enzymes in malaria and other vector-borne infectious diseases. Biofactors 2024, 50, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.A.; Rathod, P.K. Plasmodium dihydroorotate dehydrogenase: A promising target for novel anti-malarial chemotherapy. Infect. Disord. Drug Targets 2010, 10, 226–239. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gehlot, P.; Vyas, V.K. Recent advances on patents of Plasmodium falciparum dihydroorotate dehydrogenase (PfDHODH) inhibitors as antimalarial agents. Expert Opin. Ther. Pat. 2023, 33, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Lehane, A.M.; Ridgway, M.C.; Baker, E.; Kirk, K. Diverse chemotypes disrupt ion homeostasis in the Malaria parasite. Mol. Microbiol. 2014, 94, 327–339, Erratum in Mol. Microbiol. 2015, 97, 179. [Google Scholar] [CrossRef] [PubMed]
- Santos, B.M.D.; Przyborski, J.M.; Garcia, C.R.S. Changes in K+ Concentration as a Signaling Mechanism in the Apicomplexa Parasites Plasmodium and Toxoplasma. Int. J. Mol. Sci. 2023, 24, 7276. [Google Scholar] [CrossRef] [PubMed]
- Scarpelli, P.H.; Pecenin, M.F.; Garcia, C.R.S. Intracellular Ca2+ Signaling in Protozoan Parasites: An Overview with a Focus on Mitochondria. Int. J. Mol. Sci. 2021, 22, 469. [Google Scholar] [CrossRef] [PubMed]
- Kayamba, F.; Faya, M.; Pooe, O.J.; Kushwaha, B.; Kushwaha, N.D.; Obakachi, V.A.; Nyamori, V.O.; Karpoormath, R. Lactate dehydrogenase and malate dehydrogenase: Potential antiparasitic targets for drug development studies. Bioorg. Med. Chem. 2021, 50, 116458. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Li, K.; Cai, Q.; Guo, J.; Yuan, M.; Wong, Y.H.; Walkinshaw, M.D.; Fothergill-Gilmore, L.A.; Michels, P.A.M.; Dedon, P.C.; et al. Pyruvate kinase from Plasmodium falciparum: Structural and kinetic insights into the allosteric mechanism. Biochem. Biophys. Res. Commun. 2020, 532, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Dillenberger, M.; Werner, A.-D.; Velten, A.-S.; Rahlfs, S.; Becker, K.; Fritz-Wolf, K. Structural Analysis of Plasmodium falciparum Hexokinase Provides Novel Information about Catalysis Due to a Plasmodium-Specific Insertion. Int. J. Mol. Sci. 2023, 24, 12739. [Google Scholar] [CrossRef] [PubMed]
- Veiga, M.I.; Peng, W.K. Rapid phenotyping towards personalized malaria medicine. Malar. J. 2020, 19, 68. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Williams, D.; Le Roch, K.G. Chapter 14-Genomics and precision medicine for malaria: A dream come true? In Genomic and Precision Medicine, 3rd ed.; Ginsburg, G.S., Willard, H.F., Tsalik, E.L., Woods, C.W., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 223–255. ISBN 9780128014967. [Google Scholar] [CrossRef]
