Comparative Review on Cancer Pathology from Aberrant Histone Chaperone Activity
Abstract
1. Introduction
2. Results
2.1. FACT Complex
2.1.1. Human Hepatocellular Carcinoma (HCC) and FACT
2.1.2. Breast Cancer (BrCa) and FACT

2.1.3. FACT-Targeted Therapy: Curaxin
2.2. ASF1
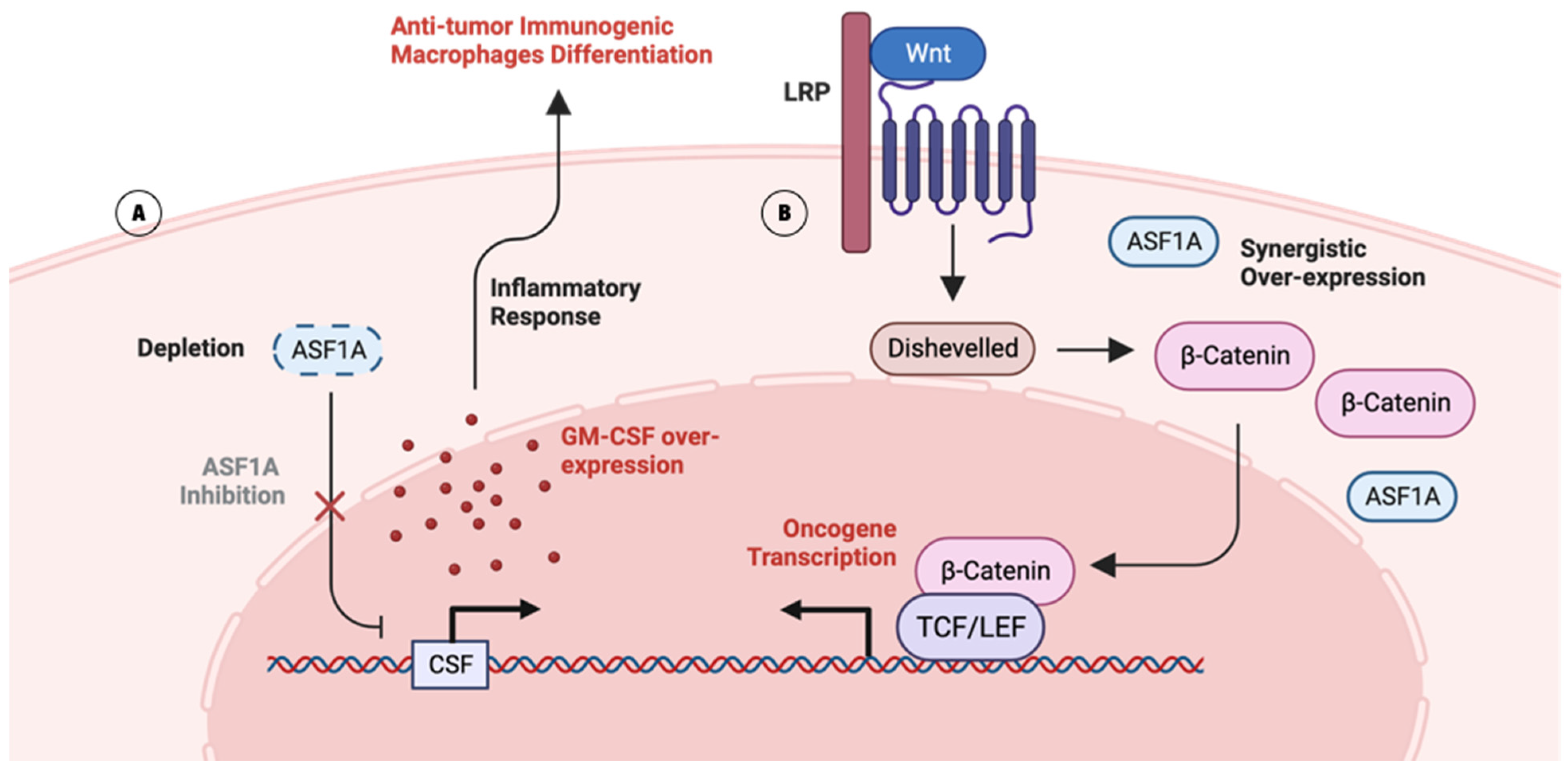
2.2.1. Lung Adenocarcinoma (LUAD) and ASF1
2.2.2. Gastrointestinal Cancer (GIC) and ASF1
2.2.3. ASF1-Targeted Therapy Approach: Chimeric Inhibitor
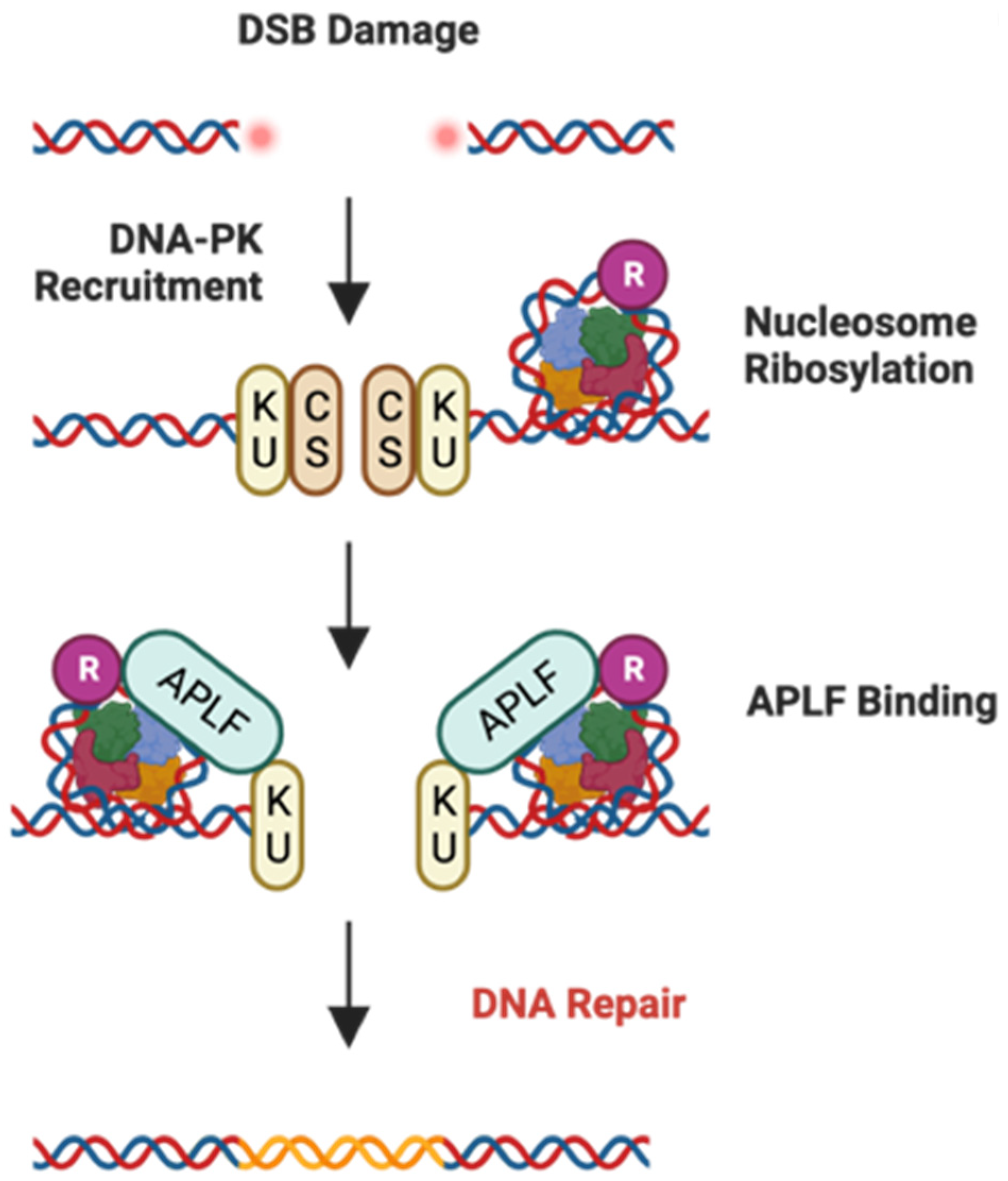
2.3. APLF
2.3.1. Glioblastoma Multiforme (GBM) and APLF
2.3.2. Bladder Cancer (BLCA) and APLF
2.3.3. APLF and Treatment Approaches
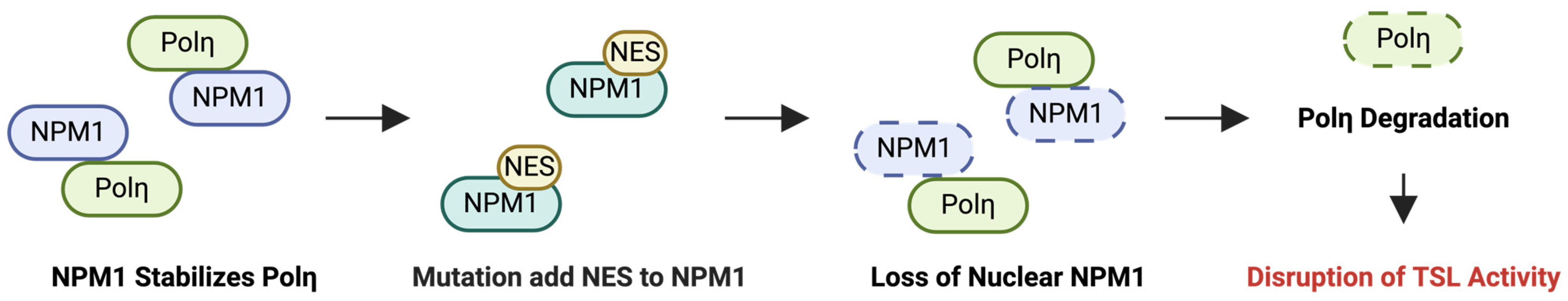
2.4. NPM1
2.4.1. Oral Squamous Cell Carcinoma (OSCC) and NPM1
2.4.2. Acute Myeloid Leukemia (AML) and NPM1
2.4.3. NPM1-Targeted Therapy: NSC348884 and RNAi
2.5. CAF-1
2.5.1. Gastric Cancer (GC) and CHAF1A
2.5.2. Diffuse Large B-Cell Lymphoma (DLBCL) and CHAF1A
2.5.3. Larynx Carcinoma, Skin Squamous Cell Carcinomas (SCCs), and CHAF1B
2.5.4. SCC and CHAF1B
2.5.5. CC and p48
2.5.6. THCC and p48
2.5.7. CAF1-Targeted Therapy Approach: CRISPR/Cas9
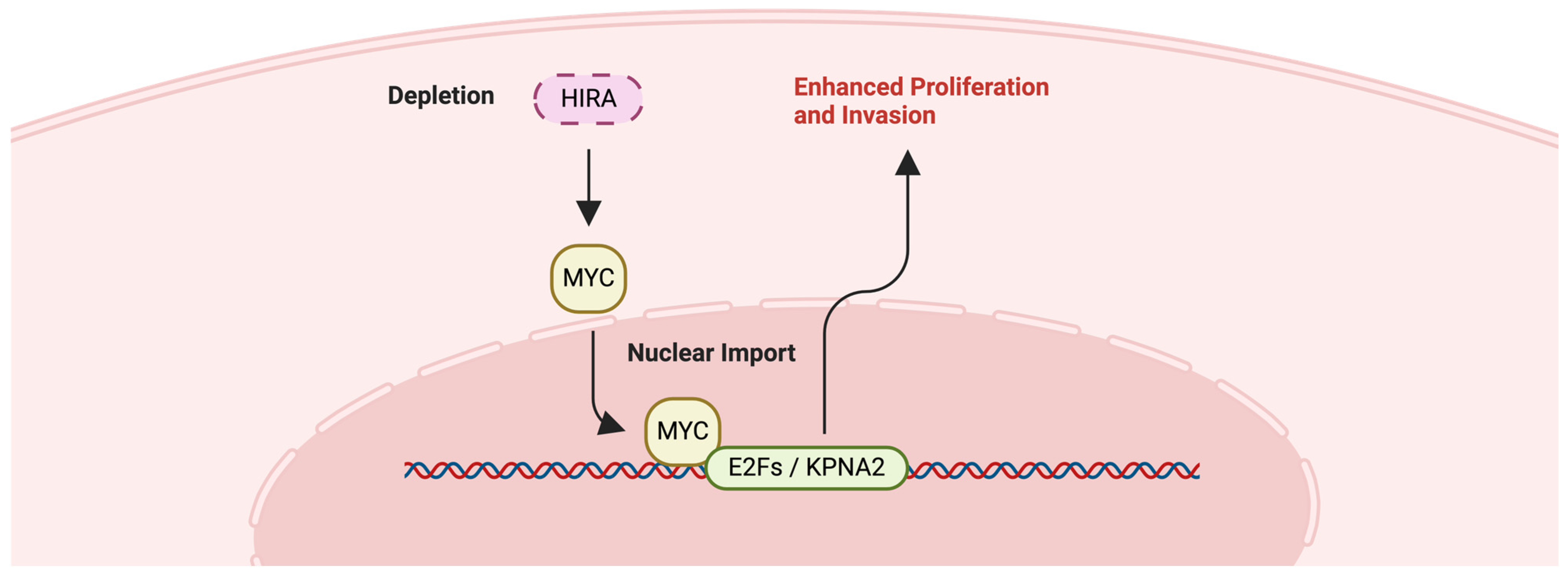
2.6. HIRA
2.6.1. Hereditary Leiomyomatosis and Renal Cell Carcinoma (HLRCC) and HIRA
2.6.2. Chronic Myeloid Leukemia (CML) and HIRA
2.6.3. HIRA-Targeted Therapy Approach: shRNA
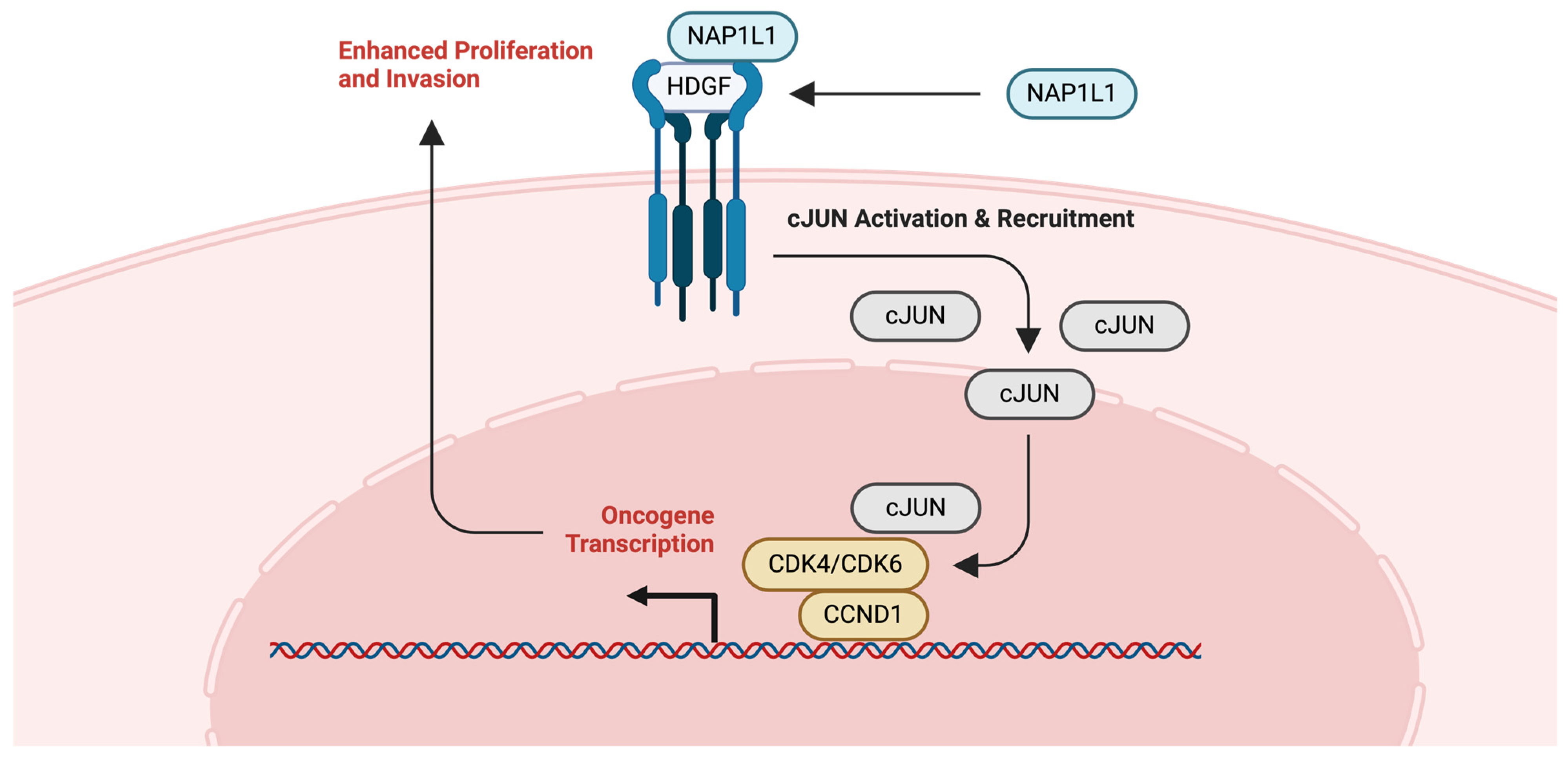
2.7. NAP1
2.7.1. Glioma and NAP1L1
2.7.2. Ovarian Cancer (OVCA) and NAP1L1
2.7.3. NAP1L1-Targeted Therapy: Gene Therapy Approach
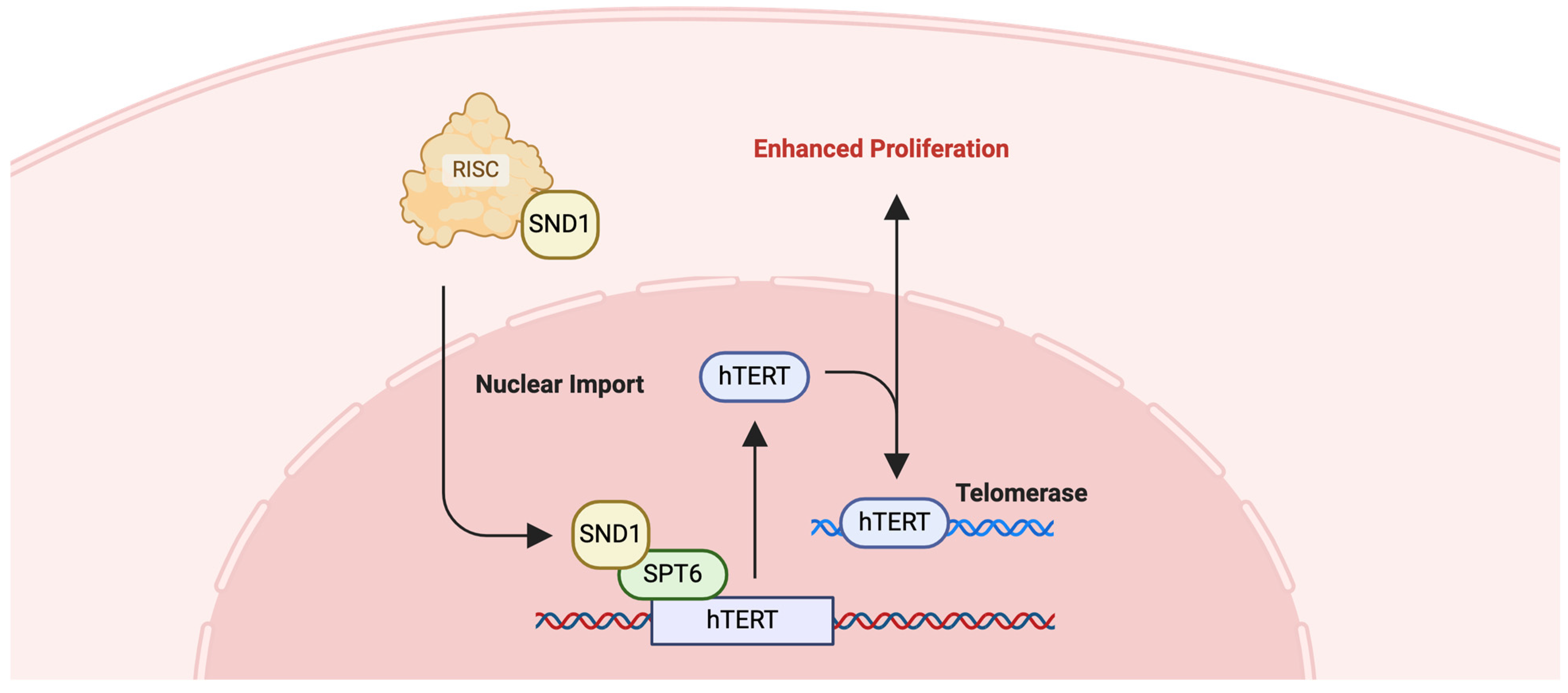
2.8. SPT6
2.8.1. Colorectal Cancer (CRC) and SPT6
2.8.2. SPT6-Targeted Therapy: Chaetocin
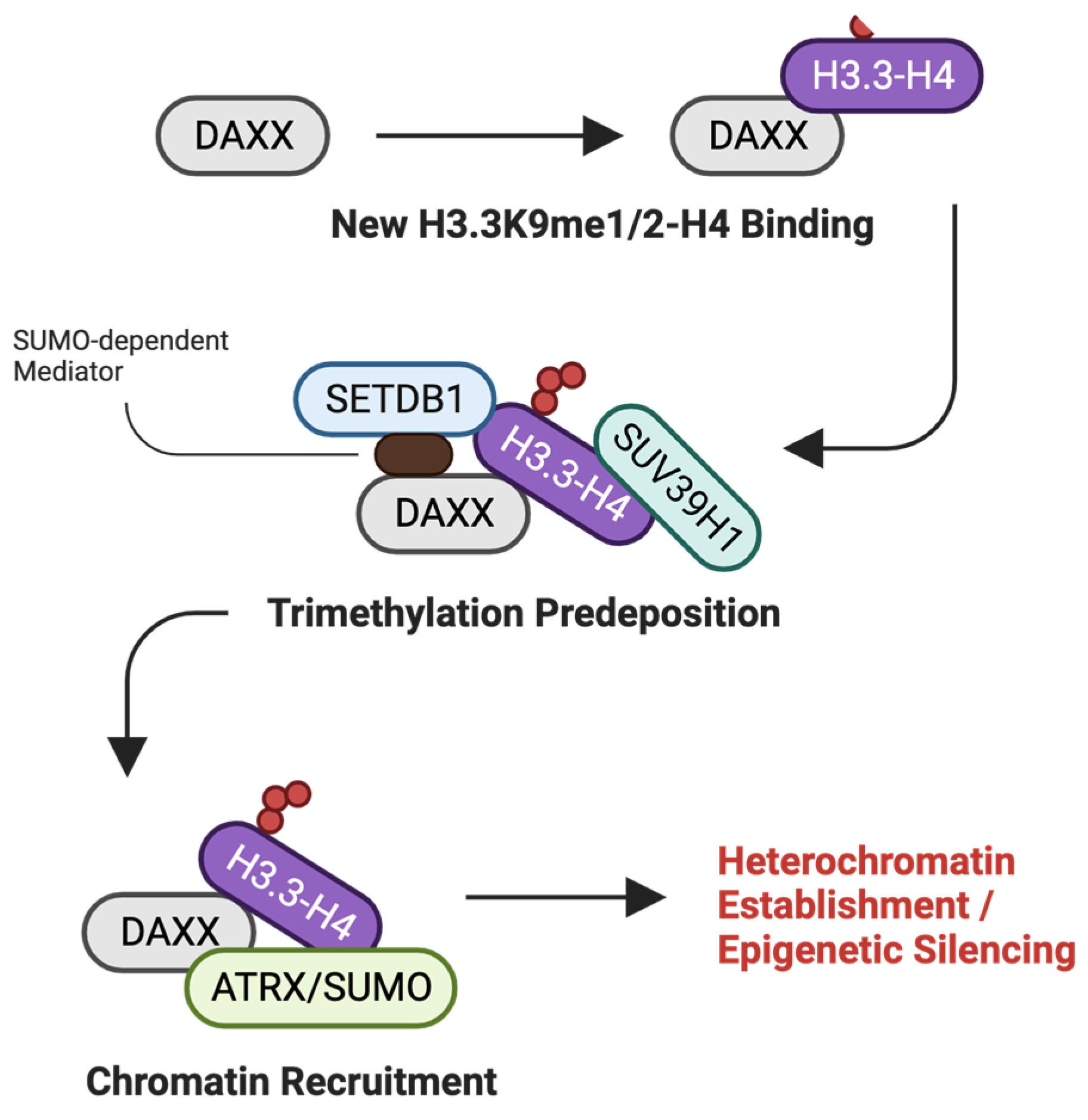
2.9. DAXX
2.9.1. Pancreatic Neuroendocrine Tumors (PanNETs) and DAXX
2.9.2. Prostate Cancer (PCa) and DAXX
2.9.3. DAXX-Targeted Therapy Approach
2.10. C1QBP
3. Discussion
4. Conclusions
Funding
Conflicts of Interest
References
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Davey, C.A.; Sargent, D.F.; Luger, K.; Maeder, A.W.; Richmond, T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002, 319, 1097–1113. [Google Scholar] [CrossRef] [PubMed]
- Bottomley, M.J. Structures of protein domains that create or recognize histone modifications. EMBO Rep. 2004, 5, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Bowman, G.D.; Poirier, M.G. Post-Translational Modifications of Histones That Influence Nucleosome Dynamics. Chem. Rev. 2015, 115, 2274–2295. [Google Scholar] [CrossRef]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Laskey, R.A.; Honda, B.M.; Mills, A.D.; Finch, J.T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 1978, 275, 416–420. [Google Scholar] [CrossRef] [PubMed]
- De Koning, L.; Corpet, A.; Haber, J.E.; Almouzni, G. Histone chaperones: An escort network regulating histone traffic. Nat. Struct. Mol. Biol. 2007, 14, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Hammond, C.M.; Stromme, C.B.; Huang, H.; Patel, D.J.; Groth, A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017, 18, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Loyola, A.; Almouzni, G. Marking histone H3 variants: How, when and why? Trends Biochem. Sci. 2007, 32, 425–433. [Google Scholar] [CrossRef]
- Scott, W.A.; Campos, E.I. Interactions With Histone H3 & Tools to Study Them. Front. Cell Dev. Biol. 2020, 8, 701. [Google Scholar] [CrossRef]
- Bobde, R.C.; Saharan, K.; Baral, S.; Gandhi, S.; Samal, A.; Sundaram, R.; Kumar, A.; Singh, A.K.; Datta, A.; Vasudevan, D. In Vitro Characterization of Histone Chaperones using Analytical, Pull-Down and Chaperoning Assays. J. Vis. Exp. 2021, 178, e63218. [Google Scholar] [CrossRef] [PubMed]
- Eitoku, M.; Sato, L.; Senda, T.; Horikoshi, M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell. Mol. Life Sci. 2008, 65, 414–444. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Y.; Johnson, S.L.; Gamarra, N.I.; Narlikar, G.J. Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annu. Rev. Biophys. 2016, 45, 153–181. [Google Scholar] [CrossRef] [PubMed]
