Long Noncoding RNAs in Response to Hyperosmolarity Stress, but Not Salt Stress, Were Mainly Enriched in the Rice Roots
Abstract
1. Introduction
2. Results
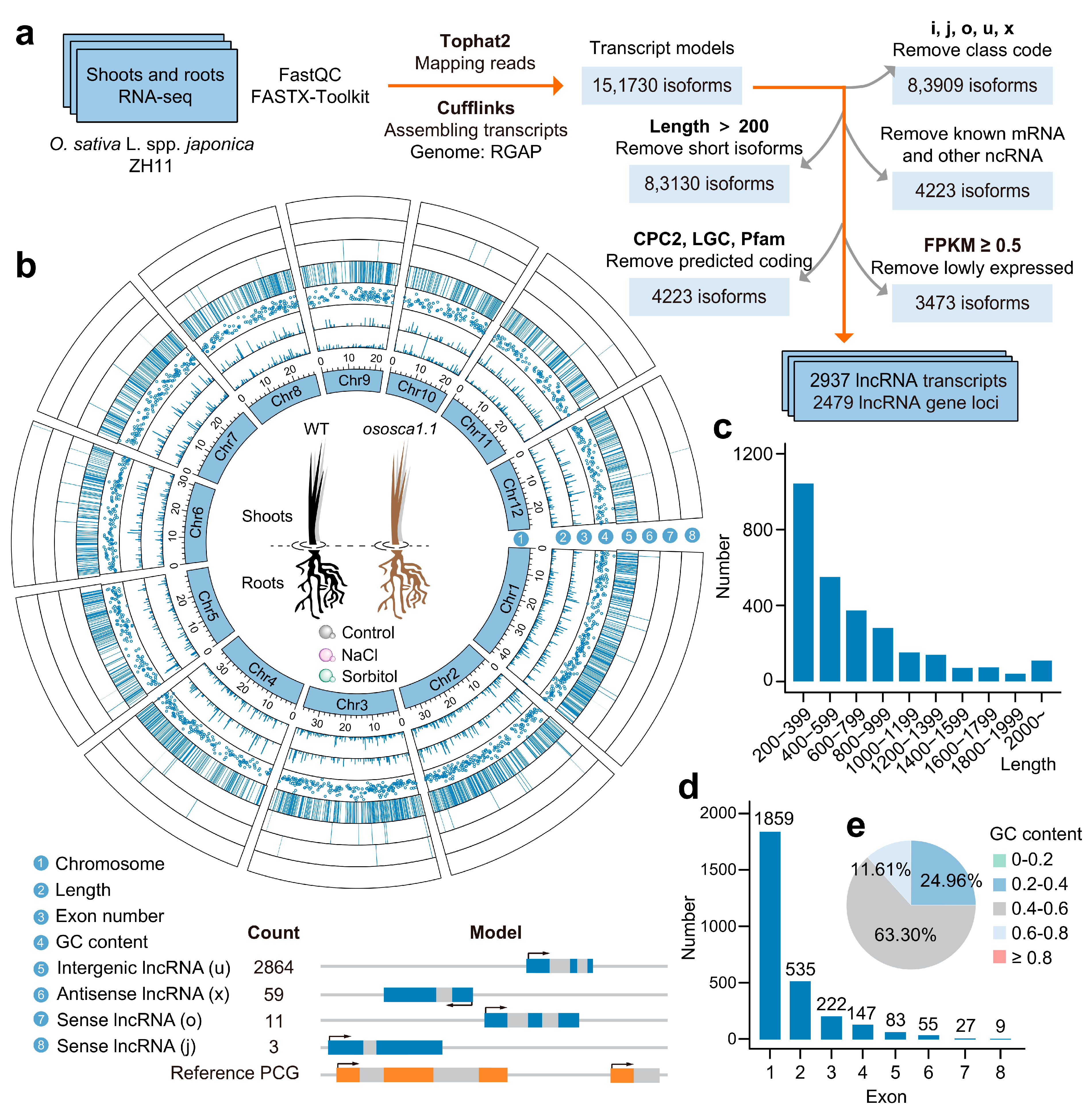
2.1. A Large Number of Intergenic lncRNAs Exist in Rice Shoots and Roots
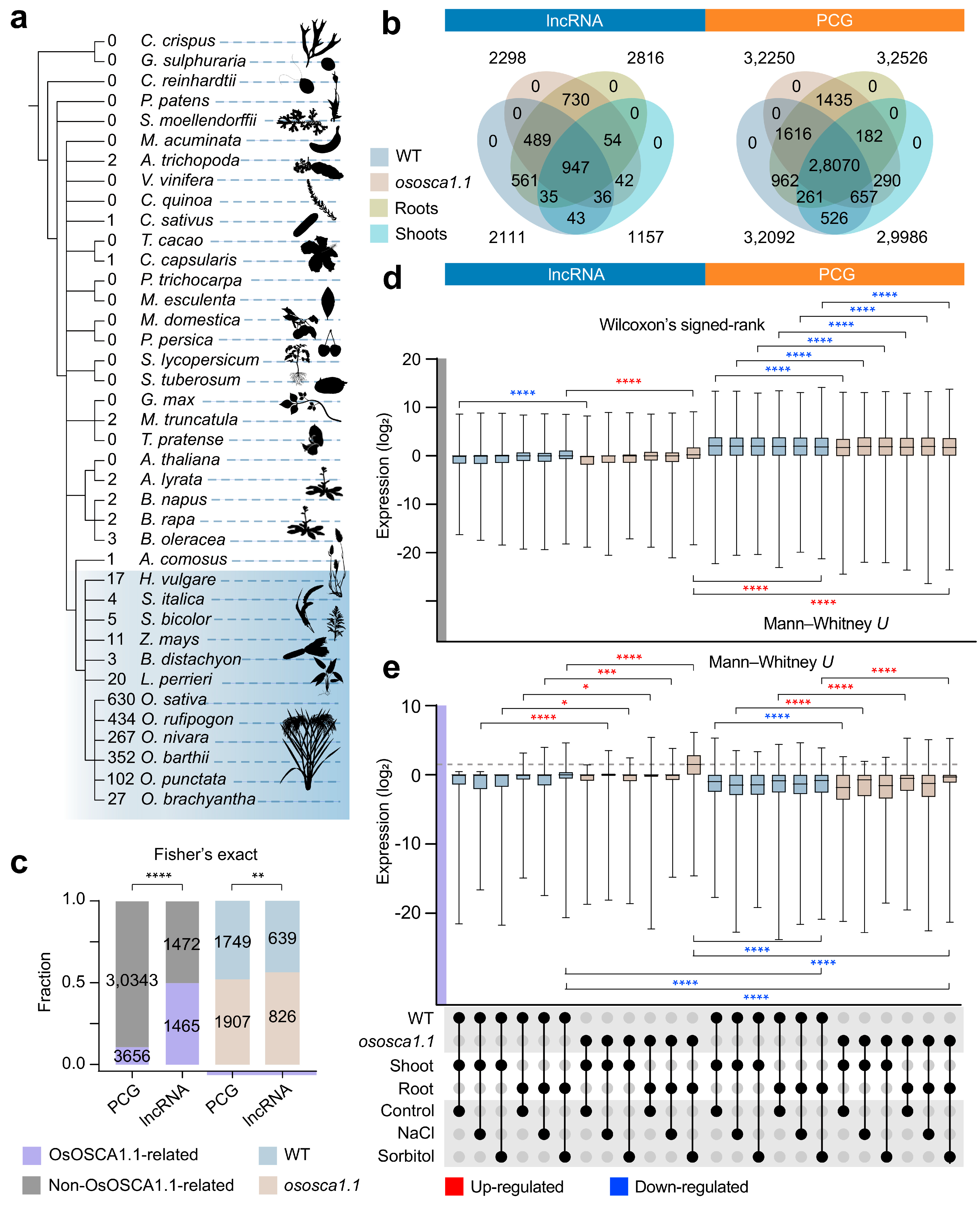
2.2. Transcriptional Levels of OsOSCA1.1-Related lncRNAs in Roots Are Greatly Enhanced under Hyperosmolality Stress
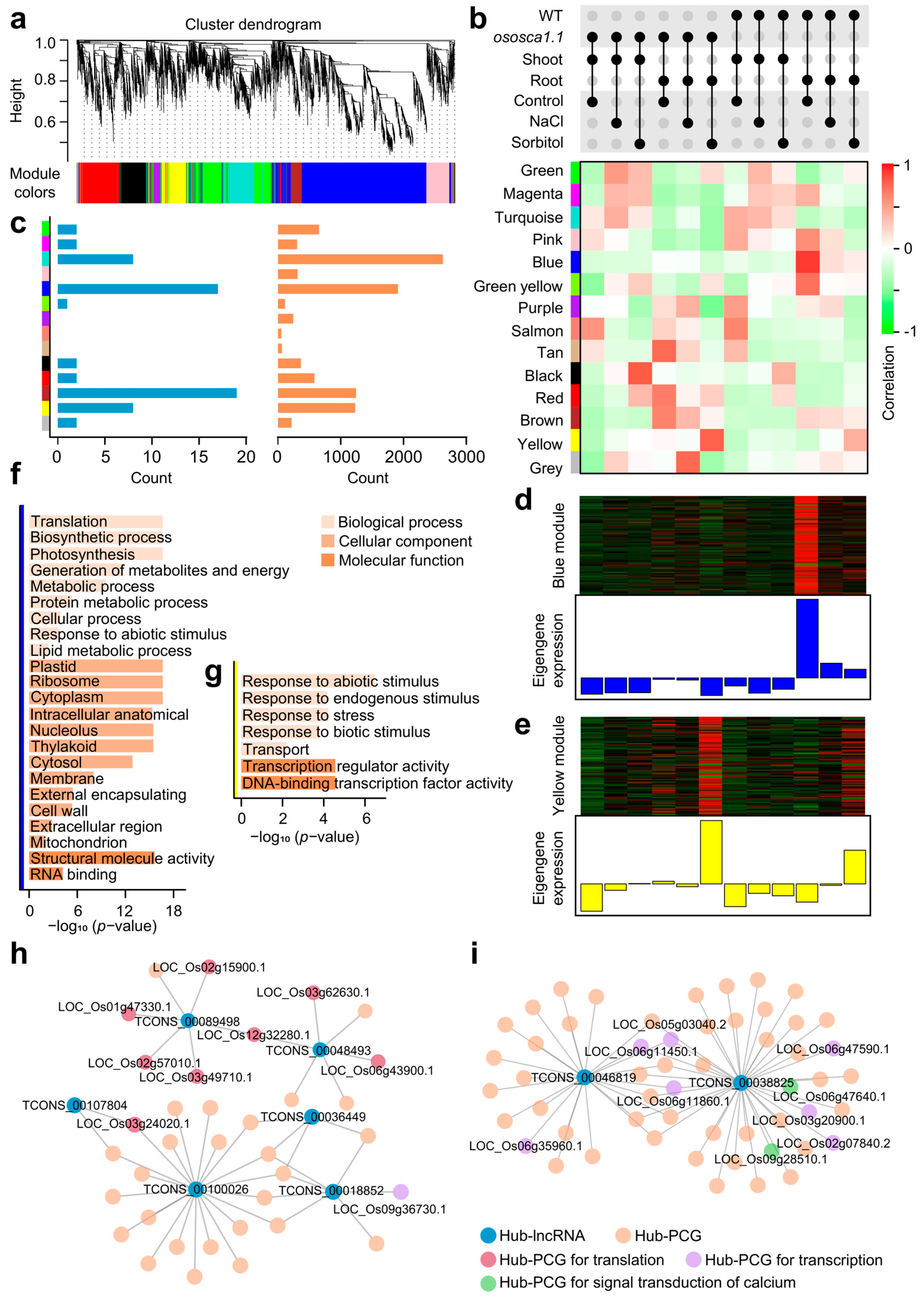
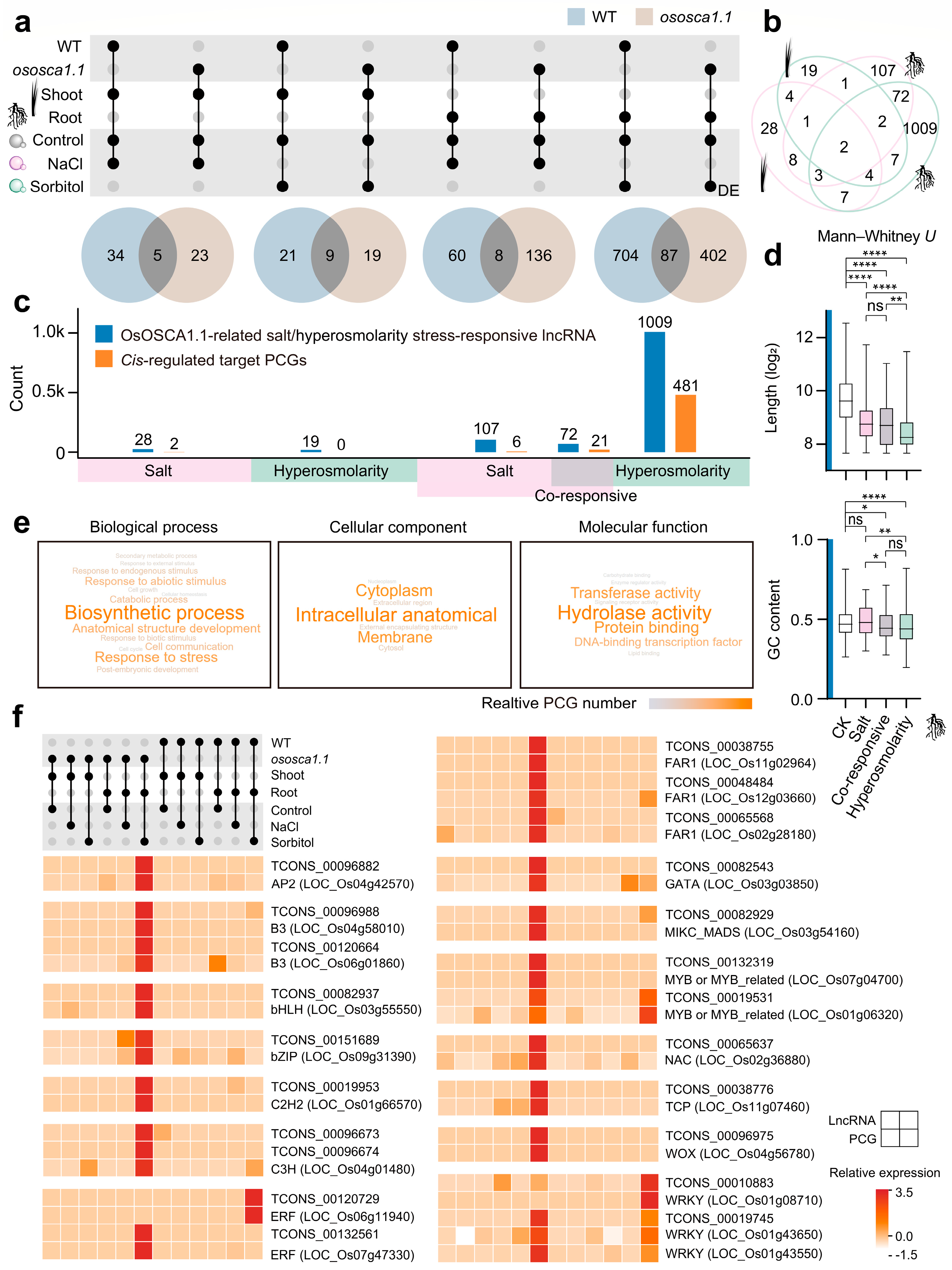
2.3. Co-Expression Network Points Out the Connection between lncRNAs and Their Trans-Regulated Target PCGs about OsOSCA1.1-Mediated Hyperosmolality Stress Sensing
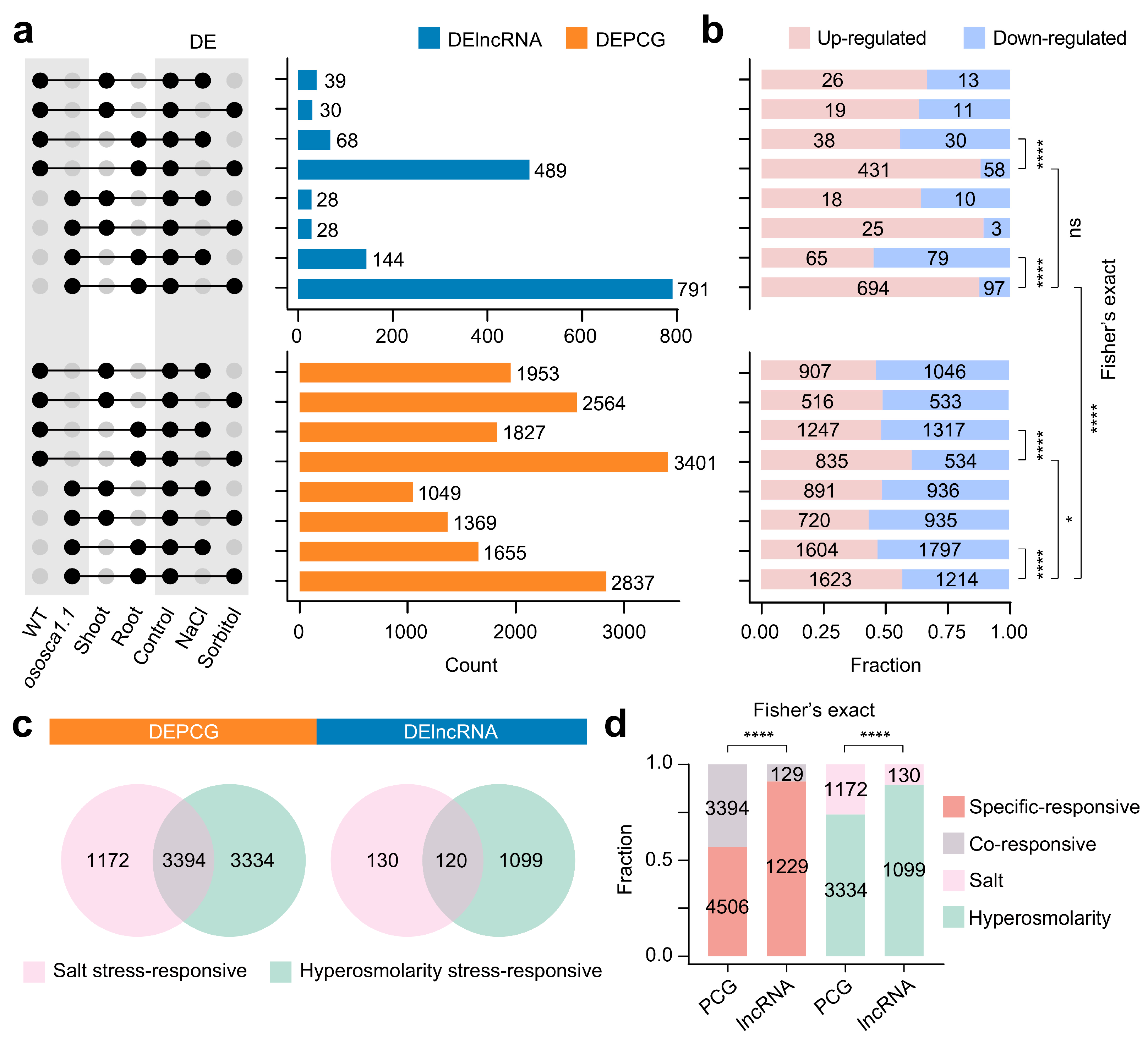
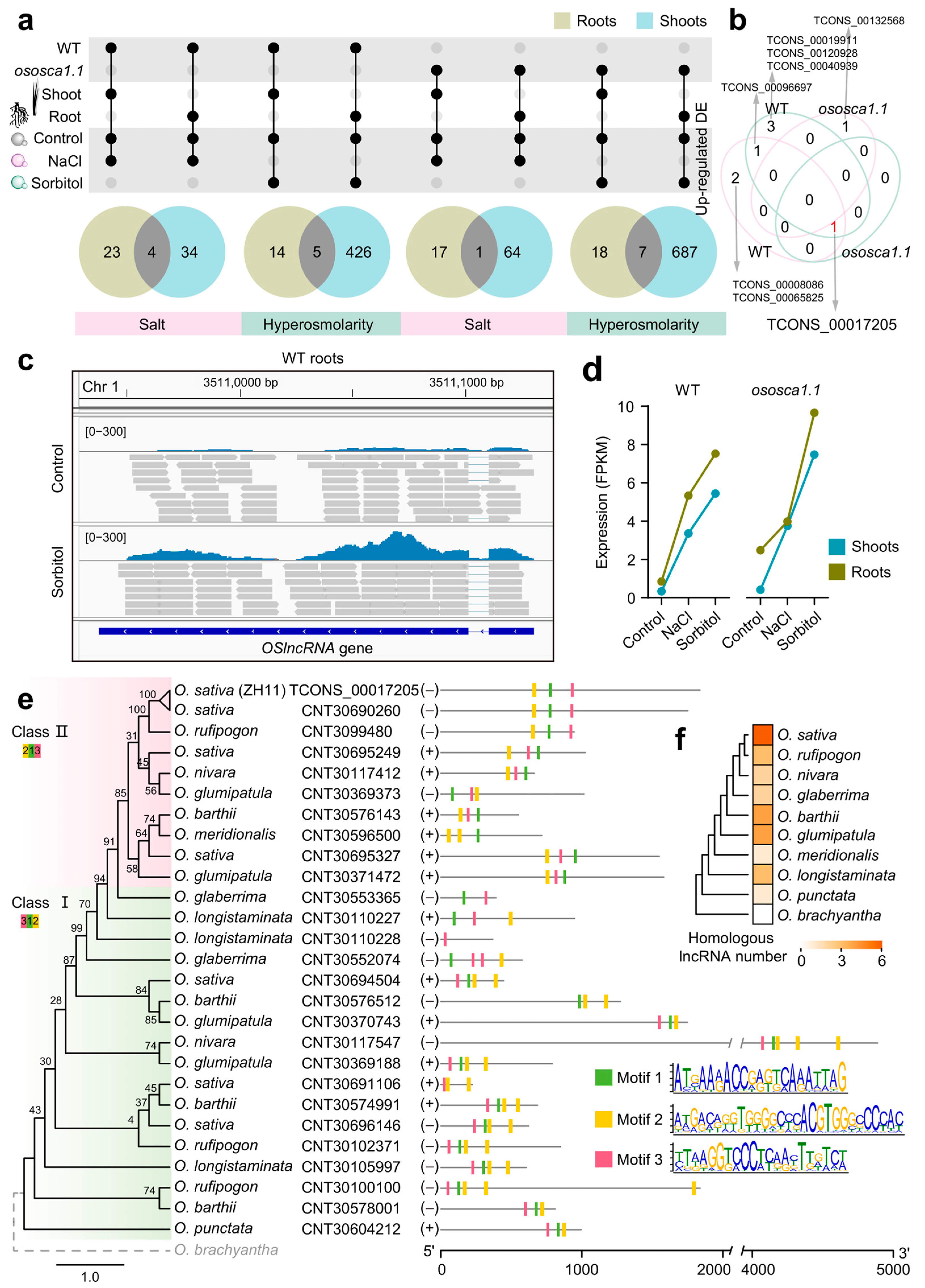
2.4. Compared with PCGs, lncRNAs Are More Sensitive to Hyperosmolarity Stress Than to Salt Stress in Rice Roots
2.5. OsOSCA1.1-Related Hyperosmolarity Stress-Responsive lncRNAs Are Enriched in Roots, and Their Potential cis-Regulated Genes Are Closely Related to Transcriptional Regulation and Signaling Transduction
2.6. An Oryza-Specific Hyperosmolarity Stress-Activated lncRNA Possesses Conserved Motif Architecture
3. Discussion
3.1. LncRNAs of cis- and trans-Regulation Enrich Regulatory Networks of Abiotic Stress
3.2. Evolutionary History of a Conserved and Hyperosmolarity Stress-Activated lncRNA Gene
3.3. Concluding Remarks and Future of Plant lncRNA Biology
4. Materials and Methods
4.1. Plant Materials
4.2. Transcriptome Assembly and lncRNA Identification
4.3. Basic Characteristics and Differential Expression Analysis of Transcripts
4.4. Prediction of Trans-Regulated Target PCGs of lncRNAs
4.5. Prediction of cis-Regulated Target Genes of DElncRNAs
4.6. Identification of Transcription Factors
4.7. Phylogenetic and Motif Structural Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sarma, B.; Kashtoh, H.; Lama Tamang, T.; Bhattacharyya, P.N.; Mohanta, Y.K.; Baek, K.H. Abiotic Stress in Rice: Visiting the Physiological Response and Its Tolerance Mechanisms. Plants 2023, 12, 3948. [Google Scholar] [CrossRef]
- Xie, Z.; Jin, L.; Sun, Y.; Zhan, C.; Tang, S.; Qin, T.; Liu, N.; Huang, J. OsNAC120 Balances Plant Growth and Drought Tolerance by Integrating GA and ABA Signaling in Rice. Plant Commun. 2024, 5, 100782. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Zhang, H.; Zhai, N.; Ma, X.; Zhou, H.; Cui, Y.; Wang, C.; Xu, G. Overexpression of OsRLCK241 Confers Enhanced Salt and Drought Tolerance in Transgenic Rice (Oryza sativa L.). Gene 2021, 768, 145278. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant Hormone-Mediated Regulation of Stress Responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Dong, Q.; Wallrad, L.; Almutairi, B.O.; Kudla, J. Ca2+ Signaling in Plant Responses to Abiotic Stresses. J. Integr. Plant Biol. 2022, 64, 287–300. [Google Scholar] [CrossRef]