- Meng, F.; Liang, Z.; Zhao, K.; Luo, C. Drug design targeting active posttranslational modification protein isoforms. Med. Res. Rev. 2021, 41, 1701–1750. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.H.; Chen, K.F.; Hao, B.B.; Tan, M.J. Proteomic characterization of post-translational modifications in drug discovery. Acta Pharmacol. Sin. 2022, 43, 3112–3129. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bah, A.; Forman-Kay, J.D. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, L.; Kashina, A. Post-translational Modifications of the Protein Termini. Front. Cell Dev. Biol. 2021, 9, 719590. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, W.; Zhang, J.; MacRaild, C.A.; Norton, R.S.; Anders, R.F.; Zhang, X. Modulation of the aggregation of an amyloidogenic sequence by flanking-disordered region in the intrinsically disordered antigen merozoite surface protein 2. Eur. Biophys. J. 2019, 48, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Ameri, M.; Nezafat, N.; Eskandari, S. The potential of intrinsically disordered regions in vaccine development. Expert Rev. Vaccines 2022, 21, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Niang, E.H.A.; Bassene, H.; Fenollar, F.; Mediannikov, O. Biological Control of Mosquito-Borne Diseases: The Potential of Wolbachia-Based Interventions in an IVM Framework. J. Trop. Med. 2018, 2018, 1470459. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mushtaq, I.; Sarwar, M.S.; Chaudhry, A.; Shah, S.A.H.; Ahmad, M.M. Updates on traditional methods for combating malaria and emerging Wolbachia-based interventions. Front. Cell. Infect. Microbiol. 2024, 14, 1330475. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yu, S.; Wang, J.; Luo, X.; Zheng, H.; Wang, L.; Yang, X.; Wang, Y. Transmission-Blocking Strategies Against Malaria Parasites During Their Mosquito Stages. Front. Cell. Infect. Microbiol. 2022, 12, 820650. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vandana, V.; Dong, S.; Sheth, T.; Sun, Q.; Wen, H.; Maldonado, A.; Xi, Z.; Dimopoulos, G. Wolbachia infection-responsive immune genes suppress Plasmodium falciparum infection in Anopheles stephensi. PLoS Pathog. 2024, 20, e1012145. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
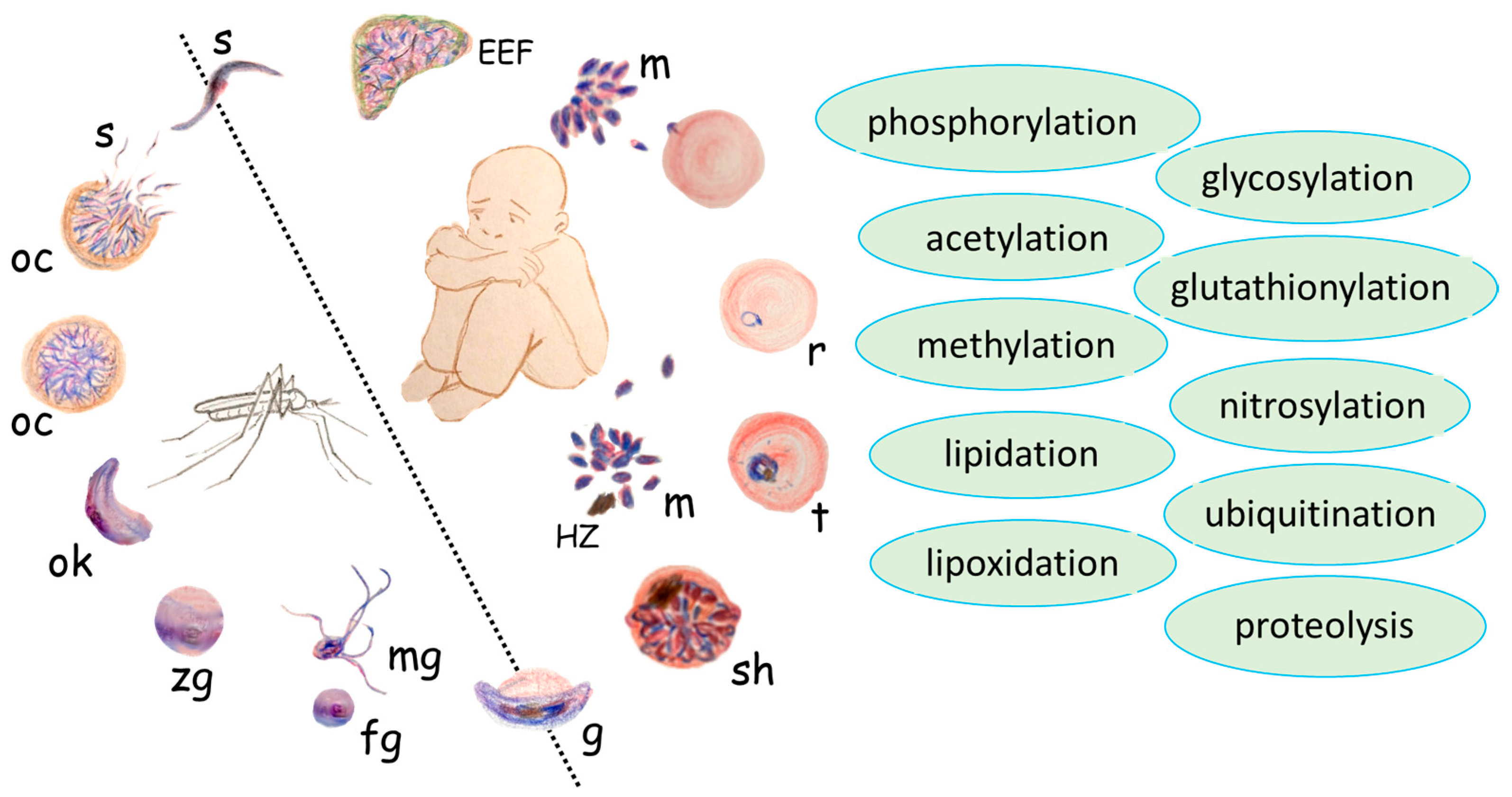
| Enzyme | Modified Protein | PTM | Targeted Function | Refs. |
|---|---|---|---|---|
| PbCDPK1 §, CDPK4, CDPK6 | NA | Phosphorylation | Development and hepatocyte invasion | [37] |
| P.f., P.b. CDPK4 NEK-2, NEK-4 MAP-2 * | Microtubule-associated proteins | Phosphorylation | Microtubule-associated functions, motility * | [15,38] |
| PbCDPK1 | MyoA | Phosphorylation | Gliding motility | [16,39] |
| PfCDPK-6 | NA | Phosphorylation | Sporozoite formation | [17,37] |
| PfeIK1, PbIK2 (eIK2) | eIF2alpha | Phosphorylation | Stress response, translational shutdown | [25,26,27,28] |
| P.b. UIS2 | eIF2alpha-P | Dephosphorylation | Activation of genes for invasion and in liver transformation | [26] |