- Das, C.; Lucia, M.S.; Hansen, K.C.; Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009, 459, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Das, C.; Tyler, J.K. Histone exchange and histone modifications during transcription and aging. Biochim. Biophys. Acta 2013, 1819, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Pchelintsev, N.A.; McBryan, T.; Rai, T.S.; van Tuyn, J.; Ray-Gallet, D.; Almouzni, G.; Adams, P.D. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 2013, 3, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Karatas, H.; Townsend, E.C.; Cao, F.; Chen, Y.; Bernard, D.; Liu, L.; Lei, M.; Dou, Y.; Wang, S. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J. Am. Chem. Soc. 2013, 135, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Seol, J.H.; Song, T.Y.; Oh, S.E.; Jo, C.; Choi, A.; Kim, B.; Park, J.; Hong, S.; Song, I.; Jung, K.Y.; et al. Identification of small molecules that inhibit the histone chaperone Asf1 and its chromatin function. BMB Rep. 2015, 48, 685–690. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Debela, D.T.; Muzazu, S.G.; Heraro, K.D.; Ndalama, M.T.; Mesele, B.W.; Haile, D.C.; Kitui, S.K.; Manyazewal, T. New approaches and procedures for cancer treatment: Current perspectives. SAGE Open Med. 2021, 9, 20503121211034366. [Google Scholar] [CrossRef]
- Marciano, G.; Da Vela, S.; Tria, G.; Svergun, D.I.; Byron, O.; Huang, D.T. Structure-specific recognition protein-1 (SSRP1) is an elongated homodimer that binds histones. J. Biol. Chem. 2018, 293, 10071–10083. [Google Scholar] [CrossRef]
- Foglia, B.; Parola, M. Of FACT complex and oxidative stress response: A KEAP1/NRF2-dependent novel mechanism sustaining hepatocellular carcinoma progression. Gut 2020, 69, 195–196. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, L.; Hong, E.; Safina, A.; Poe, D.; Gurova, K. Histone chaperone FACT is essential to overcome replication stress in mammalian cells. Oncogene 2020, 39, 5124–5137. [Google Scholar] [CrossRef] [PubMed]
- Bhakat, K.K.; Ray, S. The FAcilitates Chromatin Transcription (FACT) complex: Its roles in DNA repair and implications for cancer therapy. DNA Repair. 2022, 109, 103246. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Nizovtseva, E.V.; Razin, S.V.; Formosa, T.; Gurova, K.V.; Studitsky, V.M. Histone Chaperone FACT and Curaxins: Effects on Genome Structure and Function. J. Cancer Metastasis Treat. 2019, 5, 78. [Google Scholar] [CrossRef] [PubMed]
- Samant, H.; Amiri, H.S.; Zibari, G.B. Addressing the worldwide hepatocellular carcinoma: Epidemiology, prevention and management. J. Gastrointest. Oncol. 2021, 12, S361–S373. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Han, F.; Du, Y.; Shi, H.; Zhou, W. Hypoxic microenvironment in cancer: Molecular mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, L.; Vetrano, E.; Rinaldi, B.; Galiero, R.; Caturano, A.; Salvatore, T.; Sasso, F.C. HCC and Molecular Targeting Therapies: Back to the Future. Biomedicines 2021, 9, 1345. [Google Scholar] [CrossRef] [PubMed]
- Lukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanislawek, A. Breast Cancer-Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies—An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef] [PubMed]
- Khella, C.A.; Mehta, G.A.; Mehta, R.N.; Gatza, M.L. Recent Advances in Integrative Multi-Omics Research in Breast and Ovarian Cancer. J. Pers. Med. 2021, 11, 149. [Google Scholar] [CrossRef]
- Saldivar, J.C.; Cortez, D.; Cimprich, K.A. The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017, 18, 622–636. [Google Scholar] [CrossRef]
- Shen, J.; Yang, C.; Zhang, M.S.; Chin, D.W.; Chan, F.F.; Law, C.T.; Wang, G.; Cheng, C.L.; Chen, M.; Wan, R.T.; et al. Histone chaperone FACT complex coordinates with HIF to mediate an expeditious transcription program to adapt to poorly oxygenated cancers. Cell Rep. 2022, 38, 110304. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Cai, Z.; Jiang, T.; Han, J.; Zhang, B. Histone Chaperones and Digestive Cancer: A Review of the Literature. Cancers 2022, 14, 5584. [Google Scholar] [CrossRef]
- Volokh, O.I.; Sivkina, A.L.; Moiseenko, A.V.; Popinako, A.V.; Karlova, M.G.; Valieva, M.E.; Kotova, E.Y.; Kirpichnikov, M.P.; Formosa, T.; Studitsky, V.M.; et al. Mechanism of curaxin-dependent nucleosome unfolding by FACT. Front. Mol. Biosci. 2022, 9, 1048117. [Google Scholar] [CrossRef]
