Whole-Genome Sequencing and Analysis of Tumour-Forming Radish (Raphanus sativus L.) Line
Abstract
1. Introduction
2. Results
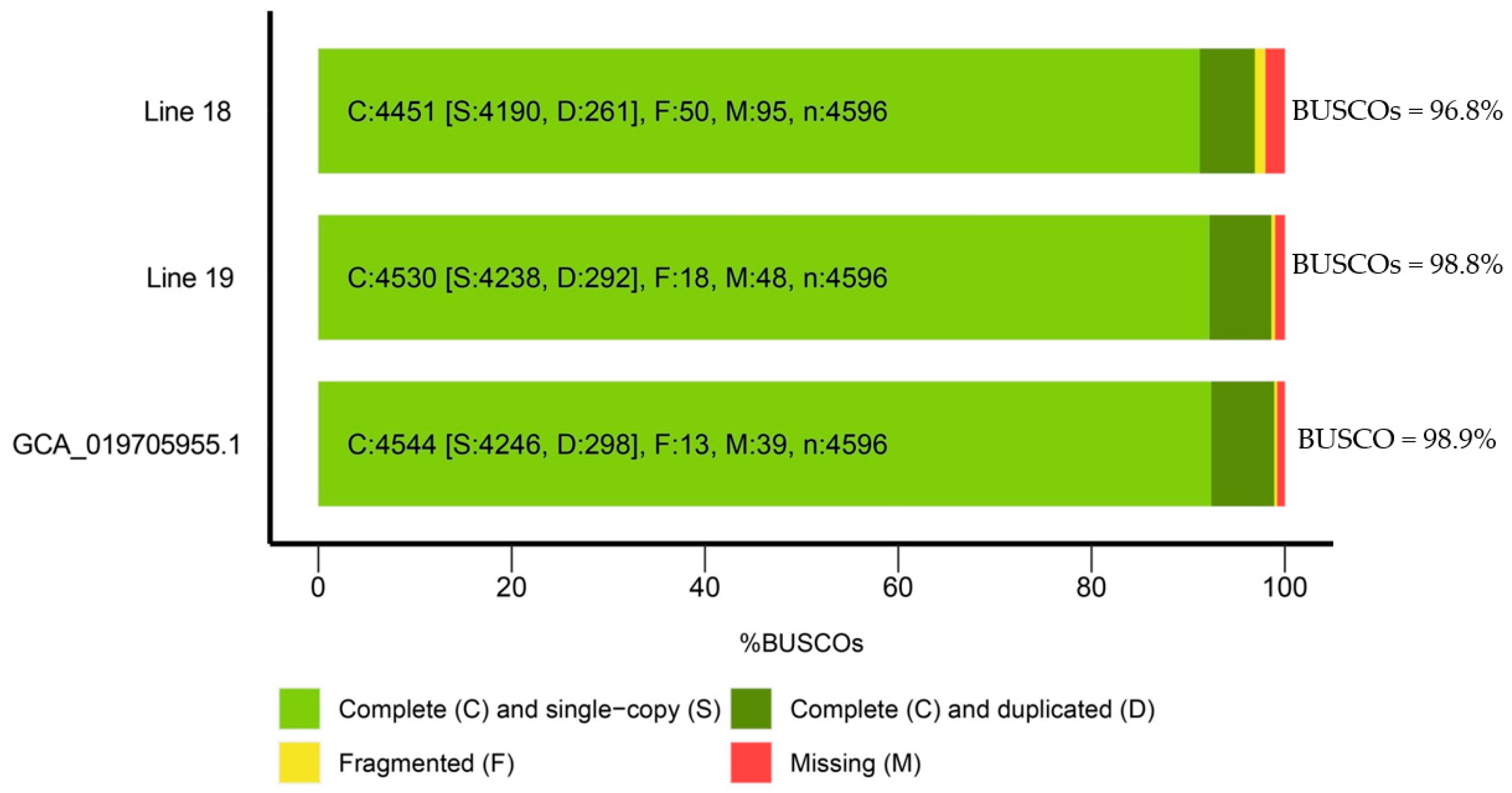
2.1. Assessment of the Assembly Quality of the Genomes of Two Radish Inbred Lines
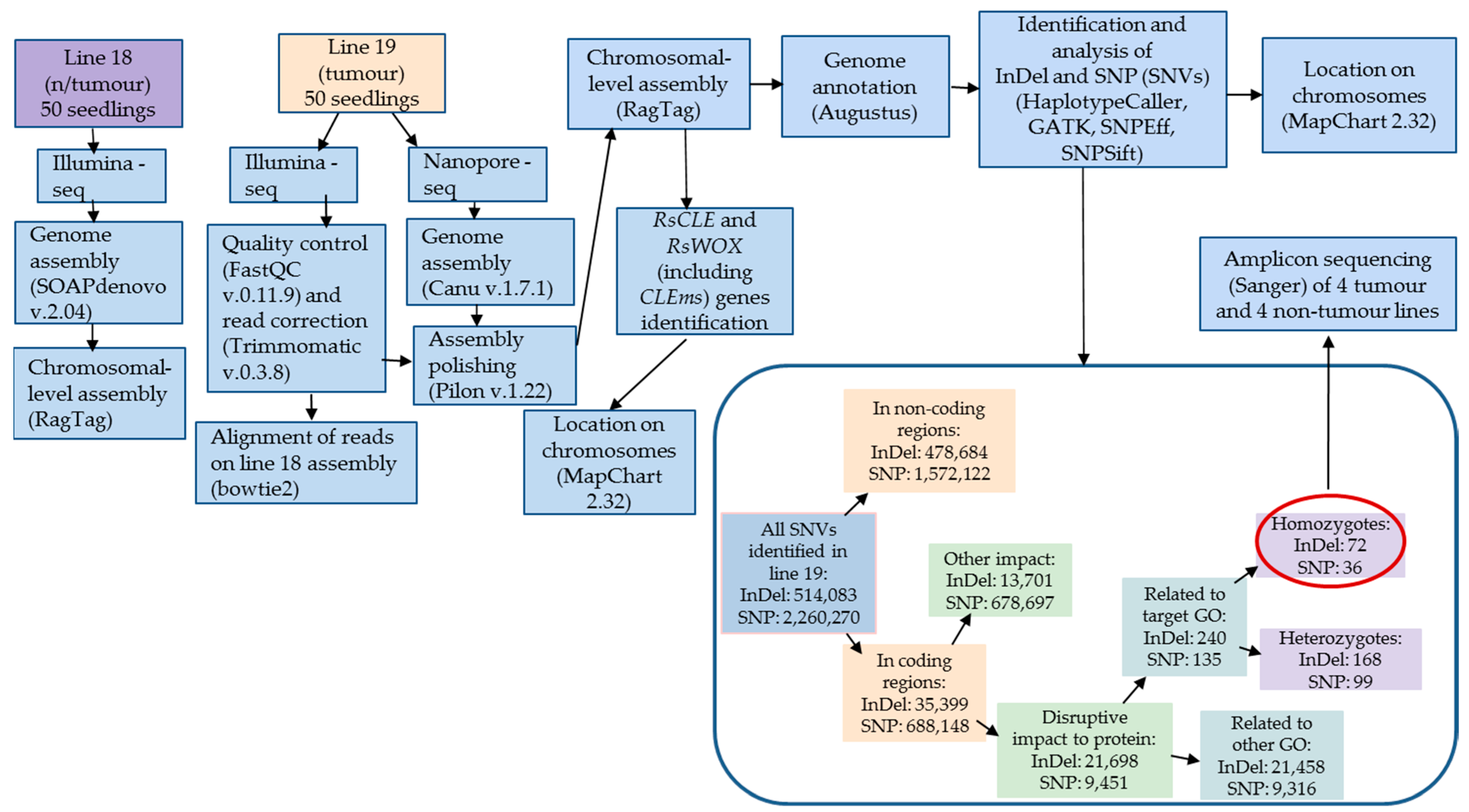
2.2. Identification of SNVs in the Protein-Coding Genes of the Tumour Radish Line
2.3. Search for the Presence of Identified SNVs in the Tumour and Non-Tumour Lines of the Radish Genetic Collection
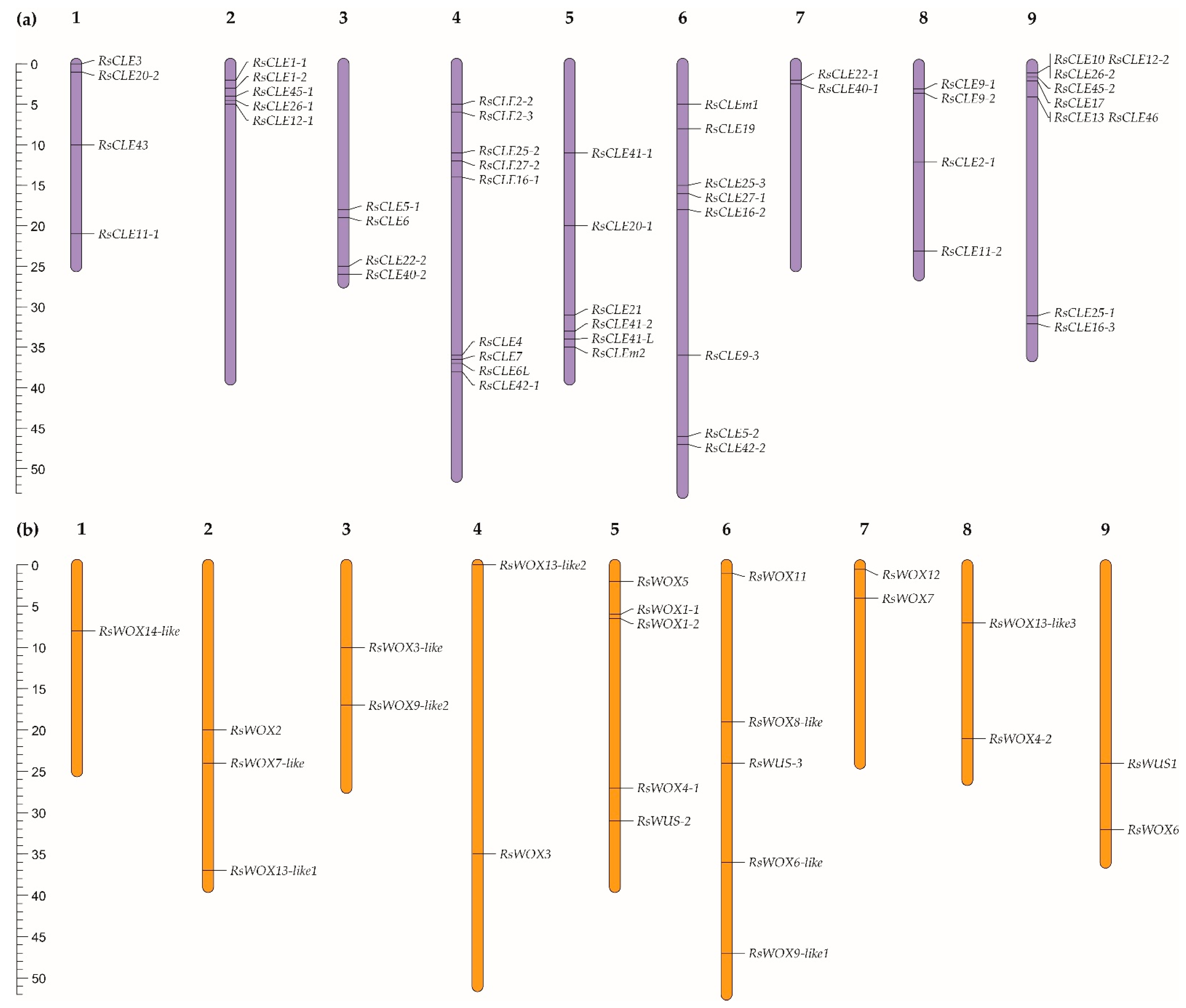
2.4. Identification and Chromosomal Localisation of WOX and CLE Genes in the Obtained Genome Assemblies of Inbred Radish Lines
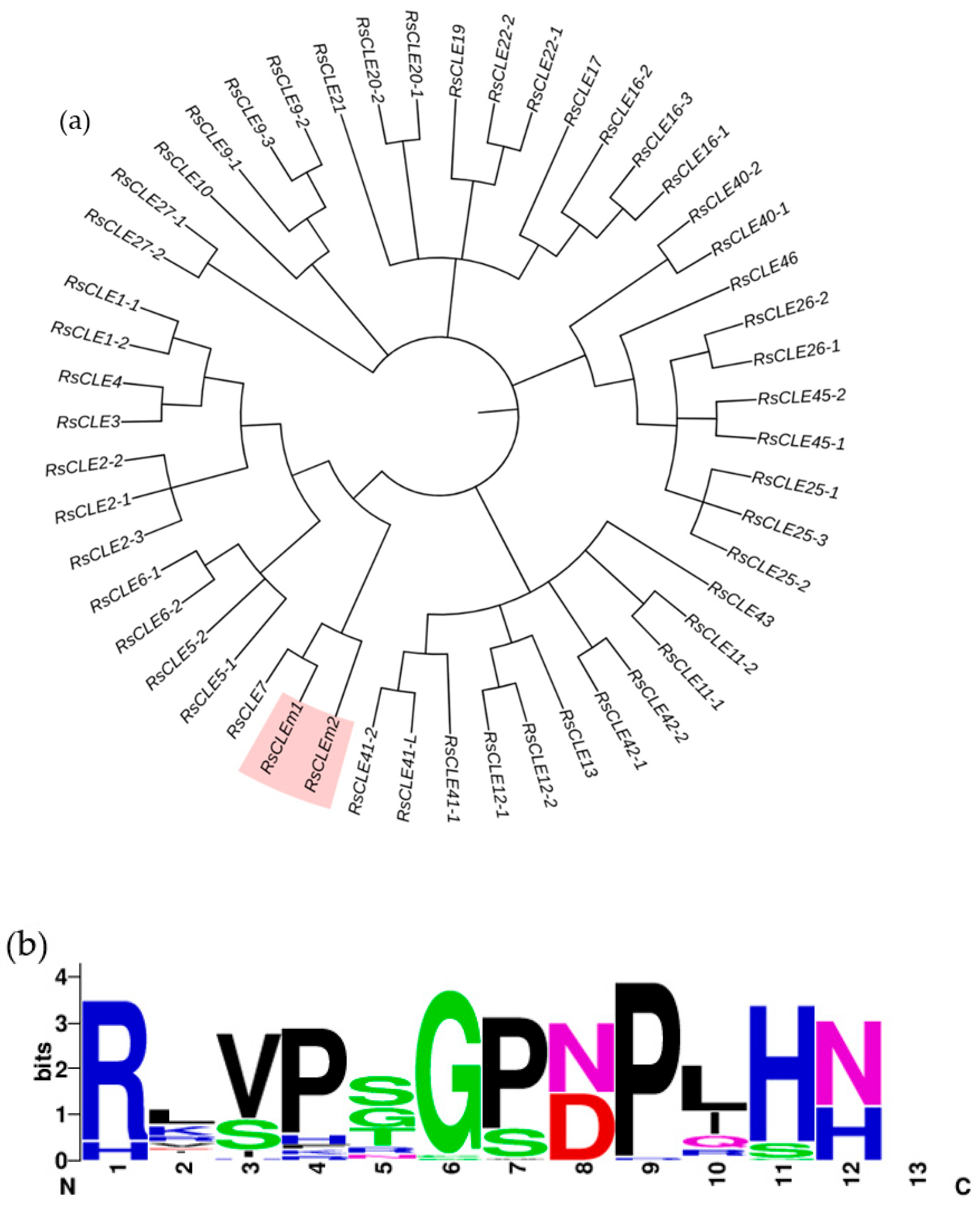
2.5. Identification of Radish CLE Genes Likely to Encode Proteins with Multiple CLE Domains
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Genomic DNA Isolation, Library Preparation, and Sequencing
4.3. Bioinformatic Processing of the Sequencing Results
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| InDel | Insertion or Deletion |
| SNP | Single Nucleotide Polymorphism |
| SNV | Single Nucleotide Variant |
References
- Doonan, J.H.; Sablowski, R. Walls around Tumours—Why Plants Do Not Develop Cancer. Nat. Rev. Cancer 2010, 10, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Harashima, H.; Sugimoto, K. Integration of Developmental and Environmental Signals into Cell Proliferation and Differentiation through RETINOBLASTOMA-RELATED 1. Curr. Opin. Plant Biol. 2016, 29, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Komaki, S.; Sugimoto, K. Control of the Plant Cell Cycle by Developmental and Environmental Cues. Plant Cell Physiol. 2012, 53, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Dodueva, I.E.; Lebedeva, M.A.; Kuznetsova, K.A.; Gancheva, M.S.; Paponova, S.S.; Lutova, L.L. Plant Tumors: A Hundred Years of Study. Planta 2020, 251, 82. [Google Scholar] [CrossRef] [PubMed]
- Krupková, E.; Immerzeel, P.; Pauly, M.; Schmülling, T. The TUMOROUS SHOOT DEVELOPMENT2 Gene of Arabidopsis Encoding a Putative Methyltransferase Is Required for Cell Adhesion and Co-Ordinated Plant Development. Plant J. 2007, 50, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Paredez, A.R.; Persson, S.; Ehrhardt, D.W.; Somerville, C.R. Genetic Evidence That Cellulose Synthase Activity Influences Microtubule Cortical Array Organization. Plant Physiol. 2008, 147, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Krupková, E.; Schmülling, T. Developmental Consequences of the Tumorous Shoot Development1 Mutation, a Novel Allele of the Cellulose-Synthesizing KORRIGAN1 Gene. Plant Mol. Biol. 2009, 71, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Iwai, H.; Masaoka, N.; Ishii, T.; Satoh, S. A Pectin Glucuronyltransferase Gene Is Essential for Intercellular Attachment in the Plant Meristem. Proc. Natl. Acad. Sci. USA 2002, 99, 16319–16324. [Google Scholar] [CrossRef] [PubMed]
- Buzovkina, I.S.; Lutova, L.A. The Genetic Collection of Radish Inbred Lines: History and Prospects. Russ. J. Genet. 2007, 43, 1181–1192. [Google Scholar] [CrossRef]
- Narbut, S.I. Genetic tumor in a radish obtained by inbreeding. Vestn. Leningr. Univ. Biol. 1967, 3, 144–149. [Google Scholar]
- Lebedeva (Osipova), M.A.; Tvorogova, V.E.; Vinogradova, A.P.; Gancheva, M.S.; Azarakhsh, M.; Ilina, E.L.; Demchenko, K.N.; Dodueva, I.E.; Lutova, L.A. Initiation of Spontaneous Tumors in Radish (Raphanus Sativus): Cellular, Molecular and Physiological Events. J. Plant Physiol. 2015, 173, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, A.; Dodueva, I.; Tvorogova, V.; Predeus, A.; Pravdina, O.; Kuznetsova, K.; Lutova, L. Transcriptomic Analysis of Radish (Raphanus Sativus L.) Spontaneous Tumor. Plants 2021, 10, 919. [Google Scholar] [CrossRef]
- Matveeva, T.V.; Frolova, N.V.; Smets, R.; Dodueva, I.E.; Buzovkina, I.S.; Van Onckelen, H.; Lutova, L.A. Hormonal Control of Tumor Formation in Radish. J. Plant Growth Regul. 2004, 23, 37–43. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. The Mechanism of Eukaryotic Translation Initiation and Principles of Its Regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Khan, M.H.U.; Yang, Y.; Zhu, K.; Li, H.; Zhu, M.; Amoo, O.; Khan, S.U.; Fan, C.; Zhou, Y. Identification and Comprehensive Analysis of the CLV3/ESR-Related (CLE) Gene Family in Brassica Napus L. Plant Biol. 2020, 22, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.-M.; Kim, N.; Ahn, B.O.; Oh, M.; Chung, W.-H.; Chung, H.; Jeong, S.; Lim, K.-B.; Hwang, Y.-J.; Kim, G.-B.; et al. Elucidating the Triplicated Ancestral Genome Structure of Radish Based on Chromosome-Level Comparison with the Brassica Genomes. Theor. Appl. Genet. 2016, 129, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, T.; Wang, J.; Wang, P.; Qiu, Y.; Zhao, W.; Pang, S.; Li, X.; Wang, H.; Song, J.; et al. Pan-Genome of Raphanus Highlights Genetic Variation and Introgression among Domesticated, Wild, and Weedy Radishes. Mol. Plant 2021, 14, 2032–2055. [Google Scholar] [CrossRef]