| SERA-8 | CSP | Proteolysis | Protein cleavage/processing | [17] |
| P.f. serine protease | PfAMA-1, TRAP | Proteolysis | Protein shedding. Hepatocyte invasion | [30] |
| Papain family cysteine protease | PbCSP | Proteolysis | Hepatocyte invasion | [40,41] |
| PbAtg4 | PbAtg8 | Proteolysis | Protein processing, liver merozoite development | [32,33,42] |
| PfSET7 | P.f. H3K4, H3K9, H3K36 | Methylation | NA | [19] |
| Transferase * | P.f., P.v. CSP, TRAP | Glycosylation | Hepatocyte invasion | [21,43] |
| NA | PfAMA-1 | Glycosylation | Host immune recognition | [31] |
| PfPOFUT2 | PfCSP | O-fucosylation | Protein stabilization and trafficking, folding quality control | [23,44] |
| PfPOFUT2 | PfTRAP | O-fucosylation | Protein stabilization and trafficking, folding quality control * | [23] |
| PfUCHL3 | NA | Deubiquitination | Development | [35] |
| PbDHHC3 | P.b. inner membrane complex proteins | Palmitoylation | Gliding motility machinery | [36] |
| Enzyme | Modified Protein | PTM | Targeted Function | Refs. |
|---|---|---|---|---|
| PfPK9 | PfUBC13 | Phosphorylation | Parasite development | [46] |
| PbCDPK2 | NA | Phosphorylation | Liver schizont development | [37] |
| PbMLFK | Proteins exported to the host | Phosphorylation | Protein export, host response | [47] |
| P.f. Src kinases Lyn, Lck, CrkL | PfTRAP | Phosphorylation | Cell signaling, hepatocyte invasion | [49] |
| PfCLK3, CDK | NA | Phosphorylation | Development | [61,62] |
| PfPKG | P.f. proteasome RPT1 | Phosphorylation with consequent proteolysis | Proteasome processing | [45] |
| PbSUB1 | PbMSP1, MSP6, MSP7 | Proteolysis | Egress from hepatocytes | [50] |
| PbSERA-3 | PbCSP * | Proteolysis | Egress from hepatocytes | [63] |
| P.b. S2P protease | P.b. transcription factors | Proteolysis | Parasite development | [54] |
| P.b. biotin ligase | P.b. ACC | Biotinylation | Apicoplast metabolism | [51] |
| PbDHHC3 | P.b. inner membrane complex proteins | Palmitoylation | Liver invasion, egress | [36] |
| Enzyme | Modified Protein | PTM | Targeted Function | Refs. |
|---|---|---|---|---|
| PfPK4 | PfeIF2α | Phosphorylation | Environmental stress response | [74] |
| PfPK5 | PfORC1 | Phosphorylation | DNA replication, var gene regulation | [73] |
| Pfmrk and PfPK5 | NA | Phosphorylation | Cell cycle control, differentiation | [97] |
| PfPK6 | NA | Phosphorylation | Cell cycle control, differentiation of trophozoites/schizonts | [93] |
| PfPK7 | 1047 different P.f. proteins | Phosphorylation | Parasite development, ubiquitin/proteasome system | [70,71,72] |
| PfNEK-1, Aurora | NA | Phosphorylation | Development | [98,231] |
| PfCLK3 | NA | Phosphorylation | Development | [61] |
| P.f., P.b. PKG | NA | Phosphorylation | Late-stage schizont development | [10,76] |