- Fleyshman, D.; Prendergast, L.; Safina, A.; Paszkiewicz, G.; Commane, M.; Morgan, K.; Attwood, K.; Gurova, K. Level of FACT defines the transcriptional landscape and aggressive phenotype of breast cancer cells. Oncotarget 2017, 8, 20525–20542. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Dou, J.; Yu, T.; Cao, P.; Fan, N.; Borjigin, U.; Nashun, B. Distinct role of histone chaperone Asf1a and Asf1b during fertilization and pre-implantation embryonic development in mice. Epigenetics Chromatin 2021, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.K.; Rabaglia, M.E.; Wang, C.Y.; Stapleton, D.S.; Leng, N.; Kendziorski, C.; Lewis, P.W.; Keller, M.P.; Attie, A.D. Histone chaperone ASF1B promotes human beta-cell proliferation via recruitment of histone H3.3. Cell Cycle 2016, 15, 3191–3202. [Google Scholar] [CrossRef] [PubMed]
- Messiaen, S.; Guiard, J.; Aigueperse, C.; Fliniaux, I.; Tourpin, S.; Barroca, V.; Allemand, I.; Fouchet, P.; Livera, G.; Vernet, M. Loss of the histone chaperone ASF1B reduces female reproductive capacity in mice. Reproduction 2016, 151, 477–489. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mbianda, J.; Bakail, M.; Andre, C.; Moal, G.; Perrin, M.E.; Pinna, G.; Guerois, R.; Becher, F.; Legrand, P.; Traore, S.; et al. Optimal anchoring of a foldamer inhibitor of ASF1 histone chaperone through backbone plasticity. Sci. Adv. 2021, 7, eabd9153. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Ouyang, H.; Pan, X.; Zhang, Z.; Tan, J.; Yu, N.; Li, M.; Zhao, Y. Increased ASF1B Expression Correlates With Poor Prognosis in Patients With Gliomas. Front. Oncol. 2022, 12, 912101. [Google Scholar] [CrossRef]
- Liang, X.; Yuan, X.; Yu, J.; Wu, Y.; Li, K.; Sun, C.; Li, S.; Shen, L.; Kong, F.; Jia, J.; et al. Histone Chaperone ASF1A Predicts Poor Outcomes for Patients With Gastrointestinal Cancer and Drives Cancer Progression by Stimulating Transcription of beta-Catenin Target Genes. eBioMedicine 2017, 21, 104–116. [Google Scholar] [CrossRef]
- Myers, D.J.; Wallen, J.M. Lung Adenocarcinoma. In StatPearls; StatPearls: Treasure Island, FL, USA, 2024. [Google Scholar]
- Li, F.; Huang, Q.; Luster, T.A.; Hu, H.; Zhang, H.; Ng, W.L.; Khodadadi-Jamayran, A.; Wang, W.; Chen, T.; Deng, J.; et al. In Vivo Epigenetic CRISPR Screen Identifies Asf1a as an Immunotherapeutic Target in Kras-Mutant Lung Adenocarcinoma. Cancer Discov. 2020, 10, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Watterson, A.; Coelho, M.A. Cancer immune evasion through KRAS and PD-L1 and potential therapeutic interventions. Cell Commun. Signal 2023, 21, 45. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Taghi Khani, A.; Sanchez Ortiz, A.; Swaminathan, S. GM-CSF: A Double-Edged Sword in Cancer Immunotherapy. Front. Immunol. 2022, 13, 901277. [Google Scholar] [CrossRef]
- Brettingham-Moore, K.H.; Sprod, O.R.; Chen, X.; Oakford, P.; Shannon, M.F.; Holloway, A.F. Determinants of a transcriptionally competent environment at the GM-CSF promoter. Nucleic Acids Res. 2008, 36, 2639–2653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, Z.; Guan, M.; Liu, N.; Meng, F.; Wang, G. ASF1B Promotes Oncogenesis in Lung Adenocarcinoma and Other Cancer Types. Front. Oncol. 2021, 11, 731547. [Google Scholar] [CrossRef] [PubMed]
- Bellelli, R.; Belan, O.; Pye, V.E.; Clement, C.; Maslen, S.L.; Skehel, J.M.; Cherepanov, P.; Almouzni, G.; Boulton, S.J. POLE3-POLE4 Is a Histone H3-H4 Chaperone that Maintains Chromatin Integrity during DNA Replication. Mol. Cell 2018, 72, 112–126.e115. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wang, S.; Zangari, M.; Xu, H.; Cao, T.M.; Xu, C.; Wu, Y.; Xiao, F.; Liu, Y.; Yang, Y.; et al. Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget 2010, 1, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Fei-Zhang, D.J.; Moazzam, Z.; Ejaz, A.; Cloyd, J.; Dillhoff, M.; Beane, J.; Bentrem, D.J.; Pawlik, T.M. The impact of digital inequities on gastrointestinal cancer disparities in the United States. J. Surg. Oncol. 2023, 128, 155–166. [Google Scholar] [CrossRef]
- Jardim, S.R.; de Souza, L.M.P.; de Souza, H.S.P. The Rise of Gastrointestinal Cancers as a Global Phenomenon: Unhealthy Behavior or Progress? Int. J. Environ. Res. Public Health 2023, 20, 3640. [Google Scholar] [CrossRef]