- Shirasawa, K.; Hirakawa, H.; Fukino, N.; Kitashiba, H.; Isobe, S. Genome Sequence and Analysis of a Japanese Radish (Raphanus Sativus) Cultivar Named ‘Sakurajima Daikon’ Possessing Giant Root. DNA Res. 2020, 27, dsaa010. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Provost, C.M.; Turner, S.R. Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling. PLoS Genet. 2012, 8, e1002997. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, G.; Jeon, E.; Sohn, E.J.; Lee, Y.; Kang, H.; Lee, D.W.; Kim, D.H.; Hwang, I. The Immediate Upstream Region of the 5′-UTR from the AUG Start Codon Has a Pronounced Effect on the Translational Efficiency in Arabidopsis thaliana. Nucleic Acids Res. 2014, 42, 485–498. [Google Scholar] [CrossRef]
- Kuznetsova, K.; Efremova, E.; Dodueva, I.; Lebedeva, M.; Lutova, L. Functional Modules in the Meristems: “Tinkering” in Action. Plants 2023, 12, 3661. [Google Scholar] [CrossRef] [PubMed]
- Tvorogova, V.E.; Krasnoperova, E.Y.; Potsenkovskaia, E.A.; Kudriashov, A.A.; Dodueva, I.E.; Lutova, L.A. What Does the WOX Say? Review of Regulators, Targets, Partners. Mol. Biol. 2021, 55, 362–391. [Google Scholar] [CrossRef] [PubMed]
- Song, X.-F.; Hou, X.-L.; Liu, C.-M. CLE Peptides: Critical Regulators for Stem Cell Maintenance in Plants. Planta 2021, 255, 5. [Google Scholar] [CrossRef] [PubMed]
- Poliushkevich, L.O.; Gancheva, M.S.; Dodueva, I.E.; Lutova, L.A. Receptors of CLE Peptides in Plants. Russ. J. Plant Physiol. 2020, 67, 1–16. [Google Scholar] [CrossRef]
- Aliaga Fandino, A.C.; Kim, H.; Rademaker, J.D.; Lee, J.-Y. Reprogramming of the Cambium Regulators during Adventitious Root Development upon Wounding of Storage Tap Roots in Radish (Raphanus Sativus L.). Biol. Open 2019, 8, bio039677. [Google Scholar] [CrossRef] [PubMed]
- Gancheva, M.S.; Dodueva, I.E.; Lebedeva, M.A.; Tvorogova, V.E.; Tkachenko, A.A.; Lutova, L.A. Identification, Expression, and Functional Analysis of CLE Genes in Radish (Raphanus Sativus L.) Storage Root. BMC Plant Biol. 2016, 16 (Suppl. S1), 7. [Google Scholar] [CrossRef] [PubMed]
- Kitashiba, H.; Li, F.; Hirakawa, H.; Kawanabe, T.; Zou, Z.; Hasegawa, Y.; Tonosaki, K.; Shirasawa, S.; Fukushima, A.; Yokoi, S.; et al. Draft Sequences of the Radish (Raphanus Sativus L.) Genome. DNA Res. 2014, 21, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Jeong, Y.-M.; Jeong, S.; Kim, G.-B.; Baek, S.; Kwon, Y.-E.; Cho, A.; Choi, S.-B.; Kim, J.; Lim, W.-J.; et al. Identification of Candidate Domestication Regions in the Radish Genome Based on High-Depth Resequencing Analysis of 17 Genotypes. Theor. Appl. Genet. 2016, 129, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, J.; Qiu, Y.; Wang, H.; Wang, P.; Zhang, X.; Li, C.; Song, J.; Gui, W.; Shen, D.; et al. SSR-Sequencing Reveals the Inter- and Intraspecific Genetic Variation and Phylogenetic Relationships among an Extensive Collection of Radish (Raphanus) Germplasm Resources. Biology 2021, 10, 1250. [Google Scholar] [CrossRef]
- Xu, L.; Wang, Y.; Dong, J.; Zhang, W.; Tang, M.; Zhang, W.; Wang, K.; Chen, Y.; Zhang, X.; He, Q.; et al. A Chromosome-Level Genome Assembly of Radish (Raphanus Sativus L.) Reveals Insights into Genome Adaptation and Differential Bolting Regulation. Plant Biotechnol. J. 2023, 21, 990–1004. [Google Scholar] [CrossRef]
- Park, H.Y.; Lim, Y.; Jung, M.; Sathiyamoorthy, S.; Heo, S.H.; Park, B.; Shin, Y. Genome of Raphanus Sativus L. Bakdal, an Elite Line of Large Cultivated Korean Radish. Front. Genet. 2024, 15, 1328050. [Google Scholar] [CrossRef]
- Xing, X.; Hu, T.; Wang, Y.; Li, Y.; Wang, W.; Hu, H.; Wei, Q.; Yan, Y.; Gan, D.; Bao, C.; et al. Construction of SNP Fingerprints and Genetic Diversity Analysis of Radish (Raphanus Sativus L.). Front. Plant Sci. 2024, 15, 1329890. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Shirasawa, K.; Fukino, N.; Hirakawa, H.; Akanuma, T.; Kitashiba, H. Identification of Genome-Wide Single-Nucleotide Polymorphisms among Geographically Diverse Radish Accessions. DNA Res. 2020, 27, dsaa001. [Google Scholar] [CrossRef]
- Smyczynski, C.; Roudier, F.; Gissot, L.; Vaillant, E.; Grandjean, O.; Morin, H.; Masson, T.; Bellec, Y.; Geelen, D.; Faure, J.-D. The C Terminus of the Immunophilin PASTICCINO1 Is Required for Plant Development and for Interaction with a NAC-like Transcription Factor. J. Biol. Chem. 2006, 281, 25475–25484. [Google Scholar] [CrossRef]
- Sieberer, T.; Hauser, M.-T.; Seifert, G.J.; Luschnig, C. PROPORZ1, a Putative Arabidopsis Transcriptional Adaptor Protein, Mediates Auxin and Cytokinin Signals in the Control of Cell Proliferation. Curr. Biol. 2003, 13, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Frémont, N.; Riefler, M.; Stolz, A.; Schmülling, T. The Arabidopsis TUMOR PRONE5 Gene Encodes an Acetylornithine Aminotransferase Required for Arginine Biosynthesis and Root Meristem Maintenance in Blue Light. Plant Physiol. 2013, 161, 1127–1140. [Google Scholar] [CrossRef]