- Ketehouli, T.; Nguyen Quoc, V.H.; Dong, J.; Do, H.; Li, X.; Wang, F. Overview of the Roles of Calcium Sensors in Plants’ Response to Osmotic Stress Signalling. Funct. Plant Biol. 2022, 49, 589–599. [Google Scholar] [CrossRef]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 Mediates Osmotic-Stress-Evoked Ca2+ Increases Vital for Osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.; Sun, L. Structure of the Hyperosmolality-Gated Calcium-Permeable Channel OSCA1.2. Nat. Commun. 2018, 9, 5060. [Google Scholar] [CrossRef]
- Maity, K.; Heumann, J.M.; McGrath, A.P.; Kopcho, N.J.; Hsu, P.K.; Lee, C.W.; Mapes, J.H.; Garza, D.; Krishnan, S.; Morgan, G.P.; et al. Cryo-EM Structure of OSCA1.2 from Oryza sativa Elucidates the Mechanical Basis of Potential Membrane Hyperosmolality Gating. Proc. Natl. Acad. Sci. USA 2019, 116, 14309–14318. [Google Scholar] [CrossRef]
- Chakraborty, S.; Gangwar, R.; Zahra, S.; Poddar, N.; Singh, A.; Kumar, S. Genome-wide Characterization and Comparative Analysis of the OSCA Gene Family and Identification of Its Potential Stress-Responsive Members in Legumes. Sci. Rep. 2023, 13, 5914. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, Y.; Ni, S.; Mu, J.; Liu, M.; Wang, Y.; Zhao, Y. Genome-wide Identification and Analysis of the OSCA Gene Family in Barley (Hordeum vulgare L.). J. Genet. 2022, 101, 34. [Google Scholar] [CrossRef]
- Tong, K.; Wu, X.; He, L.; Qiu, S.; Liu, S.; Cai, L.; Rao, S.; Chen, J. Genome-Wide Identification and Expression Profile of OSCA Gene Family Members in Triticum aestivum L. Int. J. Mol. Sci. 2022, 23, 469. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, F.; Wen, Z.; Li, Y.; Wang, F.; Zhu, T.; Zhuo, W.; Jin, X.; Wang, Y.; Zhao, H.; et al. Genome-wide Survey and Expression Analysis of the OSCA Gene Family in Rice. BMC Plant Biol. 2015, 15, 261. [Google Scholar] [CrossRef]
- Liu, C.; Wang, H.; Zhang, Y.; Cheng, H.; Hu, Z.; Pei, Z.M.; Li, Q. Systematic Characterization of the OSCA Family Members in Soybean and Validation of Their Functions in Osmotic Stress. Int. J. Mol. Sci. 2022, 23, 10570. [Google Scholar] [CrossRef]
- Han, Y.; Wang, Y.; Zhai, Y.; Wen, Z.; Liu, J.; Xi, C.; Zhao, H.; Wang, Y.; Han, S. OsOSCA1.1 Mediates Hyperosmolality and Salt Stress Sensing in Oryza sativa. Biology 2022, 11, 678. [Google Scholar] [CrossRef]
- Li, Q.; Wang, M.; Fang, L. BASIC PENTACYSTEINE2 Negatively Regulates Osmotic Stress Tolerance by Modulating LEA4-5 Expression in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 168, 373–380. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a BZIP Transcription Factor, Confers Salinity and Drought Tolerance in Rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef]