| P.f. cAMP-PKA, GSK-3 | PfAMA1, glycogen synthase | Phosphorylation | Parasite motility, egress, and invasion | [11,67,111,232] |
| PfPKA, PKB | P.f. myosin A, GAP45, CDPK1 | Phosphorylation | Glideosome function | [11,81] |
| PfCDPK1 | PfGAP45 | Phosphorylation | Schizont development | [82] |
| PfCDPK1 | P.f. RhopH3 | Phosphorylation | Invasion | [104] |
| PfCDPK1, CDPK5, CDPK7 | PfMBP, MCP, NPT1, histone H2B * | Phosphorylation | Merozoite attachment, invasion | [27,233] |
| PfPDK1 | PfPKA, PI3K | Phosphorylation | Invasion | [9,83] |
| Host casein kinase II * | PfNAPL, EMP1 | Phosphorylation | Nucleosome assembly | [84,91] |
| PfCK1, PfCK2 | PfNAPS, histones, ALBAs | Phosphorylation | Chromatin dynamics | [86,87,88] |
| PfMap-2 | P.f. transcriptional regulators * | Phosphorylation | Schizogony | [99] |
| PfFIKK3, FIKK9.1, FIKK9.5, FIKK10.1, FIKK10.2 | Host erythrocyte spectrin, ankyrin, band-3 | Phosphorylation | Trafficking of parasite proteins to the host | [14,102,103] |
| PfSRPK1 | Splicing factor PfSR1 | Phosphorylation | mRNA splicing | [105] |
| PfCDPK * | PfCRT | Phosphorylation | Drug resistance | [168] |
| P.f. CaMK | NA | Phosphorylation | Development | [37] |
| NA | PfAtg proteins | Phosphorylation, lipidation, ubiquitination | Apicoplast maintenance, membrane trafficking of proteins | [187] |
| PfGCN5, PfMYST | P.f. H3K9ac, H3K4me2, H3K4me3, H3K9me3 | Acetylation, methylation | Var gene regulation | [8,118,119,120] |
| NA | P.f. 14-3-3, 20S proteasome beta subunit, PFK, ACS, ACT1, elongation factors 1 and 2, enolase, ApiAP2, ADA2 | Acetylation | Development | [9,112,234] |
| PfSIR2 (PfSIR2A, PfSIR2B), sirtuins, PfHDAC1, PfHDA2 | P.f. H3K9ac, H3K14ac, H4K16ac | Deacetylation | Silencing of genes, antigenic variation, rDNA transcription | [8,115,116,235] |
| PfPRMT1, PfPRMT5 | P.f. H4R3me, PfSmD1, other non-histone proteins | Methylation | Development | [122,127] |
| HKMTs: PfSET2, PfSET7, PfSET10 | P.f. H3K4me, H3K36me | Methylation | Var gene regulation | [19,123,124,125] |
| NA | PfROP14, RON3, TEX1, 6-cysteine protein (p12), rifin, PfEMP1, IMC 1g, 1c | Methylation | Invasion | [126] |
| NA | PfNOP1, ALBA1, DEAD/DEAH helicase, 40S ribosomal proteins, eIF2, NAPL, MCM4-7, SNF2 helicase, RAD50, RAB1-B, RAB11-A, RAB18, AP-1 | Methylation | RNA translation, chromatin organization, DNA replication, DNA repair, protein trafficking | [121] |
| PfLSD1, JHDM, PfJmjC1,2,3 | P.f. histones | Demethylation | Development | [128,129,235] |
| PfDPY19 | PfTRAP | C-mannosylation | NA | [236] |
| PfOGT | P.f. Hsp70 and α-tubulin | O-GlcNAc-glycosilation | Survival | [237] |
| P.f. falcipain-2, -3, falcilysin PfAPP | Host Hb, P.f. proteins | Proteolysis | Intraerythrocytic development, Hb digestion | [130,131,134,144,145] |