- Lecarpentier, Y.; Schussler, O.; Hebert, J.L.; Vallee, A. Multiple Targets of the Canonical WNT/beta-Catenin Signaling in Cancers. Front. Oncol. 2019, 9, 1248. [Google Scholar] [CrossRef]
- Chen, C.; Bao, H.; Lin, W.; Chen, X.; Huang, Y.; Wang, H.; Yang, Y.; Liu, J.; Lv, X.; Teng, L. ASF1b is a novel prognostic predictor associated with cell cycle signaling pathway in gastric cancer. J. Cancer 2022, 13, 1985–2000. [Google Scholar] [CrossRef] [PubMed]
- Miknis, G.F.; Stevens, S.J.; Smith, L.E.; Ostrov, D.A.; Churchill, M.E. Development of novel Asf1-H3/H4 inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 963–968. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cherry, A.L.; Nott, T.J.; Kelly, G.; Rulten, S.L.; Caldecott, K.W.; Smerdon, S.J. Versatility in phospho-dependent molecular recognition of the XRCC1 and XRCC4 DNA-damage scaffolds by aprataxin-family FHA domains. DNA Repair. 2015, 35, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, P.V.; Ahel, D.; Ryan, D.P.; Weston, R.; Wiechens, N.; Kraehenbuehl, R.; Owen-Hughes, T.; Ahel, I. DNA repair factor APLF is a histone chaperone. Mol. Cell 2011, 41, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Corbeski, I.; Dolinar, K.; Wienk, H.; Boelens, R.; van Ingen, H. DNA repair factor APLF acts as a H2A-H2B histone chaperone through binding its DNA interaction surface. Nucleic Acids Res. 2018, 46, 7138–7152. [Google Scholar] [CrossRef] [PubMed]
- Corbeski, I.; Guo, X.; Eckhardt, B.V.; Fasci, D.; Wiegant, W.; Graewert, M.A.; Vreeken, K.; Wienk, H.; Svergun, D.I.; Heck, A.J.R.; et al. Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF. Sci. Adv. 2022, 8, eabo0517. [Google Scholar] [CrossRef] [PubMed]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee Sh, U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar] [CrossRef]
- Yalamarty, S.S.K.; Filipczak, N.; Li, X.; Subhan, M.A.; Parveen, F.; Ataide, J.A.; Rajmalani, B.A.; Torchilin, V.P. Mechanisms of Resistance and Current Treatment Options for Glioblastoma Multiforme (GBM). Cancers 2023, 15, 2116. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.; Costa, A.; Osorio, L.; Lago, R.C.; Linhares, P.; Carvalho, B.; Caeiro, C. Current Standards of Care in Glioblastoma Therapy. In Glioblastoma; De Vleeschouwer, S., Ed.; Exon Publications: Brisbane, Australia, 2017. [Google Scholar]
- Dong, W.; Li, L.; Teng, X.; Yang, X.; Si, S.; Chai, J. End Processing Factor APLF Promotes NHEJ Efficiency and Contributes to TMZ- and Ionizing Radiation-Resistance in Glioblastoma Cells. Onco Targets Ther. 2020, 13, 10593–10605. [Google Scholar] [CrossRef]
- Grundy, G.J.; Rulten, S.L.; Zeng, Z.; Arribas-Bosacoma, R.; Iles, N.; Manley, K.; Oliver, A.; Caldecott, K.W. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013, 32, 112–125. [Google Scholar] [CrossRef]
- Khochikar, M.V. Rationale for an early detection program for bladder cancer. Indian. J. Urol. 2011, 27, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Khan, S.M.; Mahajan, M.; Sharma, P.; Mahajan, A. Urinary Bladder Carcinoma in Females: A Clinico-Pathological Assessment. Cureus 2023, 15, e39753. [Google Scholar] [CrossRef] [PubMed]
- Viveiros, N.; Flores, B.C.; Lobo, J.; Martins-Lima, C.; Cantante, M.; Lopes, P.; Deantonio, C.; Palu, C.; Sainson, R.C.; Henrique, R.; et al. Detailed bladder cancer immunoprofiling reveals new clues for immunotherapeutic strategies. Clin. Transl. Immunol. 2022, 11, e1402. [Google Scholar] [CrossRef]
- Minoli, M.; Kiener, M.; Thalmann, G.N.; Kruithof-de Julio, M.; Seiler, R. Evolution of Urothelial Bladder Cancer in the Context of Molecular Classifications. Int. J. Mol. Sci. 2020, 21, 5670. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.; Marquardt, S.; Li, F.; Spitschak, A.; Murr, N.; Edelhauser, B.A.H.; Iliakis, G.; Putzer, B.M.; Logotheti, S. Rewiring E2F1 with classical NHEJ via APLF suppression promotes bladder cancer invasiveness. J. Exp. Clin. Cancer Res. 2019, 38, 292. [Google Scholar] [CrossRef] [PubMed]
- Porwit, A.; McCullough, J.; Erber, W.N. Blood and Bone Marrow Pathology; Elsevier Health Sciences: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Senapati, P.; Bhattacharya, A.; Das, S.; Dey, S.; Sudarshan, D.; Vishwakarma, J.; Sudevan, S.; Ramachandran, R.; Maliekal, T.T.; Kundu, T.K. Histone Chaperone Nucleophosmin Regulates Transcription of Key Genes Involved in Oral Tumorigenesis. Mol. Cell. Biol. 2022, 42, e0066920. [Google Scholar] [CrossRef]