- Nefissi Ouertani, R.; Arasappan, D.; Abid, G.; Ben Chikha, M.; Jardak, R.; Mahmoudi, H.; Mejri, S.; Ghorbel, A.; Ruhlman, T.A.; Jansen, R.K. Transcriptomic Analysis of Salt-stress-responsive Genes in Barley Roots and Leaves. Int. J. Mol. Sci. 2021, 22, 8155. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant Noncoding RNAs: Hidden Players in Development and Stress Responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Palos, K.; Yu, L.; Railey, C.E.; Nelson Dittrich, A.C.; Nelson, A.D.L. Linking Discoveries, Mechanisms, and Technologies to Develop a Clearer Perspective on Plant Long Noncoding RNAs. Plant Cell 2023, 35, 1762–1786. [Google Scholar] [CrossRef]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xionga, L. A Nucleus-Localized Long Non-Coding RNA Enhances Drought and Salt Stress Tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Abe, M. Regulation of Reproductive Development by Non-Coding RNA in Arabidopsis: To Flower or Not to Flower. J. Plant Res. 2012, 125, 693–704. [Google Scholar] [CrossRef]
- Zhou, Y.F.; Zhang, Y.C.; Sun, Y.M.; Yu, Y.; Lei, M.Q.; Yang, Y.W.; Lian, J.P.; Feng, Y.Z.; Zhang, Z.; Yang, L.; et al. The Parent-of-Origin LncRNA MISSEN Regulates Rice Endosperm Development. Nat. Commun. 2021, 12, 6525. [Google Scholar] [CrossRef]
- Gao, C.; Zheng, X.; Li, H.; Ussi Ali, M.; Gao, Y.; Xiong, J. Roles of LncRNAs in Rice: Advances and Challenges. Rice Sci. 2020, 27, 384–395. [Google Scholar]
- Wang, Y.; Luo, X.; Sun, F.; Hu, J.; Zha, X.; Su, W.; Yang, J. Overexpressing LncRNA LAIR Increases Grain Yield and Regulates Neighbouring Gene Cluster Expression in Rice. Nat. Commun. 2018, 9, 3516. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Liao, J.Y.; Li, Z.Y.; Yu, Y.; Zhang, J.P.; Li, Q.F.; Qu, L.H.; Shu, W.S.; Chen, Y.Q. Genome-wide Screening and Functional Analysis Identify a Large Number of Long Noncoding RNAs Involved in the Sexual Reproduction of Rice. Genome Biol. 2014, 15, 512. [Google Scholar] [CrossRef]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide Analysis Uncovers Regulation of Long Intergenic Noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef]
- Wang, H.; Niu, Q.W.; Wu, H.W.; Liu, J.; Ye, J.; Yu, N.; Chua, N.H. Analysis of Non-Coding Transcriptome in Rice and Maize Uncovers Roles of Conserved LncRNAs Associated with Agriculture Traits. Plant J. 2015, 84, 404–416. [Google Scholar] [CrossRef]
- Palos, K.; Nelson Dittrich, A.C.; Yu, L.; Brock, J.R.; Railey, C.E.; Wu, H.Y.L.; Sokolowska, E.; Skirycz, A.; Hsu, P.Y.; Gregory, B.D.; et al. Identification and Functional Annotation of Long Intergenic Non-Coding RNAs in Brassicaceae. Plant Cell 2022, 34, 3233–3260. [Google Scholar] [CrossRef]
- Zheng, K.; Wu, X.; Xue, X.; Li, W.; Wang, Z.; Chen, J.; Zhang, Y.; Qiao, F.; Zhao, H.; Zhang, F.; et al. Transcriptome Screening of Long Noncoding RNAs and Their Target Protein-Coding Genes Unmasks a Dynamic Portrait of Seed Coat Coloration Associated with Anthocyanins in Tibetan Hulless Barley. Int. J. Mol. Sci. 2023, 24, 10587. [Google Scholar] [CrossRef]
