Discovery and Characterization of the ddx41 Gene in Atlantic Salmon: Evolutionary Implications, Structural Functions, and Innate Immune Responses to Piscirickettsia salmonis and Renibacterium salmoninarum Infections
Abstract
1. Introduction
2. Results
2.1. In Silico and Experimental Characterization of ddx41 Gene and Protein of S. salar
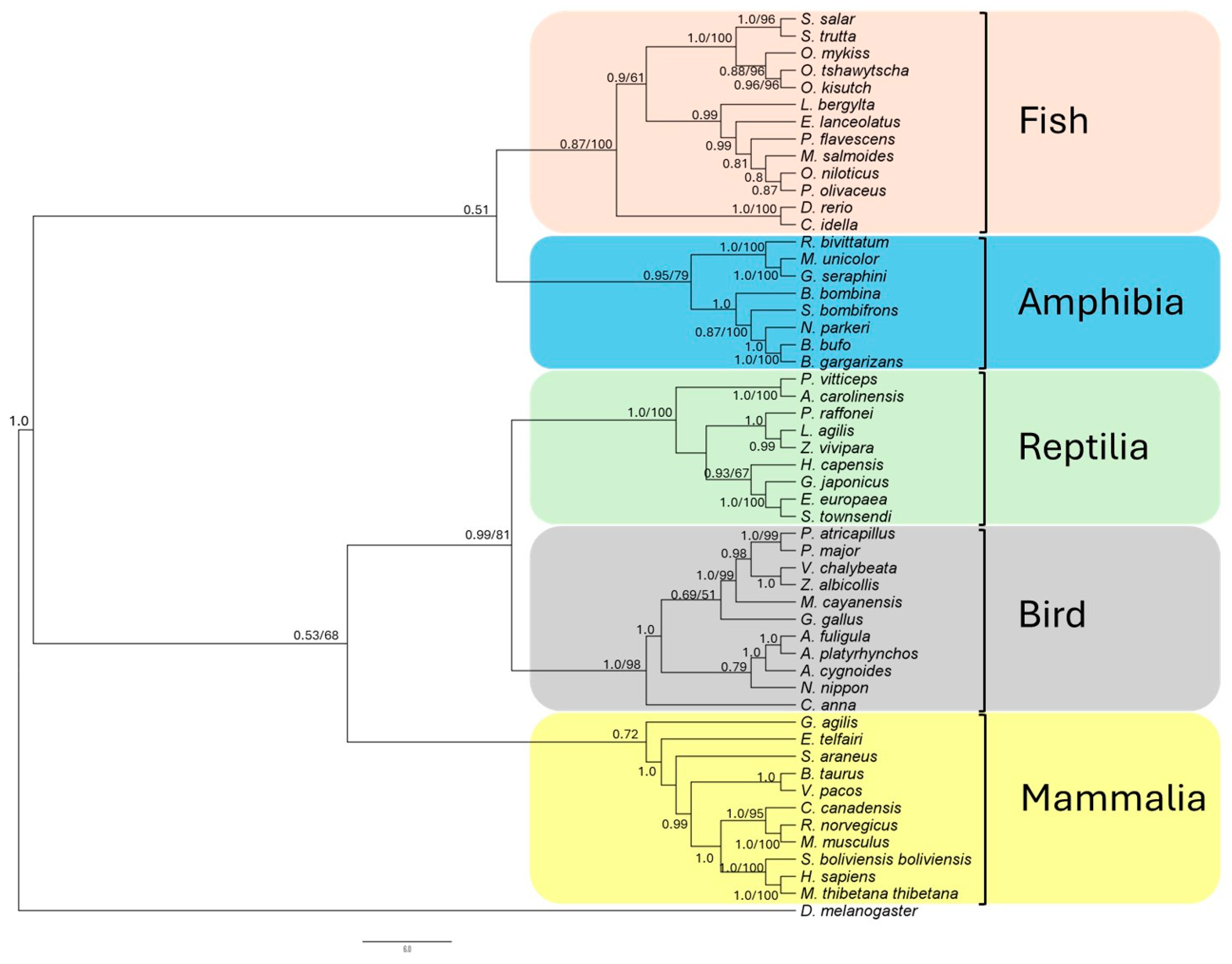
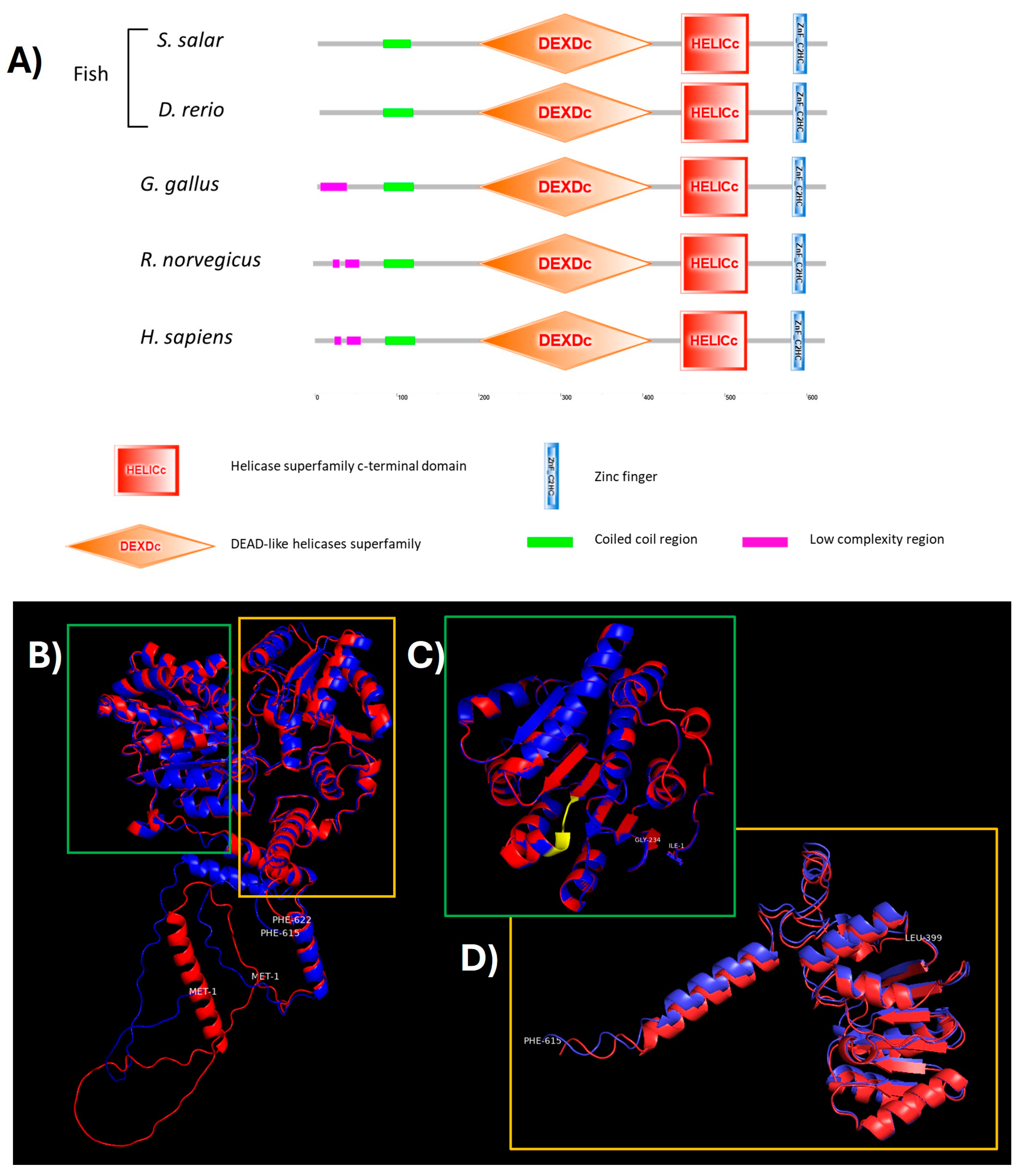
2.2. Prediction Origin, Phylogenetic, and Structural–Functional Analysis of ddx41 in S. salar
2.3. Expression Pattern of the ddx41 Gene across S. salar Tissues
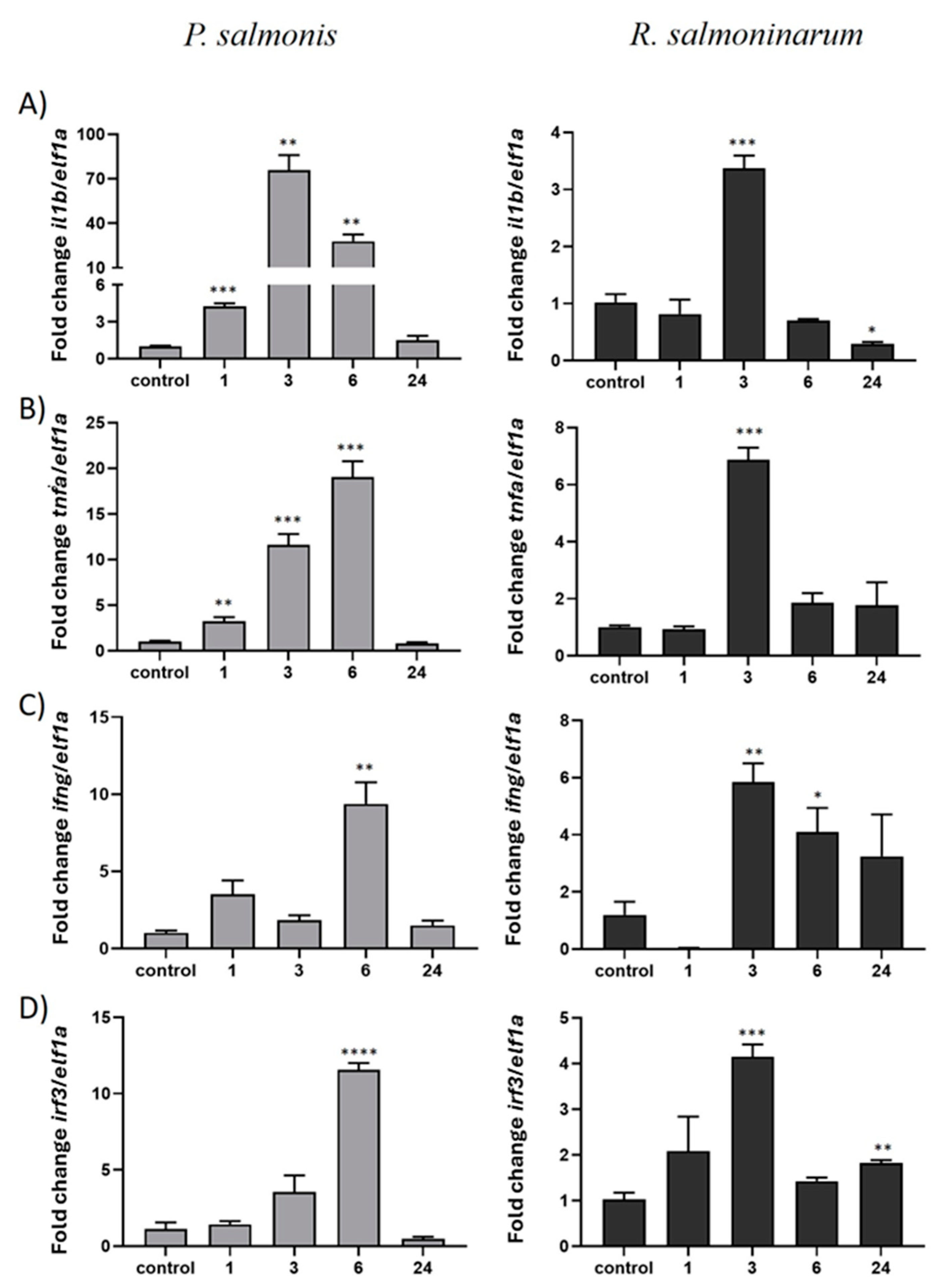
2.4. Gene Expression of ddx41 and Inflammatory Factors in SHK-1 Cells Infected with Gram-Positive and Gram-Negative S. salar Pathogens
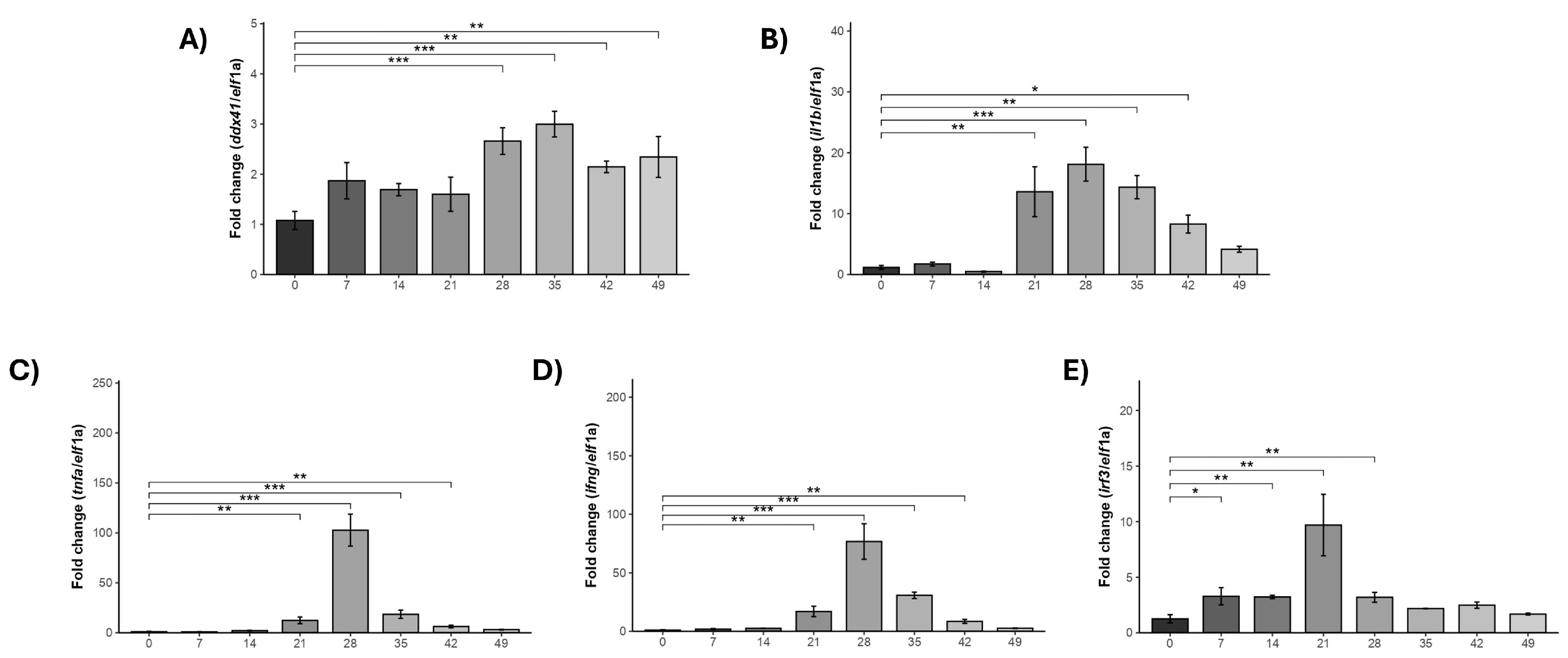
2.5. The Gene Expression of ddx41 and Immune Innate Genes in the Head Kidney of S. salar in Response to P. salmonis Challenge
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Study: Identification, Structural Analysis of Salmo salar ddx41 Gene, and Phylogenetics
4.1.1. Data Retrieval and Sequence Alignment
4.1.2. mRNA Sequence Analysis of S. salar Head Kidney Transcriptomes
4.1.3. Model Selection and Phylogenetic Analysis
4.1.4. Structural Analysis and Protein Models
4.2. Experimental Animals
4.3. Proteomic Identification and Characterization of DDX41 from S. salar Head Kidney
4.3.1. Protein Extraction for nLC-MS/MS
4.3.2. Protein Extraction and Digestion for nLC-MS/MS
4.3.3. Liquid Chromatography
4.3.4. The timsTOF Pro Mass Spectrometer
4.3.5. Database Searching
4.4. Infection Assay in SHK1 Cell Line with Pathogenic Bacteria
4.5. Cohabitation Challenge with P. salmonis
4.6. Primer Design and Functional Quantitative Real-Time PCR (qPCR) Analysis
4.7. Statistical Analysis and Graph
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Sen, R.; Nayak, L.; De, R.K. A review on host–pathogen interactions: Classification and prediction. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1581–1599. [Google Scholar] [CrossRef]