- Smith, H.H. The Inheritance of Genetic Tumors in Nicotiana Hybrids. J. Hered. 1988, 79, 277–283. [Google Scholar] [CrossRef]
- Bellec, Y.; Harrar, Y.; Butaeye, C.; Darnet, S.; Bellini, C.; Faure, J.-D. Pasticcino2 Is a Protein Tyrosine Phosphatase-like Involved in Cell Proliferation and Differentiation in Arabidopsis. Plant J. 2002, 32, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Pavangadkar, K.A.; Thomashow, M.F.; Triezenberg, S.J. Physical and Functional Interactions of Arabidopsis ADA2 Transcriptional Coactivator Proteins with the Acetyltransferase GCN5 and with the Cold-Induced Transcription Factor CBF1. Biochim. Biophys. Acta 2006, 1759, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Provost, C.M.; Mishra, L.; Turner, S.R. WOX4 and WOX14 Act Downstream of the PXY Receptor Kinase to Regulate Plant Vascular Proliferation Independently of Any Role in Vascular Organisation. Development 2013, 140, 2224–2234. [Google Scholar] [CrossRef]
- Hassani, S.B.; Trontin, J.-F.; Raschke, J.; Zoglauer, K.; Rupps, A. Constitutive Overexpression of a Conifer WOX2 Homolog Affects Somatic Embryo Development in Pinus pinaster and Promotes Somatic Embryogenesis and Organogenesis in Arabidopsis Seedlings. Front. Plant Sci. 2022, 13, 838421. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Wang, X.; Ishida, T.; Grienenberger, E.; Zheng, Q.; Wang, J.; Zhang, Y.; Chen, W.; Chen, M.; Song, X.-F.; et al. A Group of CLE Peptides Regulates de Novo Shoot Regeneration in Arabidopsis thaliana. New Phytol. 2022, 235, 2300–2312. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Dexter, R.; Qualley, A.; Kish, C.M.; Ma, C.J.; Koeduka, T.; Nagegowda, D.A.; Dudareva, N.; Pichersky, E.; Clark, D. Characterization of a Petunia Acetyltransferase Involved in the Biosynthesis of the Floral Volatile Isoeugenol. Plant J. 2007, 49, 265–275. [Google Scholar] [CrossRef]
- Randall, R.S.; Miyashima, S.; Blomster, T.; Zhang, J.; Elo, A.; Karlberg, A.; Immanen, J.; Nieminen, K.; Lee, J.-Y.; Kakimoto, T.; et al. AINTEGUMENTA and the D-Type Cyclin CYCD3;1 Regulate Root Secondary Growth and Respond to Cytokinins. Biol. Open 2015, 4, 1229–1236. [Google Scholar] [CrossRef]
- Cromer, L.; Jolivet, S.; Horlow, C.; Chelysheva, L.; Heyman, J.; De Jaeger, G.; Koncz, C.; De Veylder, L.; Mercier, R. Centromeric Cohesion Is Protected Twice at Meiosis, by SHUGOSHINs at Anaphase I and by PATRONUS at Interkinesis. Curr. Biol. 2013, 23, 2090–2099. [Google Scholar] [CrossRef] [PubMed]
- Kono, A.; Umeda-Hara, C.; Lee, J.; Ito, M.; Uchimiya, H.; Umeda, M. Arabidopsis D-Type Cyclin CYCD4;1 Is a Novel Cyclin Partner of B2-Type Cyclin-Dependent Kinase. Plant Physiol. 2003, 132, 1315–1321. [Google Scholar] [CrossRef] [PubMed]
- Day, R.C.; Müller, S.; Macknight, R.C. Identification of Cytoskeleton-Associated Genes Expressed during Arabidopsis Syncytial Endosperm Development. Plant Signal. Behav. 2009, 4, 883–886. [Google Scholar] [CrossRef] [PubMed]
- Pradillo, M.; Knoll, A.; Oliver, C.; Varas, J.; Corredor, E.; Puchta, H.; Santos, J.L. Involvement of the Cohesin Cofactor PDS5 (SPO76) During Meiosis and DNA Repair in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1034. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Choi, H.-S.; Cho, H.-T. Root Hair-Specific EXPANSIN A7 Is Required for Root Hair Elongation in Arabidopsis. Mol. Cells 2011, 31, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xu, L.; Lin, H.; Cao, J. Two Expansin Genes, AtEXPA4 and AtEXPB5, Are Redundantly Required for Pollen Tube Growth and AtEXPA4 Is Involved in Primary Root Elongation in Arabidopsis thaliana. Genes 2021, 12, 249. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, Y.; Chang, J.; Zheng, F.; Pei, H.; Yi, Y.; Chang, C.; Dong, C.-H. Regulatory Function of Arabidopsis Lipid Transfer Protein 1 (LTP1) in Ethylene Response and Signaling. Plant Mol. Biol. 2016, 91, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.; Valdivia, E.R.; Fraga, P.; Iglesias, N.; Revilla, G.; Zarra, I. Soluble and Membrane-Bound β-Glucosidases Are Involved in Trimming the Xyloglucan Backbone. Plant Physiol. 2017, 173, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Coculo, D.; Lionetti, V. The Plant Invertase/Pectin Methylesterase Inhibitor Superfamily. Front. Plant Sci. 2022, 13, 863892. [Google Scholar] [CrossRef]
- Kong, Z.; Ioki, M.; Braybrook, S.; Li, S.; Ye, Z.-H.; Julie Lee, Y.-R.; Hotta, T.; Chang, A.; Tian, J.; Wang, G.; et al. Kinesin-4 Functions in Vesicular Transport on Cortical Microtubules and Regulates Cell Wall Mechanics during Cell Elongation in Plants. Mol. Plant 2015, 8, 1011–1023. [Google Scholar] [CrossRef]