- Jin, J.; Lu, P.; Xu, Y.; Li, Z.; Yu, S.; Liu, J.; Wang, H.; Chua, N.H.; Cao, P. PLncDB V2.0: A Comprehensive Encyclopedia of Plant Long Noncoding RNAs. Nucleic Acids Res. 2021, 49, D1489–D1495. [Google Scholar] [CrossRef]
- Yang, X.; Liu, C.; Niu, X.; Wang, L.; Li, L.; Yuan, Q.; Pei, X. Research on LncRNA Related to Drought Resistance of Shanlan Upland Rice. BMC Genom. 2022, 23, 336. [Google Scholar] [CrossRef]
- Mirdar Mansuri, R.; Azizi, A.H.; Sadri, A.H.; Shobbar, Z.S. Long Non-coding RNAs as the Regulatory Hubs in Rice Response to Salt Stress. Sci. Rep. 2022, 12, 21696. [Google Scholar] [CrossRef]
- Ponting, C.P. Biological Function in the Twilight Zone of Sequence Conservation. BMC Biol. 2017, 15, 71. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Lee, J.T. Epigenetic Regulation by Long Noncoding RNAs. Science 2012, 338, 1435–1439. [Google Scholar] [CrossRef]
- Nejat, N.; Mantri, N. Plant Immune System: Crosstalk between Responses to Biotic and Abiotic Stresses the Missing Link in Understanding Plant Defence. Curr. Issues Mol. Biol. 2017, 23, 1–16. [Google Scholar] [CrossRef]
- Zhang, P.; He, R.; Yang, J.; Cai, J.; Qu, Z.; Yang, R.; Gu, J.; Wang, Z.Y.; Adelson, D.L.; Zhu, Y.; et al. The Long Non-Coding RNA DANA2 Positively Regulates Drought Tolerance by Recruiting ERF84 to Promote JMJ29-Mediated Histone Demethylation. Mol. Plant 2023, 16, 1339–1353. [Google Scholar] [CrossRef]
- Walter, N.G. Are Non-protein Coding RNAs Junk or Treasure?: An Attempt to Explain and Reconcile Opposing Viewpoints of Whether the Human Genome is Mostly Transcribed into Non-Functional or Functional RNAs. BioEssays 2024, 46, 2300201. [Google Scholar] [CrossRef]
- Song, X.; Hu, J.; Wu, T.; Yang, Q.; Feng, X.; Lin, H.; Feng, S.; Cui, C.; Yu, Y.; Zhou, R.; et al. Comparative Analysis of Long Noncoding RNAs in Angiosperms and Characterization of Long Noncoding RNAs in Response to Heat Stress in Chinese Cabbage. Hortic. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Ulitsky, I. Evolution to the Rescue: Using Comparative Genomics to Understand Long Non-Coding RNAs. Nat. Rev. Genet. 2016, 17, 601–614. [Google Scholar] [CrossRef]
- Simopoulos, C.M.A.; Weretilnyk, E.A.; Golding, G.B. Molecular Traits of Long Non-Protein Coding RNAs from Diverse Plant Species Show Little Evidence of Phylogenetic Relationships. G3 Genes Genom. Genet. 2019, 9, 2511–2520. [Google Scholar] [CrossRef]
- Morris, K.V.; Mattick, J.S. The Rise of Regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef]
- Statello, L. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Boonburapong, B.; Buaboocha, T. Genome-wide Identification and Analyses of the Rice Calmodulin and Related Potential Calcium Sensor Proteins. BMC Plant Biol. 2007, 7, 4. [Google Scholar] [CrossRef][Green Version]