- Magnadóttir, B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006, 20, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, S.; Tong, Z.; Li, L.; Wang, G. Maternal transfer and protective role of the alternative complement components in zebrafish Danio rerio. PLoS ONE 2009, 4, e4498. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Yoneyama, M.; Fujita, T. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010, 20, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Diacovich, L.; Gorvel, J.P. Bacterial manipulation of innate immunity to promote infection. Nat. Rev. Microbiol. 2010, 8, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Villanueva-Cañas, J.; Ruiz-Orera, J.; Agea, M.I.; Gallo, M.; Andreu, D.; Albà, M.M. New Genes and Functional Innovation in Mammals. Genome Biol. Evol. 2017, 9, 1886–1900. [Google Scholar] [CrossRef]
- Armingol, E.; Officer, A.; Harismendy, O.; Lewis, N.E. Deciphering cell-cell interactions and communication from gene expression. Nat. Rev. Genet. 2021, 22, 71–88. [Google Scholar] [CrossRef]
- Mills, E.; Pultz, I.S.; Kulasekara, H.D.; Miller, S.I. The bacterial second messenger c-di-GMP: Mechanisms of signaling. Cell Microbiol. 2011, 13, 1122–1129. [Google Scholar] [CrossRef]
- Woodward, J.J.; Iavarone, A.T.; Portnoy, D.A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 2010, 328, 1703–1705. [Google Scholar] [CrossRef]
- Parvatiyar, K.; Zhang, Z.; Teles, R.M.; Ouyang, S.; Jiang, Y.; Iyer, S.S.; Zaver, S.A.; Schenk, M.; Zeng, S.; Zhong, W.; et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 2012, 13, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.; Dey, R.J.; Cheung, L.S.; Pokkali, S.; Guo, H.; Lee, J.H.; Bishai, W.R. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 2015, 21, 401–406. [Google Scholar] [CrossRef]
- Luecke, S.; Paludan, S.R. Molecular requirements for sensing of intracellular microbial nucleic acids by the innate immune system. Cytokine 2017, 98, 4–14. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.; Su, N.; Liu, D.; Luo, R.; Jin, H. Molecular cloning and functional characterization of duck DDX41. Dev. Comp. Immunol. 2018, 88, 183–189. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, Y.; Liu, Z.J.; Ouyang, S. The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell 2017, 8, 83–89. [Google Scholar] [CrossRef]