| PfPMs, HAP | Host Hb, PfEMP1 | Proteolysis | Protein degradation, export and exposure, merozoite egress | [130,141,142,143] |
| PfSPP, M18 aspartyl aminopeptidase | P.f. signal peptides | Proteolysis | Parasite invasion and growth | [146,147] |
| P.f. falcipain-1, DPAP3 | P.f. proteins | Proteolysis | invasion | [131,135] |
| PfSERA-4, -5, -6 | Parasite and host proteins | Proteolysis | Egress form erythrocyte | [7,63] |
| PfSUB2 * | PfAMA-1, MSP1, MSP7, TRAMP/PTTRAMP | Proteolysis | Invasion | [238,239] |
| PfPP1 | NA | Proteolysis | Glucose metabolism, DNA synthesis for segmentation | [107,108,240] |
| P.f. calcineurin | PfHSP90 * | Dephosphorylation, proteolysis | Late-stage schizonts growth. Invasion | [109,110] |
| Pf-calpain, subtilisin-1, -2, -3, | NA | Proteolysis | Calcium modulation, trophozoite development, invasion, egress | [7,136,137] |
| P.f. ROMs | PfTRAP, CTRP, MTRAP, PFF0800c, EBA-175, BAEBL, JESEBL, MAEBL, AMA1, Rh1, Rh2a, Rh2b, Rh4 | Proteolysis | Invasion, egress | [138,139,140] |
| P.f. S-nitrosylase * | 319 potential P.f. targets, GAPDH, PfTrx1 | S-nitrosylation | Carbohydrate metabolism, parasite growth | [153] |
| NA | PfEMP1, AMA1, MSP1, MSP2 | N-linked glycosylation | Development, invasion | [155,158,241,242] |
| NA | P.f. TrxR, Trx, Tpx1, GSR, GST, Plrx, mitochondrial LipDH, GDH1, Glo1/2, OAT, LDH, GAPDH, PK, PGM | S-glutathionylation | Protein functional inhibition | [159] |
| P.f. glutaredoxin 1, thioredoxin 1, plasmoredoxin | Same as above | Deglutathionylation | Protein activation | [159] |
| PfNMT | PfGAP45, ARO, others | Myristoylation | Development, egress, invasion | [8,160,164,175] |
| NA | PfGAP45, PfCRT, PfMDP1, ARO, MTIP, Alveolin, others | Palmitoylation | Development, egress, invasion | [162,163,164,168] |
| P. yoelii and P.f. DHHC2, DHHC7 | CDPK1, GAP45 | Palmitoylation | Rhoptry-related parasite invasion | [169,170,171] |
| PfFT, GGT | PfRabs, PfYkt6p, FCP, HSP40, others | Prenylation | Food vacuole functionality, others | [9,172,176,177,178,179,243] |
| P.f. ERAD enzymes: E1, E2, E3 | NA | Ubiquitination | Cell-cycle machinery | [34,188,189] |
| P.f., P.b. Ubc13 | NA | Ubiquitination | Development | [194,195] |
| NA | major subunit of RNA polymerase II, ApiAP2 | Ubiquitination | Development | [192] |
| NA | PfREX1 | Ubiquitination | Nutrient import in trophozoites | [191] |
| PfUCHL3 | NA | Deubiquitination | Parasite development | [35,193] |
| Enzyme | Modified Protein | PTM | Targeted Function | Refs. |
|---|---|---|---|---|
| P.f. and P.b. CDPK4, NEK-2, NEK-4, MAP-2 * | P.f. and P.b. microtubule-associated proteins | Phosphorylation | Male exflagellation, motility * | [15,38,100,101] |
| PfPKG | NA | Phosphorylation | Rounding shape, exflagellation | [77,264] |
| PfPKA | NA | Phosphorylation | Deformability | [83] |