- Lindstrom, M.S. NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochem. Res. Int. 2011, 2011, 195209. [Google Scholar] [CrossRef] [PubMed]
- Chin, L.; Wong, C.Y.G.; Gill, H. Targeting and Monitoring Acute Myeloid Leukaemia with Nucleophosmin-1 (NPM1) Mutation. Int. J. Mol. Sci. 2023, 24, 3161. [Google Scholar] [CrossRef]
- Laliberte, C.; Ng, N.; Eymael, D.; Higgins, K.; Ali, A.; Kiss, A.; Bradley, G.; Magalhaes, M.A.O. Characterization of Oral Squamous Cell Carcinoma Associated Inflammation: A Pilot Study. Front. Oral. Health 2021, 2, 740469. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Warnakulasuriya, S. Global epidemiology of oral and oropharyngeal cancer. Oral. Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Vakiti, A.; Mewawalla, P. Acute Myeloid Leukemia. Available online: https://www.statpearls.com/point-of-care/25443 (accessed on 27 April 2024).
- Leukemia—Acute Myeloid—AML: Statistics. Available online: https://www.cancer.net/cancer-types/leukemia-acute-myeloid-aml/statistics (accessed on 17 January 2024).
- Medeiros, B.C.; Chan, S.M.; Daver, N.G.; Jonas, B.A.; Pollyea, D.A. Optimizing survival outcomes with post-remission therapy in acute myeloid leukemia. Am. J. Hematol. 2019, 94, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Pollyea, D.A.; Altman, J.K.; Assi, R.; Bixby, D.; Fathi, A.T.; Foran, J.M.; Gojo, I.; Hall, A.C.; Jonas, B.A.; Kishtagari, A.; et al. Acute Myeloid Leukemia, Version 3.2023, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2023, 21, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Nicoletti, I.; Martelli, M.F.; Mecucci, C. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): Biologic and clinical features. Blood 2007, 109, 874–885. [Google Scholar] [CrossRef]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Federici, L.; Falini, B. Nucleophosmin mutations in acute myeloid leukemia: A tale of protein unfolding and mislocalization. Protein Sci. 2013, 22, 545–556. [Google Scholar] [CrossRef]
- Sekhar, K.R.; Freeman, M.L. Nucleophosmin Plays a Role in Repairing DNA Damage and Is a Target for Cancer Treatment. Cancer Res. 2023, 83, 1573–1580. [Google Scholar] [CrossRef] [PubMed]
- Ziv, O.; Zeisel, A.; Mirlas-Neisberg, N.; Swain, U.; Nevo, R.; Ben-Chetrit, N.; Martelli, M.P.; Rossi, R.; Schiesser, S.; Canman, C.E.; et al. Identification of novel DNA-damage tolerance genes reveals regulation of translesion DNA synthesis by nucleophosmin. Nat. Commun. 2014, 5, 5437. [Google Scholar] [CrossRef] [PubMed]
- Rechkoblit, O.; Johnson, R.E.; Buku, A.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Structural insights into mutagenicity of anticancer nucleoside analog cytarabine during replication by DNA polymerase eta. Sci. Rep. 2019, 9, 16400. [Google Scholar] [CrossRef]
- Balusu, R.; Fiskus, W.; Rao, R.; Chong, D.G.; Nalluri, S.; Mudunuru, U.; Ma, H.; Chen, L.; Venkannagari, S.; Ha, K.; et al. Targeting levels or oligomerization of nucleophosmin 1 induces differentiation and loss of survival of human AML cells with mutant NPM1. Blood 2011, 118, 3096–3106. [Google Scholar] [CrossRef]
- Zhou, Y.; Fang, Y.; Zhou, J.; Liu, Y.; Wu, S.; Xu, B. NPM1 is a Novel Therapeutic Target and Prognostic Biomarker for Ewing Sarcoma. Front. Genet. 2021, 12, 771253. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Przegląd Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Liang, X.; Li, S.; Li, T.; Shang, W.; Ma, L.; Jia, X.; Shao, W.; Sun, P.; Chen, C.; et al. CHAF1A interacts with TCF4 to promote gastric carcinogenesis via upregulation of c-MYC and CCND1 expression. eBioMedicine 2018, 38, 69–78. [Google Scholar] [CrossRef]
- Wang, D.; Li, X.; Shen, B.; Chen, X.; Shu, Y. Histone chaperone CHAF1A impacts the outcome of fluoropyrimidines-based adjuvant therapy in gastric cancer by regulating the expression of thymidylate synthetase. Gene 2019, 716, 144034. [Google Scholar] [CrossRef]
- Padala, S.A. Diffuse Large B-Cell Lymphoma. Available online: https://www.statpearls.com/point-of-care/24581 (accessed on 24 April 2023).