- Perčulija, V.; Ouyang, S. Diverse Roles of DEAD/DEAH-Box Helicases in Innate Immunity and Diseases. In Helicases from All Domains of Life; Tuteja, R., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 141–171. [Google Scholar]
- Omura, H.; Oikawa, D.; Nakane, T.; Kato, M.; Ishii, R.; Ishitani, R.; Tokunaga, F.; Nureki, O. Structural and functional analysis of DDX41: A bispecific immune receptor for DNA and cyclic dinucleotide. Sci. Rep. 2016, 6, 34756. [Google Scholar] [CrossRef] [PubMed]
- Rocak, S.; Linder, P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004, 5, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lee, S.; Park, J.W.; Jung, H.S.; Kim, J.; Yang, H.; Lee, J.H.; Lee, D. Analysis of tissue-specific interferon regulatory factor 3 (IRF3) gene expression against viral infection in Paralichthys olivaceus. Dev. Reprod. 2021, 25, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Schröder, M.; Baran, M.; Bowie, A.G. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008, 6, 2147–2157. [Google Scholar] [CrossRef]
- Ma, J.X.; Li, J.Y.; Fan, D.D.; Feng, W.; Lin, A.F.; Xiang, L.X.; Shao, J.Z. Identification of DEAD-Box RNA Helicase DDX41 as a trafficking protein that involves in multiple innate immune signaling pathways in a Zebrafish model. Front. Immunol. 2018, 11, 1327. [Google Scholar] [CrossRef]
- Zhang, Z.; Bao, M.; Lu, N.; Weng, L.; Yuan, B.; Liu, Y.J. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 2013, 14, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Quynh, N.T.; Hikima, J.; Kim, Y.R.; Fagutao, F.F.; Kim, M.S.; Aoki, T.; Jung, T.S. The cytosolic sensor, DDX41, activates antiviral and inflammatory immunity in response to stimulation with double-stranded DNA adherent cells of the olive flounder, Paralichthys olivaceus. Fish Shellfish Immunol. 2015, 44, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, Y.; Huang, X.; Li, C.; Ni, S.W.; Yu, Y.; Qin, Q. Grouper DDX41 exerts antiviral activity against fish iridovirus and nodavirus infection. Fish Shellfish Immunol. 2019, 91, 40–49. [Google Scholar] [CrossRef]