| NA | Pfg27 | Phosphorylation | RNA oligomerization | [253] |
| PfCK2α | P.f. transcription factors, nuclear proteins * | Phosphorylation | Development | [88] |
| P.b. calcineurin | NA | Dephosphorylation, proteolysis | Male gametogenesis, fertilization | [109] |
| PfSUB2, PfDPAP2 | Pfg377 * | Proteolysis | Osmiophilic body function | [255] |
| PfGSK-3 | P.f. glycogen synthase | Proteolysis | Maturation | [232] |
| PfSUB2 | NA | Proteolysis | Egress from erythrocyte | [255] |
| P.f. plasmepsins PMX, PMV | NA | Proteolysis | Gametogenesis | [257,258] |
| NA | PfGEXPs | N-acetylation | Early-stages development | [256] |
| P.f. acetyltransferases, methyltransferases | P.f. H2A, H2A.Z, H2B, H2B.Z, H3, H3.3, H4 | Acetylation, methylation | Development | [246] |
| PfHDA2, PfSIR2 | NA | Deacetylation | Conversion by PfAP2-G | [115,116,261] |
| NA | P.f. H3BK112 | Ubiquitination | Development | [246] |
| PfDHHC9 | NA | Palmitoylation | Development | [262] |
| NA | P.f. protein phosphatase PPM2 | Acetylation, methylation, glutathionylation, nitrosylation palmitoylation, ubiquitination | Regulation of sex allocation | [12] |
| Enzyme | Modified Protein | PTM | Targeted Function | Refs. |
|---|---|---|---|---|
| PbCDPK1, CDPK2, CRK5 | NA | Phosphorylation | Development of the male gametes | [240,252,291] |
| PbARK2 | P.b. EB1, MISFIT, Myosin-K | Phosphorylation | Gametocyte spindle dynamics in chromosome segregation | [275] |
| P.f. Aurora B * | P.f. H3S10ph | Phosphorylation | PfHP1-related gametocyte differentiation | [266] |
| NA | P.b. HSP70, WD40 repeat protein msi1, enolase, actin-1, RNR, others | Phosphorylation | Gametocyte cytoskeleton, HSPs, DNA synthesis, signaling | [263,269] |
| PbSRPK | NA | Phosphorylation | Microgamete formation | [38] |
| PfCDPK1, CDPK2, CDPK3, CDPK4, CDPK6 | NA | Phosphorylation | Ookinete infectivity and development | [37] |
| PbPPM1 | NA | Dephosphorylation | Exflagellation | [12] |
| P.b. calcineurin | NA | Dephosphorylation, proteolysis | Gamete development, fertilization, ookinete-to-oocyst | [109] |
| PbSERA-3 | NA | Proteolysis | Male gametocyte egress | [63] |
| NA | P.f. H3K9ac, H3K14ac H3K18ac, H3K27ac, H3K56ac, H4K8ac, H4K16ac, H4ac4 | Acetylation | Gametocytes gene activation | [266,277] |
| NA | P.f. H2A.Z, H3K4me3, H3K9me3, H3K27me2, H3K36me3, H3K37me, H3R17mec, H4K20me | Methylation | Gametocyte differentiation | [266,277,278] |
| PfDRY19 | PfTRAP | C-mannosylation | Gametocyte egress and exflagellation | [292] |
| P.b. Skp1-Cullin1-FBXO1 complex in ubiquitin machinery | NA | Ubiquitination | Gametocyte development | [279] |
| NA | PfPCNA1 | Acetylation, methylation, glutathionylation, nitrosylation palmitoylation, ubiquitination | Male gametocyte development | [263] |