- Heyndrickx, K.S.; Vandepoele, K. Systematic Identification of Functional Plant Modules through the Integration of Complementary Data Sources. Plant Physiol. 2012, 159, 884–901. [Google Scholar] [CrossRef] [PubMed]
- Dangarh, P.; Pandey, N.; Vinod, P.K. Modeling the Control of Meiotic Cell Divisions: Entry, Progression, and Exit. Biophys. J. 2020, 119, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, T.J.; Deeks, M.J.; Wang, P.; Hussey, P.J. The Evolution of the Actin Binding NET Superfamily. Front. Plant Sci. 2014, 5, 254. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Chen, Y.; Xu, Y.; Zhang, X.; Kang, Z.; Jiao, J.; Zhao, J. Interactional Similarities and Differences in the Protein Complex of PCNA and DNA Replication Factor C between Rice and Arabidopsis. BMC Plant Biol. 2019, 19, 257. [Google Scholar] [CrossRef]
- Pfluger, J.; Wagner, D. Histone Modifications and Dynamic Regulation of Genome Accessibility in Plants. Curr. Opin. Plant Biol. 2007, 10, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Breuer, C.; Stacey, N.J.; West, C.E.; Zhao, Y.; Chory, J.; Tsukaya, H.; Azumi, Y.; Maxwell, A.; Roberts, K.; Sugimoto-Shirasu, K. BIN4, a Novel Component of the Plant DNA Topoisomerase VI Complex, Is Required for Endoreduplication in Arabidopsis. Plant Cell 2007, 19, 3655–3668. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like Genes, MYB33 and MYB65, Are microRNA-Regulated Genes That Redundantly Facilitate Anther Development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef]
- Cao, X.; Yang, K.-Z.; Xia, C.; Zhang, X.-Q.; Chen, L.-Q.; Ye, D. Characterization of DUF724 Gene Family in Arabidopsis thaliana. Plant Mol. Biol. 2010, 72, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.M.; Masiero, S.; Nobre, M.S.; Costa, M.L.; Solís, M.-T.; Testillano, P.S.; Sprunck, S.; Coimbra, S. Differential Expression Patterns of Arabinogalactan Proteins in Arabidopsis thaliana Reproductive Tissues. J. Exp. Bot. 2014, 65, 5459–5471. [Google Scholar] [CrossRef]
- Ye, L.; Wang, X.; Lyu, M.; Siligato, R.; Eswaran, G.; Vainio, L.; Blomster, T.; Zhang, J.; Mähönen, A.P. Cytokinins Initiate Secondary Growth in the Arabidopsis Root through a Set of LBD Genes. Curr. Biol. 2021, 31, 3365–3373.e7. [Google Scholar] [CrossRef]
- Dong, L.; Wang, Z.; Liu, J.; Wang, X. AtSK11 and AtSK12 Mediate the Mild Osmotic Stress-Induced Root Growth Response in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 3991. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.L.; Kibble, N.A.J.; Bacic, A.; Schultz, C.J. A Fasciclin-like Arabinogalactan-Protein (FLA) Mutant of Arabidopsis thaliana, Fla1, Shows Defects in Shoot Regeneration. PLoS ONE 2011, 6, e25154. [Google Scholar] [CrossRef] [PubMed]
- Mantegazza, O.; Gregis, V.; Mendes, M.A.; Morandini, P.; Alves-Ferreira, M.; Patreze, C.M.; Nardeli, S.M.; Kater, M.M.; Colombo, L. Analysis of the Arabidopsis REM Gene Family Predicts Functions during Flower Development. Ann. Bot. 2014, 114, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Shimotohno, A.; Heidstra, R.; Blilou, I.; Scheres, B. Root Stem Cell Niche Organizer Specification by Molecular Convergence of PLETHORA and SCARECROW Transcription Factor Modules. Genes. Dev. 2018, 32, 1085–1100. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.; Sakamoto, S.; Mitsuda, N.; Suzuki, K.; Ohme-Takagi, M.; Fujiwara, S. WUSCHEL-RELATED HOMEOBOX 2 Is a Transcriptional Repressor Involved in Lateral Organ Formation and Separation in Arabidopsis. Plant Biotechnol. 2016, 33, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Zlobin, N.; Lebedeva, M.; Monakhova, Y.; Ustinova, V.; Taranov, V. An ERF121 Transcription Factor from Brassica oleracea Is a Target for the Conserved TAL-effectors from Different Xanthomonas campestris pv. campestris Strains. Mol. Plant Pathol. 2021, 22, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Horák, J.; Brzobohatý, B.; Lexa, M. Molecular and Physiological Characterisation of an Insertion Mutant in the ARR21 Putative Response Regulator Gene from Arabidopsis thaliana. Plant Biol. 2003, 5, 245–254. [Google Scholar] [CrossRef]
- Cui, H.; Kong, D.; Liu, X.; Hao, Y. SCARECROW, SCR-LIKE 23 and SHORT-ROOT Control Bundle Sheath Cell Fate and Function in Arabidopsis Thaliana. Plant J. 2014, 78, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Lakehal, A.; Dob, A.; Rahneshan, Z.; Novák, O.; Escamez, S.; Alallaq, S.; Strnad, M.; Tuominen, H.; Bellini, C. ETHYLENE RESPONSE FACTOR 115 Integrates Jasmonate and Cytokinin Signaling Machineries to Repress Adventitious Rooting in Arabidopsis. New Phytol. 2020, 228, 1611–1626. [Google Scholar] [CrossRef]
- Jorgensen, R.A.; Dorantes-Acosta, A.E. Conserved Peptide Upstream Open Reading Frames Are Associated with Regulatory Genes in Angiosperms. Front. Plant Sci. 2012, 3, 191. [Google Scholar] [CrossRef]