- Gan, Z.; Cheng, J.; Hou, J.; Xia, H.; Chen, W.; Xia, L.; Nie, P.; Lu, Y. Molecular and functional characterization of tilapia DDX41 in IFN regulation. Fish Shellfish Immunol. 2020, 99, 386–391. [Google Scholar] [CrossRef]
- Qin, X.-W.; Luo, Z.-Y.; Pan, W.-Q.; He, J.; Li, Z.-M.; Yu, Y.; Liu, C.; Weng, S.P.; He, J.G.; Guo, C.J. The Interaction of Mandarin Fish DDX41 with STING Evokes Type I Interferon Responses Inhibiting Ranavirus Replication. Viruses 2023, 15, 58. [Google Scholar] [CrossRef]
- Rozas, M.; Enríquez, R. Piscirickettsiosis and Piscirickettsia salmonis in fish: A review. J. Fish Dis. 2014, 37, 163–188. [Google Scholar] [CrossRef]
- Figueroa, J.; Cárcamo, J.; Yañez, A.; Olavarria, V.; Ruiz, P.; Manríquez, R.; Muñoz, C.; Romero, A.; Avendaño-Herrera, R. Addressing viral and bacterial threats to salmon farming in Chile: Historical contexts and perspectives for management and control. Rev. Aquac. 2019, 11, 299–324. [Google Scholar] [CrossRef]
- SERNAPESCA. Sanitary Report with Sanitary Information of Freshwater and Sea Water. National Service of Fisheries, Subdivision of Aquaculture, Animal Health Unit, Chile. 2023. Available online: https://www.sernapesca.cl/app/uploads/2023/10/informe_sanitario_con_informacion_sanitaria_de_agua_dulce_y_mar_ano_2022.pdf (accessed on 10 October 2023).
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2022, 47, W636–W641. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed]
- Mojzesz, M.; Klak, K.; Wojtal, P.; Adamek, M.; Podlasz, P.; Chmielewska-Krzesinska, M.; Matras, M.; Reichert, M.; Chadzinska, M.; Rakus, K. Viral infection-induced changes in the expression profile of non-RLR DExD/H-box RNA helicases (DDX1, DDX3, DHX9, DDX21, and DHX36) in zebrafish and common carp. Fish Shellfish Immunol. 2020, 104, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Feng-Ying, G.; Mai-Xin, L.; Miao, W.; Zhi-Gang, L.; Xiao-Li, K.; De-Feng, Z.; Jian-Meng, C. Nile tilapia DNA sensor STING is involved in the IFN-β and AP-1 signaling pathways in the immune response dependent on DDX41. Int. J. Biol. Macromol. 