| NA | PfCAF-1 subunit C | Glutathionylation, nitrosylation, acetylation | Male gametocyte depositing histones on replicated DNA | [263,286] |
| CDPK4, others | P.f. MCM2-7 | Phosphorylation, methylation, acetylation, nitrosylation, ubiquitination | Male gametocyte development | [263] |
| NA | P.f. DNA polymerase, replication factor C, DNA ligase I, DNA topoisomerase II, ORC subunit 1 | Phosphorylation, acetylation | Male gametocyte development | [263] |
| NA | P.b. RBP: Alba 4, PABP1 | Acetylation, methylation, glutathionylation, nitrosylation, palmitoylation | Female gametocyte development | [263] |
| NA | P.b. RBP: DOZI, Musashi, Alba1-3, PABP 2/3, CITH; translational proteins | Numerous PTMs | Female gametocyte development | [263] |
| NA | P.f., P.b. aconitase, KDH | Palmitoylation | Gametocytes, oocysts mitochondrial TCA cycle | [263] |
| PbPPKL | NA | Dephosphorylation | Ookinete differentiation, motility | [276] |
| PfSUB, SOPT serine proteases * | NA | Proteolysis | Ookinete development | [273] |
| P.b. DHHC1, 2, 10 | NA | Palmitoylation | Ookinete development | [280,281,282] |
| PbMLFK, CK | NA | Phosphorylation | Sporozoite growth in oocyst | [47,272] |
| P. b. ATP-dependent protease | NA | Proteolysis | Sporozoite growth in oocyst | [272] |
| PbECP1, SERA-5 | PbCSP processing * | Proteolysis | Egress from oocyst | [29,63] |
| ROMs | P. f. adhesins | Proteolysis | Invasion, egresses | [140] |
| CDPK4 NEK-2, NEK-4 MAP-2 * | P.f., P.b. microtubule-associated proteins | Phosphorylation | Motility (sporozoites, ookinetes, oocysts) * | [15,38] |
| P.b., P.v. CDPK1 | P.b., P.v. MyoA, P.v. GAP40, GAP45, GAPM2, IMC proteins, | Phosphorylation | Motility (sporozoites, ookinetes) | [16,21,39] |
| NA | P.f., P.v. CSP, TRAP | Glycosylation | Invasion of salivary gland | [21,43] |
| DHHC3 | P.b. proteins form the inner membrane complex | Palmitoylation | Sporozoite gliding motility | [36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwarzer, E.; Skorokhod, O. Post-Translational Modifications of Proteins of Malaria Parasites during the Life Cycle. Int. J. Mol. Sci. 2024, 25, 6145. https://doi.org/10.3390/ijms25116145
Schwarzer E, Skorokhod O. Post-Translational Modifications of Proteins of Malaria Parasites during the Life Cycle. International Journal of Molecular Sciences. 2024; 25(11):6145. https://doi.org/10.3390/ijms25116145
Chicago/Turabian StyleSchwarzer, Evelin, and Oleksii Skorokhod. 2024. "Post-Translational Modifications of Proteins of Malaria Parasites during the Life Cycle" International Journal of Molecular Sciences 25, no. 11: 6145. https://doi.org/10.3390/ijms25116145
APA StyleSchwarzer, E., & Skorokhod, O. (2024). Post-Translational Modifications of Proteins of Malaria Parasites during the Life Cycle. International Journal of Molecular Sciences, 25(11), 6145. https://doi.org/10.3390/ijms25116145