- Zhao, C.; Beers, E.P. Alternative Splicing of Myb-Related Genes MYR1 and MYR2 May Modulate Activities through Changes in Dimerization, Localization, or Protein Folding. Plant Signal. Behav. 2013, 8, e27325. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Song, L.; Zhang, Y.; Zheng, Z.; Liu, D. Arabidopsis PHL2 and PHR1 Act Redundantly as the Key Components of the Central Regulatory System Controlling Transcriptional Responses to Phosphate Starvation1. Plant Physiol. 2016, 170, 499–514. [Google Scholar] [CrossRef]
- Sun, Z.; Guo, T.; Liu, Y.; Liu, Q.; Fang, Y. The Roles of Arabidopsis CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs. PLoS Genet. 2015, 11, e1005598. [Google Scholar] [CrossRef] [PubMed]
- Zubo, Y.O.; Blakley, I.C.; Yamburenko, M.V.; Worthen, J.M.; Street, I.H.; Franco-Zorrilla, J.M.; Zhang, W.; Hill, K.; Raines, T.; Solano, R.; et al. Cytokinin Induces Genome-Wide Binding of the Type-B Response Regulator ARR10 to Regulate Growth and Development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E5995–E6004. [Google Scholar] [CrossRef] [PubMed]
- Lilay, G.H.; Castro, P.H.; Campilho, A.; Assunção, A.G.L. The Arabidopsis bZIP19 and bZIP23 Activity Requires Zinc Deficiency—Insight on Regulation From Complementation Lines. Front. Plant Sci. 2018, 9, 1955. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Wang, Q.; Mao, H.; Xu, J.; Wang, Y.; Hu, H.; He, S.; Tu, J.; Cheng, C.; Tian, G.; et al. AtDIV2, an R-R-Type MYB Transcription Factor of Arabidopsis, Negatively Regulates Salt Stress by Modulating ABA Signaling. Plant Cell Rep. 2018, 37, 1499–1511. [Google Scholar] [CrossRef] [PubMed]
- Steffen, J.G.; Kang, I.-H.; Portereiko, M.F.; Lloyd, A.; Drews, G.N. AGL61 Interacts with AGL80 and Is Required for Central Cell Development in Arabidopsis. Plant Physiol. 2008, 148, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Verelst, W.; Twell, D.; de Folter, S.; Immink, R.; Saedler, H.; Münster, T. MADS-Complexes Regulate Transcriptome Dynamics during Pollen Maturation. Genome Biol. 2007, 8, R249. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, T.; Liu, M.; Liu, J.; Zhang, Z. Transcriptome Analysis of Filling Stage Seeds among Three Buckwheat Species with Emphasis on Rutin Accumulation. PLoS ONE 2017, 12, e0189672. [Google Scholar] [CrossRef]
- Nakamura, M.; Katsumata, H.; Abe, M.; Yabe, N.; Komeda, Y.; Yamamoto, K.T.; Takahashi, T. Characterization of the Class IV Homeodomain-Leucine Zipper Gene Family in Arabidopsis. Plant Physiol. 2006, 141, 1363–1375. [Google Scholar] [CrossRef]
- Zhang, Z.-L.; Ogawa, M.; Fleet, C.M.; Zentella, R.; Hu, J.; Heo, J.-O.; Lim, J.; Kamiya, Y.; Yamaguchi, S.; Sun, T. Scarecrow-like 3 Promotes Gibberellin Signaling by Antagonizing Master Growth Repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 2160–2165. [Google Scholar] [CrossRef] [PubMed]
- Torti, S.; Fornara, F. AGL24 Acts in Concert with SOC1 and FUL during Arabidopsis Floral Transition. Plant Signal. Behav. 2012, 7, 1251–1254. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.; Bian, H.; Zha, Y.; Li, F.; Sun, Y.; Bai, B.; Chen, Z.; Wang, J.; Zhu, M.; Han, N. miR396a-Mediated Basic Helix-Loop-Helix Transcription Factor bHLH74 Repression Acts as a Regulator for Root Growth in Arabidopsis Seedlings. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Willemsen, V.; Boutilier, K.; Heidstra, R. AINTEGUMENTA-LIKE Proteins: Hubs in a Plethora of Networks. Trends Plant Sci. 2014, 19, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Breuer, C.; Kawamura, A.; Clark, N.M.; Rymen, B.; Braidwood, L.; Morohashi, K.; Busch, W.; Benfey, P.N.; Sozzani, R.; et al. GTL1 and DF1 Regulate Root Hair Growth through Transcriptional Repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Development 2018, 145, dev159707. [Google Scholar] [CrossRef] [PubMed]
- Hidayati, N.A.; Yamada-Oshima, Y.; Iwai, M.; Yamano, T.; Kajikawa, M.; Sakurai, N.; Suda, K.; Sesoko, K.; Hori, K.; Obayashi, T.; et al. Lipid Remodeling Regulator 1 (LRL1) Is Differently Involved in the Phosphorus-depletion Response from PSR1 in Chlamydomonas reinhardtii. Plant J. 2019, 100, 610–626. [Google Scholar] [CrossRef]
- Lu, W.; Deng, F.; Jia, J.; Chen, X.; Li, J.; Wen, Q.; Li, T.; Meng, Y.; Shan, W. The Arabidopsis thaliana Gene AtERF019 Negatively Regulates Plant Resistance to Phytophthora parasitica by Suppressing PAMP-triggered Immunity. Mol. Plant Pathol. 2020, 21, 1179–1193. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cheng, Y.; Wang, M.; Cui, S.; Guan, J. Weighted Gene Coexpression Correlation Network Analysis Reveals a Potential Molecular Regulatory Mechanism of Anthocyanin Accumulation under Different Storage Temperatures in “Friar” Plum. BMC Plant Biol. 2021, 21, 576. [Google Scholar] [CrossRef]