2023, 225, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Rauta, P.R.; Nayak, B.; Das, S. Immune system and immune responses in fish and their role in comparative immunity study: A model for higher organisms. Immunol. Lett. 2012, 148, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Brocker, C.; Thompson, D.; Matsumoto, A.; Nebert, D.W.; Vasiliou, V. Evolutionary divergence and functions of the human interleukin (IL) gene family. Hum. Genomics 2010, 5, 30–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chu, D.; Kalantar-Zadeh, K.; George, J.; Young, H.A.; Liu, G. Cytokines: From Clinical Significance to Quantification. Adv. Sci. 2021, 8, e2004433. [Google Scholar] [CrossRef] [PubMed]
- Secombes, C.J.; Wang, T.H.; Bird, S. The interleukins of fish. Dev. Comp. Immunol. 2011, 35, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Secombes, C.J. The Function of Fish Cytokines. Biology 2016, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Sverdlov, A.V.; Rogozin, I.B.; Babenko, V.N.; Koonin, E.V. Conservation versus parallel gains in intron evolution. Nucleic Acids Res. 2005, 33, 1741–1748. [Google Scholar] [CrossRef][Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2020, 38, 420–423. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Brann, T.W.; Zhou, M.; Yang, J.; Oguariri, R.M.; Lidie, K.B.; Imamichi, H.; Huang, D.W.; Lempicki, R.A.; Baseler, M.W.; et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 2011, 186, 4541–4545. [Google Scholar] [CrossRef] [PubMed]
- Andreou, A.Z. DDX41: A multifunctional DEAD-box protein involved in pre-mRNA splicing and innate immunity. Bio. Chem. 2021, 402, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Krasteva, P.V.; Giglio, K.M.; Sondermann, H. Sensing the messenger: The diverse ways that bacteria signal through c-di-GMP. Protein Sci. 2012, 21, 929–948. [Google Scholar] [CrossRef] [PubMed]