- McCormack, E.; Braam, J. Calmodulins and Related Potential Calcium Sensors of Arabidopsis. New Phytol. 2003, 159, 585–598. [Google Scholar] [CrossRef]
- Bergey, D.R.; Kandel, R.; Tyree, B.K.; Dutt, M.; Dhekney, S.A. The Role of Calmodulin and Related Proteins in Plant Cell Function: An Ever-Thickening Plot. Springer Sci. Rev. 2014, 2, 145–159. [Google Scholar] [CrossRef]
- Bazin, J.; Baerenfaller, K.; Gosai, S.J.; Gregory, B.D.; Crespi, M.; Bailey-Serres, J. Global Analysis of Ribosome-Associated Noncoding RNAs Unveils New Modes of Translational Regulation. Proc. Natl. Acad. Sci. USA 2017, 114, E10018–E10027. [Google Scholar] [CrossRef]
- Zeng, C.; Fukunaga, T.; Hamada, M. Identification and Analysis of Ribosome-Associated LncRNAs Using Ribosome Profiling Data. BMC Genom. 2018, 19, 414. [Google Scholar] [CrossRef]
- Yin, H.; Li, M.; Li, D.; Khan, S.A.; Hepworth, S.R.; Wang, S.M. Transcriptome Analysis Reveals Regulatory Framework for Salt and Osmotic Tolerance in a Succulent Xerophyte. BMC Plant Biol. 2019, 19, 88. [Google Scholar] [CrossRef]
- Chung, P.J.; Jung, H.; Jeong, D.H.; Ha, S.H.; Choi, Y.D.; Kim, J.K. Transcriptome Profiling of Drought Responsive Noncoding RNAs and Their Target Genes in Rice. BMC Genom. 2016, 17, 563. [Google Scholar] [CrossRef]
- Ding, Z.; Wu, C.; Tie, W.; Yan, Y.; He, G.; Hu, W. Strand-specific RNA-Seq Based Identification and Functional Prediction of LncRNAs in Response to Melatonin and Simulated Drought Stresses in Cassava. Plant Physiol. Biochem. 2019, 140, 96–104. [Google Scholar] [CrossRef]
- Pang, J.; Zhang, X.; Ma, X.; Zhao, J. Spatio-Temporal Transcriptional Dynamics of Maize Long Non-Coding RNAs Responsive to Drought Stress. Genes 2019, 10, 138. [Google Scholar] [CrossRef]
- Zhang, C.; Tang, G.; Peng, X.; Sun, F.; Liu, S.; Xi, Y. Long Non-Coding RNAs of Switchgrass (Panicum virgatum L.) in Multiple Dehydration Stresses. BMC Plant Biol. 2018, 18, 79. [Google Scholar] [CrossRef]
- Kumar, N.; Bharadwaj, C.; Sahu, S.; Shiv, A.; Shrivastava, A.K.; Reddy, S.P.P.; Soren, K.R.; Patil, B.S.; Pal, M.; Soni, A.; et al. Genome-wide Identification and Functional Prediction of Salt- Stress Related Long Non-Coding RNAs (LncRNAs) in Chickpea (Cicer arietinum L.). Physiol. Mol. Biol. Plants 2021, 27, 2605–2619. [Google Scholar] [CrossRef]
- Rehman, O.U.; Uzair, M.; Farooq, M.S.; Saleem, B.; Attacha, S.; Attia, K.A.; Farooq, U.; Fiaz, S.; El-Kallawy, W.H.; Kimiko, I.; et al. Comprehensive Insights into the Regulatory Mechanisms of LncRNA in Alkaline-Salt Stress Tolerance in Rice. Mol. Biol. Rep. 2023, 50, 7381–7392. [Google Scholar] [CrossRef]
- Deng, F.; Zhang, X.; Wang, W.; Yuan, R.; Shen, F. Identification of Gossypium hirsutum Long Non-Coding RNAs (LncRNAs) under Salt Stress. BMC Plant Biol. 2018, 18, 79. [Google Scholar] [CrossRef]