- Wang, L.; Li, L.R.; Young, K.H. New agents and regimens for diffuse large B cell lymphoma. J. Hematol. Oncol. 2020, 13, 175. [Google Scholar] [CrossRef]
- Yan, W.; Shi, X.; Wang, H.; Liao, A.; Yang, W. Aberrant SPOP-CHAF1A ubiquitination axis triggers tumor autophagy that endows a therapeutical vulnerability in diffuse large B cell lymphoma. J. Transl. Med. 2022, 20, 296. [Google Scholar] [CrossRef] [PubMed]
- Koroulakis, A. Laryngeal Cancer. Available online: https://www.statpearls.com/point-of-care/24035 (accessed on 7 May 2024).
- Zhang, W.; Zeng, W.; Jiang, A.; He, Z.; Shen, X.; Dong, X.; Feng, J.; Lu, H. Global, regional and national incidence, mortality and disability-adjusted life-years of skin cancers and trend analysis from 1990 to 2019: An analysis of the Global Burden of Disease Study 2019. Cancer Med. 2021, 10, 4905–4922. [Google Scholar] [CrossRef] [PubMed]
- Mascolo, M.; Ilardi, G.; Merolla, F.; Russo, D.; Vecchione, M.L.; De Rosa, G.; Staibano, S. Tissue microarray-based evaluation of Chromatin Assembly Factor-1 (CAF-1)/p60 as tumour prognostic marker. Int. J. Mol. Sci. 2012, 13, 11044–11062. [Google Scholar] [CrossRef]
- Dotto, G.P.; Rustgi, A.K. Squamous Cell Cancers: A Unified Perspective on Biology and Genetics. Cancer Cell 2016, 29, 622–637. [Google Scholar] [CrossRef]
- Sun, Q.; Fang, Q.; Guo, S. A comparison of oral squamous cell carcinoma between young and old patients in a single medical center in China. Int. J. Clin. Exp. Med. 2015, 8, 12418–12423. [Google Scholar]
- Staibano, S.; Mignogna, C.; Lo Muzio, L.; Mascolo, M.; Salvatore, G.; Di Benedetto, M.; Califano, L.; Rubini, C.; De Rosa, G. Chromatin assembly factor-1 (CAF-1)-mediated regulation of cell proliferation and DNA repair: A link with the biological behaviour of squamous cell carcinoma of the tongue? Histopathology 2007, 50, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Fowler, J.R.; Maani, E.V.; Dunton, C.J.; Gasalberti, D.P.; Jack, B.W. Cervical Cancer. In StatPearls; StatPearls: Treasure Island, FL, USA, 2024. [Google Scholar]
- Stelzle, D.; Tanaka, L.F.; Lee, K.K.; Ibrahim Khalil, A.; Baussano, I.; Shah, A.S.V.; McAllister, D.A.; Gottlieb, S.L.; Klug, S.J.; Winkler, A.S.; et al. Estimates of the global burden of cervical cancer associated with HIV. Lancet Glob. Health 2021, 9, e161–e169. [Google Scholar] [CrossRef]
- Kong, L.; Yu, X.P.; Bai, X.H.; Zhang, W.F.; Zhang, Y.; Zhao, W.M.; Jia, J.H.; Tang, W.; Zhou, Y.B.; Liu, C.J. RbAp48 is a critical mediator controlling the transforming activity of human papillomavirus type 16 in cervical cancer. J. Biol. Chem. 2007, 282, 26381–26391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nicolas, E.; Ait-Si-Ali, S.; Trouche, D. The histone deacetylase HDAC3 targets RbAp48 to the retinoblastoma protein. Nucleic Acids Res. 2001, 29, 3131–3136. [Google Scholar] [CrossRef] [PubMed]
- Johansson, C.; Jamal Fattah, T.; Yu, H.; Nygren, J.; Mossberg, A.K.; Schwartz, S. Acetylation of intragenic histones on HPV16 correlates with enhanced HPV16 gene expression. Virology 2015, 482, 244–259. [Google Scholar] [CrossRef]
- Lee, K.; Anastasopoulou, C.; Chandran, C.; Cassaro, S. Thyroid Cancer. In StatPearls; StatPearls: Treasure Island, FL, USA, 2024. [Google Scholar]
- Lee, H.; Kim, S.Y.; Kim, S.M.; Chang, H.J.; Lee, Y.S.; Park, C.S.; Chang, H.S. Long-term survival of patients with anaplastic thyroid cancer after multimodal treatment. Transl. Cancer Res. 2020, 9, 5430–5436. [Google Scholar] [CrossRef]
- Pozdeyev, N.; Berlinberg, A.; Zhou, Q.; Wuensch, K.; Shibata, H.; Wood, W.M.; Haugen, B.R. Targeting the NF-kappaB Pathway as a Combination Therapy for Advanced Thyroid Cancer. PLoS ONE 2015, 10, e0134901. [Google Scholar] [CrossRef]
- Smith, M. Chapter 14—The regulatory genome and complex common diseases. In The Regulatory Genome in Adaptation, Evolution, Development, and Disease; Smith, M., Ed.; Academic Press: Cambridge, MA, USA, 2023; pp. 359–388. [Google Scholar]
- Lamour, V.; Lecluse, Y.; Desmaze, C.; Spector, M.; Bodescot, M.; Aurias, A.; Osley, M.A.; Lipinski, M. A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum. Mol. Genet. 1995, 4, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef]
- Rai, T.S.; Cole, J.J.; Nelson, D.M.; Dikovskaya, D.; Faller, W.J.; Vizioli, M.G.; Hewitt, R.N.; Anannya, O.; McBryan, T.; Manoharan, I.; et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 2014, 28, 2712–2725. [Google Scholar] [CrossRef]
- Schmidt, L.S.; Linehan, W.M. Hereditary leiomyomatosis and renal cell carcinoma. Int. J. Nephrol. Renov. Dis. 2014, 7, 253–260. [Google Scholar] [CrossRef]
- Menko, F.H.; Maher, E.R.; Schmidt, L.S.; Middelton, L.A.; Aittomaki, K.; Tomlinson, I.; Richard, S.; Linehan, W.M. Hereditary leiomyomatosis and renal cell cancer (HLRCC): Renal cancer risk, surveillance and treatment. Fam. Cancer 2014, 13, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Valcarcel-Jimenez, L.; Rogerson, C.; Yong, C.; Schmidt, C.; Yang, M.; Cremades-Rodelgo, M.; Harle, V.; Offord, V.; Wong, K.; Mora, A.; et al. HIRA loss transforms FH-deficient cells. Sci. Adv. 2022, 8, eabq8297. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, Metabolism, and Cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- PDQ Adult Treatment Editorial Board. Chronic Myelogenous Leukemia Treatment (PDQ(R)): Health Professional Version. In PDQ Cancer Information Summaries; National Cancer Institute (US): Bethesda, MD, USA, 2002. [Google Scholar]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2020 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2020, 95, 691–709. [Google Scholar] [CrossRef] [PubMed]
- Uhm, J. Treatment after failure of frontline therapy of chronic myeloid leukemia in chronic phase including allogeneic hematopoietic stem cell transplantation. Blood Res. 2023, 58, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Majumder, A.; Dharan, A.T.; Baral, I.; Varghese, P.C.; Mukherjee, A.; Subhadradevi, L.; Narayanan, G.; Dutta, D. Histone chaperone HIRA dictate proliferation vs differentiation of chronic myeloid leukemia cells. FASEB Bioadv. 2019, 1, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Al-Bassam, J.; Corbett, K.D. alpha-Tubulin acetylation from the inside out. Proc. Natl. Acad. Sci. USA 2012, 109, 19515–19516. [Google Scholar] [CrossRef]
- Andrews, A.J.; Chen, X.; Zevin, A.; Stargell, L.A.; Luger, K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 2010, 37, 834–842. [Google Scholar] [CrossRef]
- Chen, Z.; Xie, Y.; Luo, H.; Song, Y.; Que, T.; Hu, R.; Huang, H.; Luo, K.; Li, C.; Qin, C.; et al. NAP1L1 promotes proliferation and chemoresistance in glioma by inducing CCND1/CDK4/CDK6 expression through its interaction with HDGF and activation of c-Jun. Aging 2021, 13, 26180–26200. [Google Scholar] [CrossRef]
- Friedmann, D.R.; Aguilar, A.; Fan, J.; Nachury, M.V.; Marmorstein, R. Structure of the alpha-tubulin acetyltransferase, alphaTAT1, and implications for tubulin-specific acetylation. Proc. Natl. Acad. Sci. USA 2012, 109, 19655–19660. [Google Scholar] [CrossRef] [PubMed]
- Taal, W.; Bromberg, J.E.; van den Bent, M.J. Chemotherapy in glioma. CNS Oncol. 2015, 4, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Arora, T.; Mullangi, S. Epithelial Ovarian Cancer. Available online: https://www.statpearls.com/point-of-care/95586 (accessed on 6 May 2024).