- Schütz, P.; Karlberg, T.; van den Berg, S.; Collins, R.; Lehtiö, L.; Högbom, M.; Holmberg-Schiavone, L.; Tempel, W.; Park, H.W.; Hammarström, M.; et al. Comparative structural analysis of human DEAD-box RNA helicases. PLoS ONE 2010, 5, e12791. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Song, X.; Wang, Y.; Ru, H.; Shaw, N.; Jiang, Y.; Niu, F.; Zhu, Y.; Qiu, W.; Parvatiyar, K.; et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 2012, 36, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.S.; Vidhyasagar, V.; Yang, S.; Arna, A.B.; Yadav, M.; Aggarwal, A.; Aguilera, A.N.; Shinriki, S.; Bhanumathy, K.K.; Pandey, K.; et al. DDX41 is required for cGAS-STING activation against DNA virus infection. Cell Rep. 2022, 39, 110856. [Google Scholar] [CrossRef] [PubMed]
- Tamassia, N.; Cassatella, M.A. Cytoplasmic receptors recognizing nucleic acids and mediating immune functions in neutrophils. Curr. Opin. Pharmacol. 2013, 13, 547–554. [Google Scholar] [CrossRef]
- Briard, B.; Place, D.E.; Kanneganti, T.D. DNA Sensing in the Innate Immune Response. Physiology 2020, 35, 112–124. [Google Scholar] [CrossRef]
- Guimarães, E.S.; Marinho, F.V.; de Queiroz, N.M.G.P.; Antunes, M.M.; Oliveira, S.C. Impact of STING Inflammatory Signaling during Intracellular Bacterial Infections. Cells 2021, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhu, Y.; Qiu, W.; Liu, Y.J.; Cheng, G.; Liu, Z.J.; Ouyang, S. Structural and functional analyses of human DDX41 DEAD domain. Protein Cell 2017, 8, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, Y.; Wang, Y.; Niu, Q.; Gao, Q.; Fu, Q.; Ma, J.; Wang, H.; Yan, Y.; Ding, C.; et al. Chicken DNA virus sensor DDX41 activates IFN-β signaling pathway dependent on STING. Dev. Comp. Immunol. 2017, 76, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Kadono, M.; Kanai, A.; Nagamachi, A.; Shinriki, S.; Kawata, J.; Iwato, K.; Kyo, T.; Oshima, K.; Yokoyama, A.; Kawamura, T.; et al. Biological implications of somatic DDX41 p.R525H mutation in acute myeloid leukemia. Exp. Hematol. 2016, 44, 745–754.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yuan, B.; Bao, M.; Lu, N.; Kim, T.; Liu, Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011, 12, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.A. MrModeltest v2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree v1.4.4. 2012. Available online: https://github.com/rambaut/figtree/releases/tag/v1.4.4 (accessed on 23 August 2023).