- Li, B.; Feng, C.; Zhang, W.; Sun, S.; Yue, D.; Zhang, X.; Yang, X. Comprehensive Non-Coding RNA Analysis Reveals Specific LncRNA/CircRNA–MiRNA–MRNA Regulatory Networks in the Cotton Response to Drought Stress. Int. J. Biol. Macromol. 2023, 253, 126558. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, H.; Xu, G.; Zhang, P.; Zhai, N.; Zheng, Q.; Liu, P.; Jin, L.; Bai, G.; Zhang, H. Genome-wide Analysis of Long Noncoding RNAs in Response to Salt Stress in Nicotiana tabacum. BMC Plant Biol. 2023, 23, 646. [Google Scholar] [CrossRef]
- Teng, W.; Liao, B.; Chen, M.; Shu, W. Genomic Legacies of Ancient Adaptation Illuminate GC-Content Evolution in Bacteria. Microbiol. Spectr. 2023, 11, e0214522. [Google Scholar] [CrossRef]
- Wan, S.; Zhang, Y.; Duan, M.; Huang, L.; Wang, W.; Xu, Q.; Yang, Y.; Yu, Y. Integrated Analysis of Long Non-Coding RNAs (LncRNAs) and mRNAs Reveals the Regulatory Role of LncRNAs Associated with Salt Resistance in Camellia sinensis. Front. Plant Sci. 2020, 11, 218. [Google Scholar] [CrossRef]
- Rnas, L.; Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long Non-Coding RNAs: Insights into Functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar]
- Tan, X.; Li, S.; Hu, L.; Zhang, C. Genome-wide Analysis of Long Non-Coding RNAs (LncRNAs) in Two Contrasting Rapeseed (Brassica napus L.) Genotypes Subjected to Drought Stress and Re-watering. BMC Plant Biol. 2020, 20, 81. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, Y.; Zou, C.; Yang, C.; Pan, G.; Ma, L.; Shen, Y. Integrated Analysis of Long Non-Coding RNAs and mRNAs Reveals the Regulatory Network of Maize Seedling Root Responding to Salt Stress. BMC Genom. 2022, 23, 50. [Google Scholar] [CrossRef]
- Ackah, M.; Jin, X.; Zhang, Q.; Amoako, F.K.; Wang, L.; Attaribo, T.; Zhao, M.; Yuan, F.; Herman, R.A.; Qiu, C.; et al. Long Noncoding RNA Transcriptome Analysis Reveals Novel LncRNAs in Morus Alba ‘Yu-711’ Response to Drought Stress. Plant Genome 2024, 17, e20273. [Google Scholar] [CrossRef]
- Traubenik, S.; Charon, C.; Blein, T. From Environmental Responses to Adaptation: The Roles of Plant LncRNAs. Plant Physiol. 2024, 195, 232–244. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, H.; Tian, S.; Chang, X.; Jing, R. TaSnRK2.4, an SNF1-Type Serine/Threonine Protein Kinase of Wheat (Triticum aestivum L.), Confers Enhanced Multistress Tolerance in Arabidopsis. J. Exp. Bot. 2010, 61, 683–696. [Google Scholar] [CrossRef]
- Wang, T.Z.; Liu, M.; Zhao, M.G.; Chen, R.; Zhang, W.H. Identification and Characterization of Long Non-Coding RNAs Involved in Osmotic and Salt Stress in Medicago truncatula Using Genome-Wide High-Throughput Sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef]
- Medina, C.A.; Samac, D.A.; Yu, L.X. Pan-Transcriptome Identifying Master Genes and Regulation Network in Response to Drought and Salt Stresses in Alfalfa (Medicago sativa L.). Sci. Rep. 2021, 11, 17203. [Google Scholar] [CrossRef]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of Long Noncoding RNA Evolution Derived from Direct Comparison of Transcriptomes in 17 Species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef]