- Cutcliffe, J.W.; Hellmann, E.; Heyl, A.; Rashotte, A.M. CRFs Form Protein-Protein Interactions with Each Other and with Members of the Cytokinin Signalling Pathway in Arabidopsis via the CRF Domain. J. Exp. Bot. 2011, 62, 4995–5002. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, M.; Ren, G.; Yu, B. CDC5, a DNA Binding Protein, Positively Regulates Posttranscriptional Processing and/or Transcription of Primary microRNA Transcripts. Proc. Natl. Acad. Sci. USA 2013, 110, 17588–17593. [Google Scholar] [CrossRef]
- Plesch, G.; Störmann, K.; Torres, J.T.; Walden, R.; Somssich, I.E. Developmental and Auxin-Induced Expression of the Arabidopsis prha Homeobox Gene. Plant J. 1997, 12, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-W.; Shao, C.-R.; Zhang, C.-J.; Zhou, J.-X.; Zhang, S.-W.; Li, L.; Chen, S.; Huang, H.-W.; Cai, T.; He, X.-J. The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci. PLoS Genet. 2014, 10, e1003948. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, M.; White, C.I.; Franklin, F.C.H.; Sanchez-Moran, E. The Role of Topoisomerase II in DNA Repair and Recombination in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 13115. [Google Scholar] [CrossRef] [PubMed]
- Lyu, G.; Li, D.; Li, S. Bioinformatics Analysis of BBX Family Genes and Its Response to UV-B in Arabidopsis thaliana. Plant Signal. Behav. 2020, 15, 1782647. [Google Scholar] [CrossRef] [PubMed]
- Nibau, C.; Gibbs, D.J.; Bunting, K.A.; Moody, L.A.; Smiles, E.J.; Tubby, J.A.; Bradshaw, S.J.; Coates, J.C. ARABIDILLO Proteins Have a Novel and Conserved Domain Structure Important for the Regulation of Their Stability. Plant Mol. Biol. 2011, 75, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Araya, T.; von Wirén, N.; Takahashi, H. CLE Peptides Regulate Lateral Root Development in Response to Nitrogen Nutritional Status of Plants. Plant Signal. Behav. 2014, 9, e29302. [Google Scholar] [CrossRef]
- Li, M.; Liu, C.; Hepworth, S.R.; Ma, C.; Li, H.; Li, J.; Wang, S.-M.; Yin, H. SAUR15 Interaction with BRI1 Activates Plasma Membrane H+-ATPase to Promote Organ Development of Arabidopsis. Plant Physiol. 2022, 189, 2454–2466. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Kim, Y.-S.; Yoon, H.-K.; Park, C.-M. Functional Characterization of a Small Auxin-up RNA Gene in Apical Hook Development in Arabidopsis. Plant Sci. 2007, 172, 150–157. [Google Scholar] [CrossRef]
- Qiu, T.; Chen, Y.; Li, M.; Kong, Y.; Zhu, Y.; Han, N.; Bian, H.; Zhu, M.; Wang, J. The Tissue-Specific and Developmentally Regulated Expression Patterns of the SAUR41 Subfamily of Small Auxin up RNA Genes: Potential Implications. Plant Signal. Behav. 2013, 8, e25283. [Google Scholar] [CrossRef]
- Sakai, M.; Sakamoto, T.; Saito, T.; Matsuoka, M.; Tanaka, H.; Kobayashi, M. Expression of Novel Rice Gibberellin 2-Oxidase Gene Is under Homeostatic Regulation by Biologically Active Gibberellins. J. Plant Res. 2003, 116, 161–164. [Google Scholar] [CrossRef]
- Desikan, R.; Horák, J.; Chaban, C.; Mira-Rodado, V.; Witthöft, J.; Elgass, K.; Grefen, C.; Cheung, M.-K.; Meixner, A.J.; Hooley, R.; et al. The Histidine Kinase AHK5 Integrates Endogenous and Environmental Signals in Arabidopsis Guard Cells. PLoS ONE 2008, 3, e2491. [Google Scholar] [CrossRef] [PubMed]
- Schott-Verdugo, S.; Müller, L.; Classen, E.; Gohlke, H.; Groth, G. Structural Model of the ETR1 Ethylene Receptor Transmembrane Sensor Domain. Sci. Rep. 2019, 9, 8869. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Furlan, C.; Campos, R.; Toth, J.N.; Van Norman, J.M. Distinct Mechanisms Orchestrate the Contra-Polarity of IRK and KOIN, Two LRR-Receptor-Kinases Controlling Root Cell Division. Nat. Commun. 2022, 13, 235. [Google Scholar] [CrossRef] [PubMed]
- Diepold, A.; Li, G.; Lennarz, W.J.; Nürnberger, T.; Brunner, F. The Arabidopsis AtPNG1 Gene Encodes a Peptide: N-Glycanase. Plant J. 2007, 52, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Ronceret, A.; Guilleminot, J.; Lincker, F.; Gadea-Vacas, J.; Delorme, V.; Bechtold, N.; Pelletier, G.; Delseny, M.; Chabouté, M.-E.; Devic, M. Genetic Analysis of Two Arabidopsis DNA Polymerase Epsilon Subunits during Early Embryogenesis. Plant J. 2005, 44, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Vatamaniuk, O.K.; Mari, S.; Lang, A.; Chalasani, S.; Demkiv, L.O.; Rea, P.A. Phytochelatin Synthase, a Dipeptidyltransferase That Undergoes Multisite Acylation with Gamma-Glutamylcysteine during Catalysis: Stoichiometric and Site-Directed Mutagenic Analysis of Arabidopsis thaliana PCS1-Catalyzed Phytochelatin Synthesis. J. Biol. Chem. 2004, 279, 22449–22460. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, S.; Cavet, G.; Oakenfull, E.A.; Carmichael, J.P.; Shah, Z.H.; Soni, R.; Murray, J.A. A Novel and Highly Divergent Arabidopsis Cyclin Isolated by Complementation in Budding Yeast. Biochim. Biophys. Acta 2001, 1539, 1–6. [Google Scholar] [CrossRef] [PubMed]