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments, and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; von den Driesch, L.; Galiez, C.; Martin, M.J.; Söding, J.; Steinegger, M. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 2017, 45, D170–D176. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Steinegger, M.; Söding, J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 2019, 35, 2856–2858. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Almeida, A.; Beracochea, M.; Boland, M.; Burgin, J.; Cochrane, G.; Crusoe, M.R.; Kale, V.; Potter, S.C.; Richardson, L.J.; et al. MGnify: The microbiome analysis resource in 2020. Nucleic Acids Res. 2019, 48, D570–D578. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 5, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A Method to Identify Protein Sequences That Fold into a Known Three-Dimensional Structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Schrödinger, LLC. The PyMOL Molecular Graphics System; Version 1.8; Schrödinger LLC: New York, NY, USA, 2015. [Google Scholar]
- Herrera, V.; Olavarría, N.; Saavedra, J.; Yuivar, Y.; Bustos, P.; Almarza, O.; Mancilla, M. Complete lipopolysaccharide of Piscirickettsia salmonis is required for full virulence in the intraperitoneally challenged Atlantic Salmon, Salmo salar, model. Front. Cell. Infect. Microbiol. 2022, 12, 845661. [Google Scholar] [CrossRef] [PubMed]
- Dannevig, B.H.; Brudeseth, B.E.; GjØen, T.; Rode, M.; Wergeland, H.I.; Evensen, Ø.; Press, C.M. Characterisation of a long-term cell line (SHK-1) developed from the head kidney of Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 1997, 7, 213–226. [Google Scholar] [CrossRef]
- Levican-Asenjo, J.; Soto-Rifo, R.; Aguayo, F.; Gaggero, A.; Leon, O. Salmon cells SHK-1 internalize infectious pancreatic necrosis virus by macropinocytosis. J. Fish Dis. 2019, 42, 1035–1046. [Google Scholar] [CrossRef]
- Yañez, A.J.; Silva, H.; Valenzuela, K.; Pontigo, J.P.; Godoy, M.; Troncoso, J.; Romero, A.; Figueroa, J.; Carcamo, J.G.; Avendaño-Herrera, R. Two novel blood-free solid media for the culture of the salmonid pathogen Piscirickettsia salmonis. J. Fish Dis. 2013, 36, 587–591. [Google Scholar] [CrossRef]
- Evelyn, T.P.T. An improved growth medium for the kidney disease bacterium and some notes on using the medium. Bull. Off. Int. Epiz. 1977, 87, 511–513. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Tapia, D.; Eissler, Y.; Torres, P.; Jorquera, E.; Espinoza, J.C.; Kuznar, J. Detection and phylogenetic analysis of infectious pancreatic necrosis virus in Chile. Dis. Aquat. Org. 2015, 116, 173–184. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Santibañez, N.; Vega, M.; Pérez, T.; Yáñez, A.; González-Stegmaier, R.; Figueroa, J.; Enríquez, R.; Oliver, C.; Romero, A. Biofilm produced In Vitro by Piscirickettsia salmonis generates differential cytotoxicity levels and expression patterns of immune genes in the Atlantic Salmon cell line SHK-1. Microorganisms 2020, 8, 1609. [Google Scholar] [CrossRef]
- Fredriksen, B.; Sævareid, K.; McAuley, L.; Lane, M.; Bøgwald, J.; Dalmo, R. Early immune responses in Atlantic salmon (Salmo salar L.) after immunization with PLGA nanoparticles loaded with a model antigen and β-glucan. Vaccine 2011, 29, 8338–8349. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Kim, T.K. Understanding one-way ANOVA using conceptual figures. Korean J. Anesthesiol. 2017, 70, 22–26. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020. [Google Scholar]