- Zhu, J.W.; Charkhchi, P.; Akbari, M.R. Potential clinical utility of liquid biopsies in ovarian cancer. Mol. Cancer 2022, 21, 114. [Google Scholar] [CrossRef] [PubMed]
- Rampes, S.; Choy, S.P. Early diagnosis of symptomatic ovarian cancer in primary care in the UK: Opportunities and challenges. Prim. Health Care Res. Dev. 2022, 23, e52. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.L.; Leung, C.S.; Yip, K.P.; Au Yeung, C.L.; Wong, S.T.; Mok, S.C. Cellular and molecular processes in ovarian cancer metastasis. A Review in the Theme: Cell and Molecular Processes in Cancer Metastasis. Am. J. Physiol. Cell Physiol. 2015, 309, C444–C456. [Google Scholar] [CrossRef] [PubMed]
- Xiaohua, Z.; Xie, Y.; Huang, W.; Chen, Z.; Guo, S. NAP1L1 promotes tumor proliferation through HDGF/C-JUN signaling in ovarian cancer. BMC Cancer 2022, 22, 339. [Google Scholar] [CrossRef] [PubMed]
- Dronamraju, R.; Hepperla, A.J.; Shibata, Y.; Adams, A.T.; Magnuson, T.; Davis, I.J.; Strahl, B.D. Spt6 Association with RNA Polymerase II Directs mRNA Turnover During Transcription. Mol. Cell 2018, 70, 1054–1066.e1054. [Google Scholar] [CrossRef]
- McCullough, L.; Connell, Z.; Petersen, C.; Formosa, T. The Abundant Histone Chaperones Spt6 and FACT Collaborate to Assemble, Inspect, and Maintain Chromatin Structure in Saccharomyces cerevisiae. Genetics 2015, 201, 1031–1045. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef]
- Hossain, M.S.; Karuniawati, H.; Jairoun, A.A.; Urbi, Z.; Ooi, J.; John, A.; Lim, Y.C.; Kibria, K.M.K.; Mohiuddin, A.K.M.; Ming, L.C.; et al. Colorectal Cancer: A Review of Carcinogenesis, Global Epidemiology, Current Challenges, Risk Factors, Preventive and Treatment Strategies. Cancers 2022, 14, 1732. [Google Scholar] [CrossRef]
- Zhou, H.; Zhu, L.; Song, J.; Wang, G.; Li, P.; Li, W.; Luo, P.; Sun, X.; Wu, J.; Liu, Y.; et al. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol. Cancer 2022, 21, 86. [Google Scholar] [CrossRef] [PubMed]
- Diao, C.; Guo, P.; Yang, W.; Sun, Y.; Liao, Y.; Yan, Y.; Zhao, A.; Cai, X.; Hao, J.; Hu, S.; et al. SPT6 recruits SND1 to co-activate human telomerase reverse transcriptase to promote colon cancer progression. Mol. Oncol. 2021, 15, 1180–1202. [Google Scholar] [CrossRef] [PubMed]
- Drane, P.; Ouararhni, K.; Depaux, A.; Shuaib, M.; Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010, 24, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Rapkin, L.M.; Ahmed, K.; Dulev, S.; Li, R.; Kimura, H.; Ishov, A.M.; Bazett-Jones, D.P. The histone chaperone DAXX maintains the structural organization of heterochromatin domains. Epigenetics Chromatin 2015, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Carraro, M.; Hendriks, I.A.; Hammond, C.M.; Solis-Mezarino, V.; Volker-Albert, M.; Elsborg, J.D.; Weisser, M.B.; Spanos, C.; Montoya, G.; Rappsilber, J.; et al. DAXX adds a de novo H3.3K9me3 deposition pathway to the histone chaperone network. Mol. Cell 2023, 83, 1075–1092.e1079. [Google Scholar] [CrossRef] [PubMed]
- Radu, E.C.; Saizu, A.I.; Grigorescu, R.R.; Croitoru, A.E.; Gheorghe, C. Metastatic neuroendocrine pancreatic tumor—Case report. J. Med. Life 2018, 11, 57–61. [Google Scholar] [PubMed]
- Ro, C.; Chai, W.; Yu, V.E.; Yu, R. Pancreatic neuroendocrine tumors: Biology, diagnosis, and treatment. Chin. J. Cancer 2013, 32, 312–324. [Google Scholar] [CrossRef]
- Ueda, H.; Akiyama, Y.; Shimada, S.; Mogushi, K.; Serizawa, M.; Matsumura, S.; Mitsunori, Y.; Aihara, A.; Ban, D.; Ochiai, T.; et al. Tumor suppressor functions of DAXX through histone H3.3/H3K9me3 pathway in pancreatic NETs. Endocr.-Relat. Cancer 2018, 25, 619–631. [Google Scholar] [CrossRef]
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef]
- Crawford, E.D.; Heidenreich, A.; Lawrentschuk, N.; Tombal, B.; Pompeo, A.C.L.; Mendoza-Valdes, A.; Miller, K.; Debruyne, F.M.J.; Klotz, L. Androgen-targeted therapy in men with prostate cancer: Evolving practice and future considerations. Prostate Cancer Prostatic Dis. 2019, 22, 24–38. [Google Scholar] [CrossRef]
- Buskin, A.; Scott, E.; Nelson, R.; Gaughan, L.; Robson, C.N.; Heer, R.; Hepburn, A.C. Engineering prostate cancer in vitro: What does it take? Oncogene 2023, 42, 2417–2427. [Google Scholar] [CrossRef]
- Puto, L.A.; Benner, C.; Hunter, T. The DAXX co-repressor is directly recruited to active regulatory elements genome-wide to regulate autophagy programs in a model of human prostate cancer. Oncoscience 2015, 2, 362–372. [Google Scholar] [CrossRef][Green Version]
- Wang, J.; Huang, C.L.; Zhang, Y. Complement C1q Binding Protein (C1QBP): Physiological Functions, Mutation-Associated Mitochondrial Cardiomyopathy and Current Disease Models. Front. Cardiovasc. Med. 2022, 9, 843853. [Google Scholar] [CrossRef] [PubMed]
- Barna, J.; Dimen, D.; Puska, G.; Kovacs, D.; Csikos, V.; Olah, S.; Udvari, E.B.; Pal, G.; Dobolyi, A. Complement component 1q subcomponent binding protein in the brain of the rat. Sci. Rep. 2019, 9, 4597. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Bao, X.; Li, X.D. A tri-functional amino acid enables mapping of binding sites for posttranslational-modification-mediated protein-protein interactions. Mol. Cell 2021, 81, 2669–2681.e2669. [Google Scholar] [CrossRef]
- Scully, O.J.; Shyamasundar, S.; Matsumoto, K.; Dheen, S.T.; Yip, G.W.; Bay, B.H. C1QBP Mediates Breast Cancer Cell Proliferation and Growth via Multiple Potential Signalling Pathways. Int. J. Mol. Sci. 2023, 24, 1343. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fu, D.; Su, J.; Chen, Y.; Qi, C.; Sun, Y.; Niu, Y.; Zhang, N.; Yue, D. C1QBP suppresses cell adhesion and metastasis of renal carcinoma cells. Sci. Rep. 2017, 7, 999. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Zhang, X.; Wang, Y.; Gan, H.; Xu, X.; Lv, X.; Hua, X.; Que, J.; Ordog, T.; Zhang, Z. Chromatin Assembly Factor 1 (CAF-1) facilitates the establishment of facultative heterochromatin during pluripotency exit. Nucleic Acids Res. 2019, 47, 11114–11131. [Google Scholar] [CrossRef]