- d’Erfurth, I.; Jolivet, S.; Froger, N.; Catrice, O.; Novatchkova, M.; Simon, M.; Jenczewski, E.; Mercier, R. Mutations in AtPS1 (Arabidopsis thaliana Parallel Spindle 1) Lead to the Production of Diploid Pollen Grains. PLoS Genet. 2008, 4, e1000274. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, D.; De Rybel, B.; Gudesblat, G.; Demidov, D.; Grunewald, W.; De Smet, I.; Houben, A.; Beeckman, T.; Russinova, E. Arabidopsis α Aurora Kinases Function in Formative Cell Division Plane Orientation. Plant Cell 2011, 23, 4013–4024. [Google Scholar] [CrossRef]
- Romanowski, A.; Furniss, J.J.; Hussain, E.; Halliday, K.J. Phytochrome Regulates Cellular Response Plasticity and the Basic Molecular Machinery of Leaf Development. Plant Physiol. 2021, 186, 1220–1239. [Google Scholar] [CrossRef]
- Draeger, C.; Ndinyanka Fabrice, T.; Gineau, E.; Mouille, G.; Kuhn, B.M.; Moller, I.; Abdou, M.-T.; Frey, B.; Pauly, M.; Bacic, A.; et al. Arabidopsis Leucine-Rich Repeat Extensin (LRX) Proteins Modify Cell Wall Composition and Influence Plant Growth. BMC Plant Biol. 2015, 15, 155. [Google Scholar] [CrossRef] [PubMed]
- Farquharson, K.L. Mirror, Mirror on the Wall: A Role for AGP18 in Functional Megaspore Selection. Plant Cell 2013, 25, 1190. [Google Scholar] [CrossRef]
- Li, S.; Ge, F.-R.; Xu, M.; Zhao, X.-Y.; Huang, G.-Q.; Zhou, L.-Z.; Wang, J.-G.; Kombrink, A.; McCormick, S.; Zhang, X.S.; et al. Arabidopsis COBRA-LIKE 10, a GPI-Anchored Protein, Mediates Directional Growth of Pollen Tubes. Plant J. 2013, 74, 486–497. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant Development Is Regulated by a Family of Auxin Receptor F Box Proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.J.; Choi, S.H.; Kim, J.H.; Kim, J.E.; Lee, M.H.; Chung, B.Y.; Woo, H.R.; Kim, J.-H. A Mutation in Plant-Specific SWI2/SNF2-Like Chromatin-Remodeling Proteins, DRD1 and DDM1, Delays Leaf Senescence in Arabidopsis thaliana. PLoS ONE 2016, 11, e0146826. [Google Scholar] [CrossRef] [PubMed]
- Yadav, N.S.; Khadka, J.; Domb, K.; Zemach, A.; Grafi, G. CMT3 and SUVH4/KYP Silence the Exonic Evelknievel Retroelement to Allow for Reconstitution of CMT1 mRNA. Epigenetics Chromatin 2018, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Fernández, R.; Barrero-Sicilia, C.; Carrillo-Barral, N.; Oñate-Sánchez, L.; Carbonero, P. Arabidopsis thaliana bZIP44: A Transcription Factor Affecting Seed Germination and Expression of the Mannanase-Encoding Gene AtMAN7. Plant J. 2013, 74, 767–780. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Guilfoyle, T. Auxin-Responsive Gene Expression: Genes, Promoters and Regulatory Factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Li, L.; De Clercq, I.; Narsai, R.; Xu, Y.; Hartmann, A.; Claros, D.L.; Custovic, E.; Lewsey, M.G.; Whelan, J.; et al. ANAC017 Coordinates Organellar Functions and Stress Responses by Reprogramming Retrograde Signaling. Plant Physiol. 2019, 180, 634–653. [Google Scholar] [CrossRef]
- Oh, S.-A.; Hoai, T.N.T.; Park, H.-J.; Zhao, M.; Twell, D.; Honys, D.; Park, S.-K. MYB81, a Microspore-Specific GAMYB Transcription Factor, Promotes Pollen Mitosis I and Cell Lineage Formation in Arabidopsis. Plant J. 2020, 101, 590–603. [Google Scholar] [CrossRef]
- Wang, J.; Li, G.; Li, C.; Zhang, C.; Cui, L.; Ai, G.; Wang, X.; Zheng, F.; Zhang, D.; Larkin, R.M.; et al. NF-Y Plays Essential Roles in Flavonoid Biosynthesis by Modulating Histone Modifications in Tomato. New Phytol. 2021, 229, 3237–3252. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cui, X.; Yang, B.; Xu, S.; Wei, X.; Zhao, P.; Niu, F.; Sun, M.; Wang, C.; Cheng, H.; et al. WRKY55 Transcription Factor Positively Regulates Leaf Senescence and the Defense Response by Modulating the Transcription of Genes Implicated in the Biosynthesis of Reactive Oxygen Species and Salicylic Acid in Arabidopsis. Development 2020, 147, dev189647. [Google Scholar] [CrossRef] [PubMed]
- Seyfferth, C.; Wessels, B.; Jokipii-Lukkari, S.; Sundberg, B.; Delhomme, N.; Felten, J.; Tuominen, H. Ethylene-Related Gene Expression Networks in Wood Formation. Front. Plant Sci. 2018, 9, 272. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Simm, S.; Mirus, O.; Scharf, K.-D.; Fragkostefanakis, S.; Schleiff, E. The Complexity of Vesicle Transport Factors in Plants Examined by Orthology Search. PLoS ONE 2014, 9, e97745. [Google Scholar] [CrossRef] [PubMed]