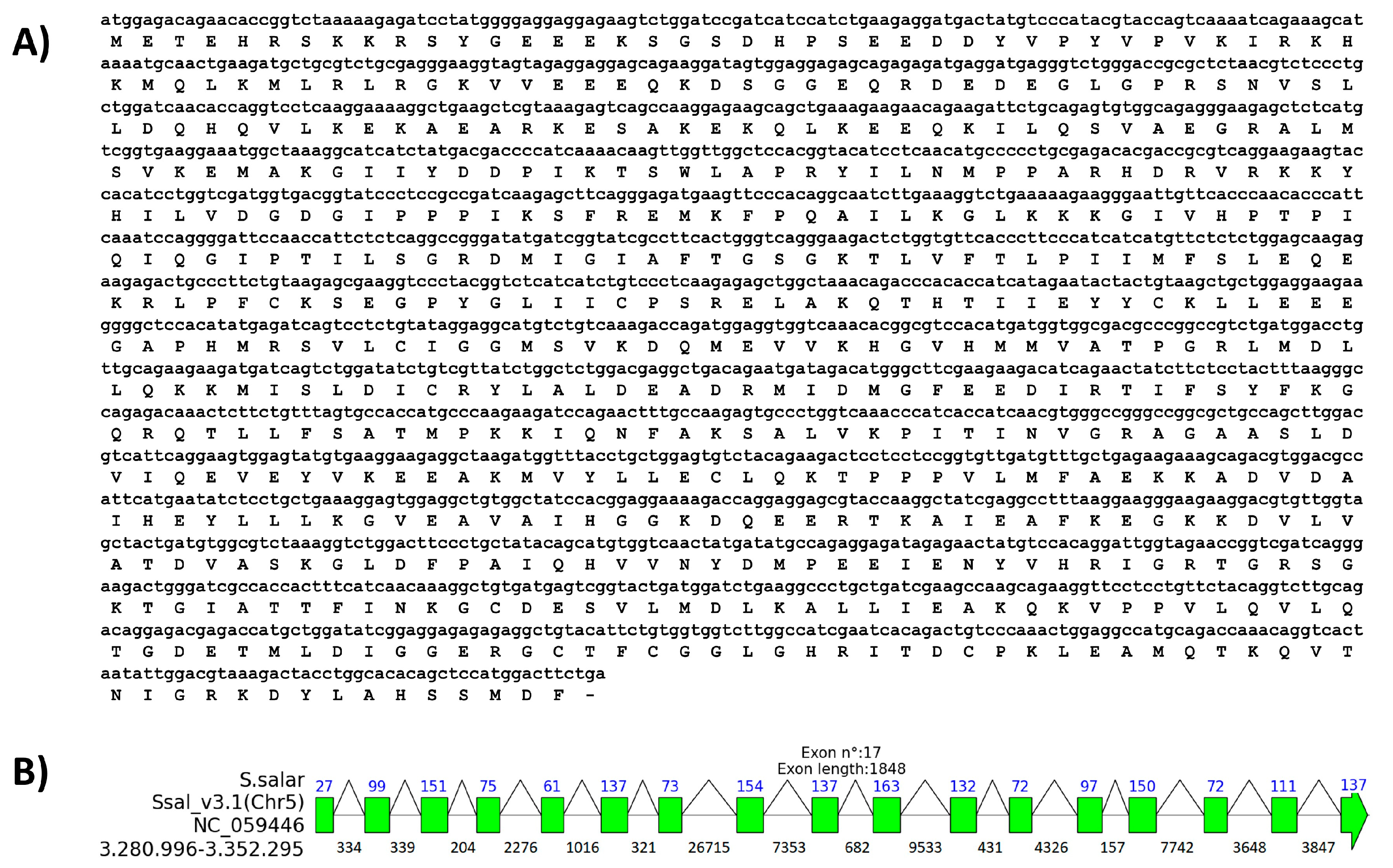



| Organism | Accession Number | BLASTn Results | BLASTp Results | |||||
|---|---|---|---|---|---|---|---|---|
| Nucleotide | Protein | Query (%) | E-Value | Per. Ident. * | Query (%) | E-Value | Per. Ident. * | |
| H. sapiens | AK222598.1 | BAD96318.1 | 100 | 0.0 | 100 | 100 | 0.0 | 100 |
| R. norvegicus | NM_001108046.2 | NP_001101516.1 | 93 | 0.0 | 89.32 | 100 | 0.0 | 98.87 |
| G. gallus | NM_001349708.2 | NP_001336637.2 | 61 | 0.0 | 83.70 | 99 | 0.0 | 92.56 |
| D. rerio | KR559928.1 | ANB32180.1 | 83 | 0.0 | 78.93 | 100 | 0.0 | 85.69 |
| S. salar | This study | 91 | 0.0 | 78.61 | 100 | 0.0 | 83.92 | |
| Organism | Domain | ERRAT | VERIFY 3D (Residues over Score Threshold) | PROCHECK (Error/Warning/Pass) |
|---|---|---|---|---|
| S. salar | Complete | 84.6702 | 71.06% | 4/3/2 |
| H. sapiens | Complete | 88.6029 | 63.90% | 4/3/2 |
| S. salar | Helicase ATP binding | 94.2478 | 73.50% | 0/3/6 |
| H. sapiens | Helicase ATP binding | 93.3628 | 83.76% | 0/3/6 |
| S. salar | Helicase C-terminal | 96.1538 | 75.61% | 2/3/4 |
| H. sapiens | Helicase C-terminal | 96.1538 | 79.88% | 2/3/4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yañez, A.J.; Barrientos, C.A.; Isla, A.; Aguilar, M.; Flores-Martin, S.N.; Yuivar, Y.; Ojeda, A.; Ibieta, P.; Hernández, M.; Figueroa, J.; et al. Discovery and Characterization of the ddx41 Gene in Atlantic Salmon: Evolutionary Implications, Structural Functions, and Innate Immune Responses to Piscirickettsia salmonis and Renibacterium salmoninarum Infections. Int. J. Mol. Sci. 2024, 25, 6346. https://doi.org/10.3390/ijms25126346
Yañez AJ, Barrientos CA, Isla A, Aguilar M, Flores-Martin SN, Yuivar Y, Ojeda A, Ibieta P, Hernández M, Figueroa J, et al. Discovery and Characterization of the ddx41 Gene in Atlantic Salmon: Evolutionary Implications, Structural Functions, and Innate Immune Responses to Piscirickettsia salmonis and Renibacterium salmoninarum Infections. International Journal of Molecular Sciences. 2024; 25(12):6346. https://doi.org/10.3390/ijms25126346
Chicago/Turabian StyleYañez, Alejandro J., Claudia A. Barrientos, Adolfo Isla, Marcelo Aguilar, Sandra N. Flores-Martin, Yassef Yuivar, Adriana Ojeda, Pablo Ibieta, Mauricio Hernández, Jaime Figueroa, and et al. 2024. "Discovery and Characterization of the ddx41 Gene in Atlantic Salmon: Evolutionary Implications, Structural Functions, and Innate Immune Responses to Piscirickettsia salmonis and Renibacterium salmoninarum Infections" International Journal of Molecular Sciences 25, no. 12: 6346. https://doi.org/10.3390/ijms25126346
APA StyleYañez, A. J., Barrientos, C. A., Isla, A., Aguilar, M., Flores-Martin, S. N., Yuivar, Y., Ojeda, A., Ibieta, P., Hernández, M., Figueroa, J., Avendaño-Herrera, R., & Mancilla, M. (2024). Discovery and Characterization of the ddx41 Gene in Atlantic Salmon: Evolutionary Implications, Structural Functions, and Innate Immune Responses to Piscirickettsia salmonis and Renibacterium salmoninarum Infections. International Journal of Molecular Sciences, 25(12), 6346. https://doi.org/10.3390/ijms25126346