- Rouillon, C.; Eckhardt, B.V.; Kollenstart, L.; Gruss, F.; Verkennis, A.E.E.; Rondeel, I.; Krijger, P.H.L.; Ricci, G.; Biran, A.; van Laar, T.; et al. CAF-1 deposits newly synthesized histones during DNA replication using distinct mechanisms on the leading and lagging strands. Nucleic Acids Res. 2023, 51, 3770–3792. [Google Scholar] [CrossRef]
- Ahmad, A.; Karim, H. Mutation on WD Dipeptide Motifs of the p48 Subunit of Chromatin Assembly Factor-1 Causing Viability and Growth of DT40 Chicken B Cell Line. Indones. J. Chem. 2010, 10, 245–250. [Google Scholar] [CrossRef]
- Kadyrova, L.Y.; Rodriges Blanko, E.; Kadyrov, F.A. Human CAF-1-dependent nucleosome assembly in a defined system. Cell Cycle 2013, 12, 3286–3297. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Geis, F.K.; Sabo, Y.; Chen, X.; Li, Y.; Lu, C.; Goff, S.P. CHAF1A/B mediate silencing of unintegrated HIV-1 DNAs early in infection. Proc. Natl. Acad. Sci. USA 2022, 119, e2116735119. [Google Scholar] [CrossRef] [PubMed]
- Sykaras, A.G.; Pergaris, A.; Theocharis, S. Challenging, Accurate and Feasible: CAF-1 as a Tumour Proliferation Marker of Diagnostic and Prognostic Value. Cancers 2021, 13, 2575. [Google Scholar] [CrossRef] [PubMed]
- Pacifico, F.; Paolillo, M.; Chiappetta, G.; Crescenzi, E.; Arena, S.; Scaloni, A.; Monaco, M.; Vascotto, C.; Tell, G.; Formisano, S.; et al. RbAp48 is a target of nuclear factor-kappaB activity in thyroid cancer. J. Clin. Endocrinol. Metab. 2007, 92, 1458–1466. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zheng, X.; Zhang, L.; Zhou, B.; Hu, H.; Li, Z.; Zhang, L.; Lin, Y.; Wang, X. Histone H4 expression is cooperatively maintained by IKKbeta and Akt1 which attenuates cisplatin-induced apoptosis through the DNA-PK/RIP1/IAPs signaling cascade. Sci. Rep. 2017, 7, 41715. [Google Scholar] [CrossRef] [PubMed]
- Verreault, A.; Kaufman, P.D.; Kobayashi, R.; Stillman, B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 1996, 87, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.F.; Wong, C.M. PP039 Inhibition of CAF-1 histone chaperone complex triggers cytosolic DNA and dsRNA sensing pathways and induces intrinsic immunity of hepatocellular carcinoma. ESMO Open 2022, 7, 100725. [Google Scholar] [CrossRef]
- Gomes, A.P.; Ilter, D.; Low, V.; Rosenzweig, A.; Shen, Z.J.; Schild, T.; Rivas, M.A.; Er, E.E.; McNally, D.R.; Mutvei, A.P.; et al. Dynamic Incorporation of Histone H3 Variants into Chromatin Is Essential for Acquisition of Aggressive Traits and Metastatic Colonization. Cancer Cell 2019, 36, 402–417.e413. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, X.; Zhang, Y.; Tang, Y.; Fang, W.; Liu, X.; Liu, Z. NAP1L1 targeting suppresses the proliferation of nasopharyngeal carcinoma. Biomed. Pharmacother. 2021, 143, 112096. [Google Scholar] [CrossRef]
- Obara, E.A.A.; Aguilar-Morante, D.; Rasmussen, R.D.; Frias, A.; Vitting-Serup, K.; Lim, Y.C.; Elbaek, K.J.; Pedersen, H.; Vardouli, L.; Jensen, K.E.; et al. SPT6-driven error-free DNA repair safeguards genomic stability of glioblastoma cancer stem-like cells. Nat. Commun. 2020, 11, 4709. [Google Scholar] [CrossRef]
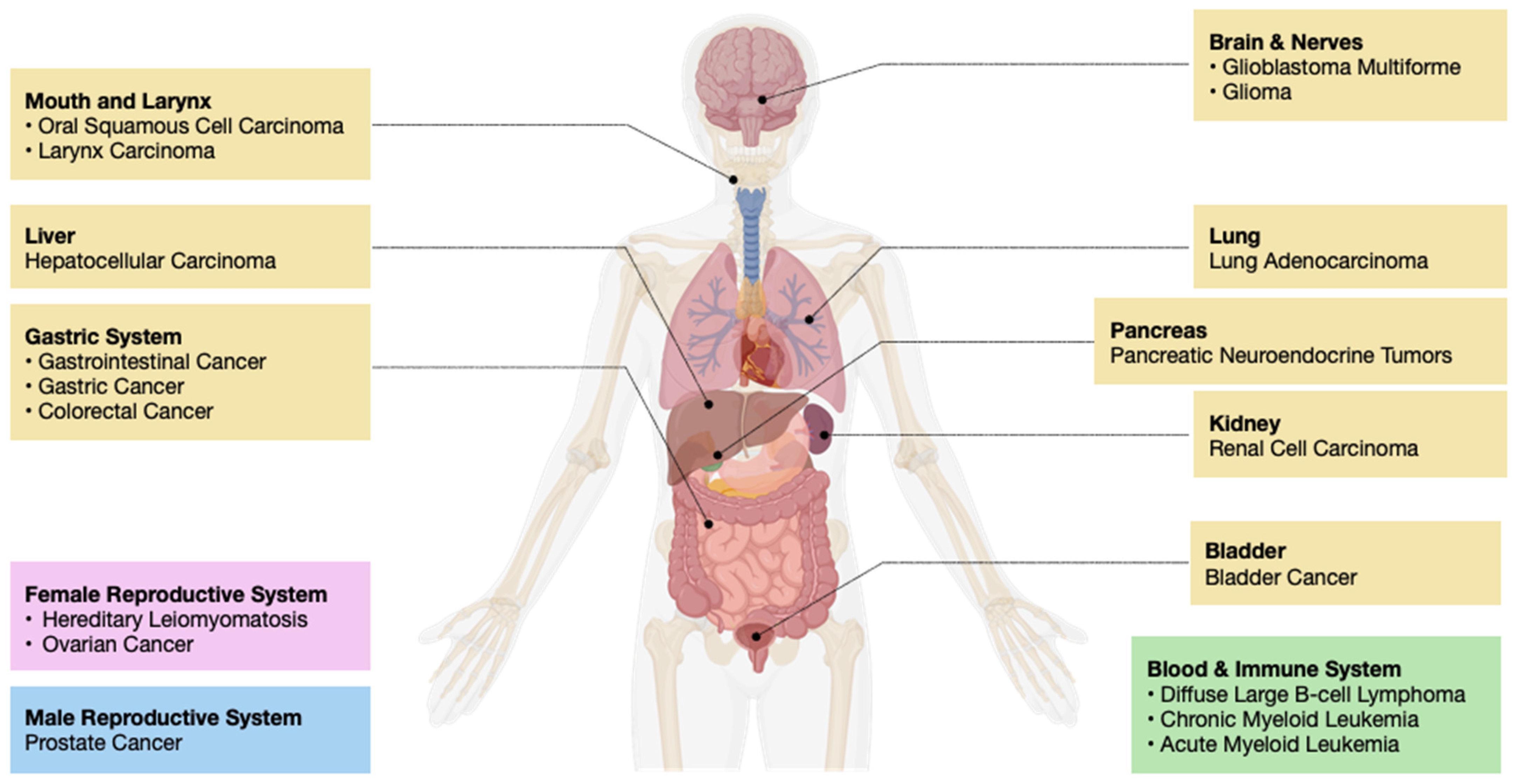

| Histone Chaperones | Functions and Associated Pathways | Cancer Implications | Therapy Approaches | References |
|---|---|---|---|---|
| FACT Complex | NRF2/KEAP1 pathway, ATR/CHK1 pathway | Breast Cancer, Hepatocellular Carcinoma, etc. | Curaxin | [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34] |
| ASF1 | GM-CSF dependent inflammatory pathway, Wnt signaling pathway | Lung Adenocarcinoma, Gastrointestinal Cancer, etc. | Chimeric Inhibitor | [35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53] |
| APLF | NHEJ pathway | Glioblastoma Multiforme, Bladder Cancer, etc. | N/A | [54,55,56,57,58,59,60,61,62,63,64,65,66,67] |
| NPM1 | DDT pathway | Oral Squamous Cell Carcinoma, Acute Myeloid Leukemia, etc. | NSC348884 and RNAi | [68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86] |
| CHAF1A | Folate metabolism pathway | Diffuse Large B-Cell Carcinoma, Gastric Cancer, etc. | Knockout | [87,88,89,90,91,92] |
| CHAF1B | ??? | Squamous Cell Carcinoma, Larynx Carcinoma, etc. | [93,94,95,96,97,98] | |
| P48 | NF-κB pathway, HDAC pathway | Cervical Cancer, Anaplastic Thyroid Carcinoma, etc. | [99,100,101,102,103,104,105,106] | |
| HIRA | MYC Pathway, Cell cycle pathway | Hereditary Leiomyomatosis and Renal Cell Carcinoma, Chronic Myeloid Leukemia, etc. | N/A | [107,108,109,110,111,112,113,114,115,116,117,118] |
| NAP1 | JNK signaling pathway | Glioma, Ovarian Cancer, etc. | Gene Therapy | [119,120,121,122,123,124,125,126,127,128] |
| SPT6 | hTERT signaling pathway | Colorectal Cancer, etc. | Chaetocin | [129,130,131,132,133,134] |
| DAXX | Epigenetic silencing pathway, Autophagy pathway | Pancreatic Neuroendocrine Tumors, Prostate Cancer, etc. | N/A | [135,136,137,138,139,140,141,142,143,144] |
| C1QBP | ??? | breast cancer, renal cancer, etc. | ??? | [145,146,147,148,149] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Bao, X. Comparative Review on Cancer Pathology from Aberrant Histone Chaperone Activity. Int. J. Mol. Sci. 2024, 25, 6403. https://doi.org/10.3390/ijms25126403
Lee J, Bao X. Comparative Review on Cancer Pathology from Aberrant Histone Chaperone Activity. International Journal of Molecular Sciences. 2024; 25(12):6403. https://doi.org/10.3390/ijms25126403
Chicago/Turabian StyleLee, Jiho, and Xiucong Bao. 2024. "Comparative Review on Cancer Pathology from Aberrant Histone Chaperone Activity" International Journal of Molecular Sciences 25, no. 12: 6403. https://doi.org/10.3390/ijms25126403
APA StyleLee, J., & Bao, X. (2024). Comparative Review on Cancer Pathology from Aberrant Histone Chaperone Activity. International Journal of Molecular Sciences, 25(12), 6403. https://doi.org/10.3390/ijms25126403