- Radoeva, T.; Lokerse, A.S.; Llavata-Peris, C.I.; Wendrich, J.R.; Xiang, D.; Liao, C.-Y.; Vlaar, L.; Boekschoten, M.; Hooiveld, G.; Datla, R.; et al. A Robust Auxin Response Network Controls Embryo and Suspensor Development through a Basic Helix Loop Helix Transcriptional Module. Plant Cell 2019, 31, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Wolff, P.; Weinhofer, I.; Seguin, J.; Roszak, P.; Beisel, C.; Donoghue, M.T.A.; Spillane, C.; Nordborg, M.; Rehmsmeier, M.; Köhler, C. High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm. PLoS Genet. 2011, 7, e1002126. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Beltran, E.; Elander, P.H.; Dalman, K.; Dayhoff, G.W.; Moschou, P.N.; Uversky, V.N.; Crespo, J.L.; Bozhkov, P.V. Tudor Staphylococcal Nuclease Is a Docking Platform for Stress Granule Components and Is Essential for SnRK1 Activation in Arabidopsis. EMBO J. 2021, 40, e105043. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Figueroa, J.; Carrillo, L.; Renau-Morata, B.; Yang, L.; Molina, R.-V.; Marino, D.; Canales, J.; Weih, M.; Vicente-Carbajosa, J.; Nebauer, S.G.; et al. The Arabidopsis Transcription Factor CDF3 Is Involved in Nitrogen Responses and Improves Nitrogen Use Efficiency in Tomato. Front. Plant Sci. 2020, 11, 601558. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Sotomayor, P.; Chávez Montes, R.A.; Silvestre-Vañó, M.; Herrera-Ubaldo, H.; Greco, R.; Pablo-Villa, J.; Galliani, B.M.; Diaz-Ramirez, D.; Weemen, M.; Boutilier, K.; et al. Altered Expression of the bZIP Transcription Factor DRINK ME Affects Growth and Reproductive Development in Arabidopsis thaliana. Plant J. 2016, 88, 437–451. [Google Scholar] [CrossRef]
- Mouchel, C.F.; Briggs, G.C.; Hardtke, C.S. Natural Genetic Variation in Arabidopsis Identifies BREVIS RADIX, a Novel Regulator of Cell Proliferation and Elongation in the Root. Genes. Dev. 2004, 18, 700–714. [Google Scholar] [CrossRef]
- Showalter, A.M.; Keppler, B.; Lichtenberg, J.; Gu, D.; Welch, L.R. A Bioinformatics Approach to the Identification, Classification, and Analysis of Hydroxyproline-Rich Glycoproteins. Plant Physiol. 2010, 153, 485–513. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Strickler, S.R.; Richards, E.J. Loss of CRWN Nuclear Proteins Induces Cell Death and Salicylic Acid Defense Signaling. Plant Physiol. 2019, 179, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Xun, Q.; Wu, Y.; Li, H.; Chang, J.; Ou, Y.; He, K.; Gou, X.; Tax, F.E.; Li, J. Two Receptor-like Protein Kinases, MUSTACHES and MUSTACHES-LIKE, Regulate Lateral Root Development in Arabidopsis thaliana. New Phytol. 2020, 227, 1157–1173. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Qiao, Z.; Muchero, W.; Chen, J.-G. Lectin Receptor-Like Kinases: The Sensor and Mediator at the Plant Cell Surface. Front. Plant Sci. 2020, 11, 596301. [Google Scholar] [CrossRef] [PubMed]
- Stührwohldt, N.; Ehinger, A.; Thellmann, K.; Schaller, A. Processing and Formation of Bioactive CLE40 Peptide Are Controlled by Posttranslational Proline Hydroxylation. Plant Physiol. 2020, 184, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Kosarev, P.; Mayer, K.F.X.; Hardtke, C.S. Evaluation and Classification of RING-Finger Domains Encoded by the Arabidopsis Genome. Genome Biol. 2002, 3, RESEARCH0016. [Google Scholar] [CrossRef] [PubMed]
- D’Ippólito, S.; Arias, L.A.; Casalongué, C.A.; Pagnussat, G.C.; Fiol, D.F. The DC1-Domain Protein VACUOLELESS GAMETOPHYTES Is Essential for Development of Female and Male Gametophytes in Arabidopsis. Plant J. 2017, 90, 261–275. [Google Scholar] [CrossRef]
- Giuntoli, B.; Lee, S.C.; Licausi, F.; Kosmacz, M.; Oosumi, T.; van Dongen, J.T.; Bailey-Serres, J.; Perata, P. A Trihelix DNA Binding Protein Counterbalances Hypoxia-Responsive Transcriptional Activation in Arabidopsis. PLoS Biol. 2014, 12, e1001950. [Google Scholar] [CrossRef]




| Variant | Impact | Functional Class | |||
|---|---|---|---|---|---|
| Type | Total | Type | Number | Type | Number |
| SNP | 2,260,270 | HIGH | 9451 | MISSENSE | 292,963 |
| LOW | 432,159 | NONSENSE | 5618 | ||
| MODERATE | 291,254 | SILENT | 393,274 | ||
| MODIFIER | 4,334,386 | ||||
| INDEL | 514,083 | HIGH | 12,234 | ||
| LOW | 15,755 | ||||
| MODERATE | 15,497 | ||||
| MODIFIER | 1,227,445 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuznetsova, X.; Dodueva, I.; Afonin, A.; Gribchenko, E.; Danilov, L.; Gancheva, M.; Tvorogova, V.; Galynin, N.; Lutova, L. Whole-Genome Sequencing and Analysis of Tumour-Forming Radish (Raphanus sativus L.) Line. Int. J. Mol. Sci. 2024, 25, 6236. https://doi.org/10.3390/ijms25116236
Kuznetsova X, Dodueva I, Afonin A, Gribchenko E, Danilov L, Gancheva M, Tvorogova V, Galynin N, Lutova L. Whole-Genome Sequencing and Analysis of Tumour-Forming Radish (Raphanus sativus L.) Line. International Journal of Molecular Sciences. 2024; 25(11):6236. https://doi.org/10.3390/ijms25116236
Chicago/Turabian StyleKuznetsova, Xenia, Irina Dodueva, Alexey Afonin, Emma Gribchenko, Lavrentii Danilov, Maria Gancheva, Varvara Tvorogova, Nikita Galynin, and Lyudmila Lutova. 2024. "Whole-Genome Sequencing and Analysis of Tumour-Forming Radish (Raphanus sativus L.) Line" International Journal of Molecular Sciences 25, no. 11: 6236. https://doi.org/10.3390/ijms25116236
APA StyleKuznetsova, X., Dodueva, I., Afonin, A., Gribchenko, E., Danilov, L., Gancheva, M., Tvorogova, V., Galynin, N., & Lutova, L. (2024). Whole-Genome Sequencing and Analysis of Tumour-Forming Radish (Raphanus sativus L.) Line. International Journal of Molecular Sciences, 25(11), 6236. https://doi.org/10.3390/ijms25116236