- Prakash, P.T.; Chebotarov, D.; Zhang, J.; Kudrna, D.A.; Torres, R.O.; Natividad, M.A.; Quintana, M.R.; Song, J.; Maldonado, C.E.; Hechanova, S.L.; et al. Oryza glumaepatula: A Wild Relative to Improve Drought Tolerance in Cultivated Rice. Plant Physiol. 2023, 193, 2381–2397. [Google Scholar] [CrossRef]
- Bechara, S.T.; Kabbani, L.E.S.; Maurer-Alcalá, X.X.; Nowacki, M. Identification of Novel, Functional Long Non-Coding RNAs Involved in Programmed, Large-Scale Genome Rearrangements. RNA 2022, 28, 1110–1127. [Google Scholar] [CrossRef]
- Freeling, M. Bias in Plant Gene Content Following Different Sorts of Duplication: Tandem, Whole-genome, Segmental, or by Transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Zhang, Q.J.; Zhu, T.; Xia, E.H.; Shi, C.; Liu, Y.L.; Zhang, Y.; Liu, Y.; Jiang, W.K.; Zhao, Y.J.; Mao, S.Y.; et al. Rapid Diversification of Five Oryza AA Genomes Associated with Rice Adaptation. Proc. Natl. Acad. Sci. USA 2014, 111, E4954–E4962. [Google Scholar] [CrossRef]
- Baskaran, P.; Rödelsperger, C.; Prabh, N.; Serobyan, V.; Markov, G.V.; Hirsekorn, A.; Dieterich, C. Ancient Gene Duplications Have Shaped Developmental Stage-Specific Expression in Pristionchus pacificus. BMC Evol. Biol. 2015, 15, 185. [Google Scholar] [CrossRef]
- Jin, J.; Huang, W.; Gao, J.P.; Yang, J.; Shi, M.; Zhu, M.Z.; Luo, D.; Lin, H.X. Genetic Control of Rice Plant Architecture under Domestication. Nat. Genet. 2008, 40, 1365–1369. [Google Scholar] [CrossRef]
- Werner, A.; Kanhere, A.; Wahlestedt, C.; Mattick, J.S. Natural Antisense Transcripts as Versatile Regulators of Gene Expression. Nat. Rev. Genet. 2024. [Google Scholar] [CrossRef]
- Ulitsky, I.; Shkumatava, A.; Jan, C.H.; Sive, H.; Bartel, D.P. Conserved Function of LincRNAs in Vertebrate Embryonic Development despite Rapid Sequence Evolution. Cell 2011, 147, 1537–1550. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, Evolution, and Mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Li, J.; Zhang, Y.B.; Liu, X.; Cai, M.; Liu, M.; Li, M.; Ma, C.; Yue, R.; Zhu, Y.; et al. A Functional Motif of Long Noncoding RNA Nron against Osteoporosis. Nat. Commun. 2021, 12, 3319. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.J.; Ulitsky, I. Discovering Functional Motifs in Long Noncoding RNAs. Wiley Interdiscip. Rev. RNA 2022, 13, e1708. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Ferreira, B.T.; Yuan, Y. The Molecular Basis of Phenotypic Evolution: Beyond the Usual Suspects. Trends Genet. 2024. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, C.; Wu, C.; Xiong, L.; Chen, G.; Zhang, Q.; Wang, S. RMD: A Rice Mutant Database for Functional Analysis of the Rice Genome. Nucleic Acids Res. 2006, 34, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; De la Bastide, M.; Hamilton, J.P.; Kanamori, H.; Mccombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare Reference Genome Using next Generation Sequence and Optical Map Data. Rice 2013, 6, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Wang, G.; Yin, H.; Li, B.; Yu, C.; Wang, F.; Xu, X.; Cao, J.; Bao, Y.; Wang, L.; Abbasi, A.A.; et al. Characterization and Identification of Long Non-Coding RNAs Based on Feature Relationship. Bioinformatics 2019, 35, 2949–2956. [Google Scholar] [CrossRef]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.L.; et al. The Pfam Protein Families Database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Xia, R. A Painless Way to Customize Circos Plot: From Data Preparation to Visualization Using TBtools. iMeta 2022, 1, e35. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Szcześniak, M.W.; Rosikiewicz, W.; Makałowska, I. CANTATAdb: A Collection of Plant Long Non-Coding RNAs. Plant Cell Physiol. 2016, 57, e8. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhang, T.; Liang, Q.; Li, C.; Fu, S.; Kundu, J.K.; Zhou, X.; Wu, J. Transcriptome Analysis of Rice Reveals the LncRNA–mRNA Regulatory Network in Response to Rice Black-Streaked Dwarf Virus Infection. Viruses 2020, 12, 951. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A Free Online Platform for Data Visualization and Graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting Functional Regulatory Maps in Plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genome Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, Y.; Zheng, K.; Min, Q.; Wang, Y.; Xue, X.; Li, W.; Zhao, H.; Qiao, F.; Han, S. Long Noncoding RNAs in Response to Hyperosmolarity Stress, but Not Salt Stress, Were Mainly Enriched in the Rice Roots. Int. J. Mol. Sci. 2024, 25, 6226. https://doi.org/10.3390/ijms25116226
Pang Y, Zheng K, Min Q, Wang Y, Xue X, Li W, Zhao H, Qiao F, Han S. Long Noncoding RNAs in Response to Hyperosmolarity Stress, but Not Salt Stress, Were Mainly Enriched in the Rice Roots. International Journal of Molecular Sciences. 2024; 25(11):6226. https://doi.org/10.3390/ijms25116226
Chicago/Turabian StylePang, Yanrong, Kaifeng Zheng, Qinyue Min, Yinxing Wang, Xiuhua Xue, Wanjie Li, Heping Zhao, Feng Qiao, and Shengcheng Han. 2024. "Long Noncoding RNAs in Response to Hyperosmolarity Stress, but Not Salt Stress, Were Mainly Enriched in the Rice Roots" International Journal of Molecular Sciences 25, no. 11: 6226. https://doi.org/10.3390/ijms25116226
APA StylePang, Y., Zheng, K., Min, Q., Wang, Y., Xue, X., Li, W., Zhao, H., Qiao, F., & Han, S. (2024). Long Noncoding RNAs in Response to Hyperosmolarity Stress, but Not Salt Stress, Were Mainly Enriched in the Rice Roots. International Journal of Molecular Sciences, 25(11), 6226. https://doi.org/10.3390/ijms25116226






