An Efficient Method for Vault Nanoparticle Conjugation with Finely Adjustable Amounts of Antibodies and Small Molecules
Abstract
:1. Introduction
2. Results
2.1. The Production and Purification of the Vault-Z Variant
2.2. Investigating Trastuzumab Binding Mode to Either Vault-Z or the Sole Z Domain by Surface Plasmon Resonance Analysis
2.3. Binding Stoichiometry to Vault-Z of Tz and Saturation-Dependent Affinity Decline Are Assessed by Densitometric and Mass Spectrometry Analyses
2.4. Trastuzumab and Cetuximab Conjugation with Vault-Z Result in Substantially Enhanced Endocytosis by Target Breast Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Strain and Growth Media
4.2. MVP-Z Gene Cloning and Selection of Positive Clones
4.3. Cell Growth, Collection, and Cell-Free Extract Production
4.4. Vault Purification
4.5. Electrophoretic Analyses
4.6. Transmission Electron Microscopy Analysis
4.7. Dynamic Light Scattering Analysis
4.8. Surface Plasmon Resonance for the Determination of Z Peptide-Antibody and Vault-Z-Antibody Binding Affinities
4.9. Densitometric and Mass Spectrometric Determination of Vault-Z-Antibody Binding Stoichiometry
4.10. Vault-Z Conjugation with MoAbs and Fluorescently Labeled Fc
4.11. Analysis of Vault-Z Endocytic Uptake after MoAb Conjugation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Avvakumova, S.; Colombo, M.; Tortora, P.; Prosperi, D. Biotechnological approaches toward nanoparticle biofunctionalization. Trends Biotechnol. 2014, 32, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Choi, D.W.; Kim, H.N.; Park, C.G.; Lee, W.; Park, H.H. Protein-Based Nanoparticles as Drug Delivery Systems. Pharmaceutics 2020, 12, 604. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Huang, L. Nanoparticle delivery of a peptide targeting EGFR signaling. J. Control. Release 2012, 157, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Lohcharoenkal, W.; Wang, L.; Chen, Y.C.; Rojanasakul, Y. Protein nanoparticles as drug delivery carriers for cancer therapy. BioMed Res. Int. 2014, 2014, 180549. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.D.; Claypool, S.E.; Liu, R. The smart targeting of nanoparticles. Curr. Pharm. Des. 2013, 19, 6315–6329. [Google Scholar]
- Spicer, C.D.; Jumeaux, C.; Gupta, B.; Stevens, M.M. Peptide and protein nanoparticle conjugates: Versatile platforms for biomedical applications. Chem. Soc. Rev. 2018, 47, 3574–3620. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.C.; Costa, P.J.; Velho, S.; Amaral, M.H. Functionalizing nanoparticles with cancer-targeting antibodies: A comparison of strategies. J. Control. Release 2020, 320, 180–200. [Google Scholar] [CrossRef] [PubMed]
- Schrama, D.; Reisfeld, R.A.; Becker, J.C. Antibody targeted drugs as cancer therapeutics. Nat. Rev. Drug. Discov. 2006, 5, 147–159. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Rome, L.H. Isolation and characterization of a novel ribonucleoprotein particle: Large structures contain a single species of small RNA. J. Cell Biol. 1986, 103, 699–709. [Google Scholar] [CrossRef]
- Frascotti, G.; Galbiati, E.; Mazzucchelli, M.; Pozzi, M.; Salvioni, L.; Vertemara, J.; Tortora, P. The vault nanoparticle: A gigantic ribonucleoprotein assembly involved in diverse physiological and pathological phenomena and an ideal nanovector for drug delivery and therapy. Cancers 2021, 13, 707. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Heuser, J.E.; Chugani, D.C.; Rome, L.H. Vaults. III. Vault ribonucleoprotein particles open into flower-like structures with octagonal symmetry. J. Cell Biol. 1991, 112, 225–235. [Google Scholar] [CrossRef]
- Anderson, D.H.; Kickhoefer, V.A.; Slevers, S.A.; Rome, L.H.; Eisenberg, D. Draft crystal structure of the vault shell at 9-Å resolution. PLoS Biol. 2007, 5, e318. [Google Scholar] [CrossRef] [PubMed]
- Kickhoefer, V.A.; Siva, A.C.; Kedersha, N.L.; Inman, E.M.; Ruland, C.; Streuli, M.; Rome, L.H. Vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 1999, 146, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Kickhoefer, V.A.; Stephen, A.G.; Harrington, L.; Robinson, M.O.; Rome, L.H. Vaults and telomerase share a common subunit, TEP1. J. Biol. Chem. 1999, 274, 32712–32717. [Google Scholar] [CrossRef]
- Kickhoefer, V.A.; Searles, R.P.; Kedersha, N.L.; Garber, M.E.; Johnson, D.L.; Rome, L.H. Vault ribonucleoprotein particles from rat and bullfrog contain a related small RNA that is transcribed by RNA polymerase III. J. Biol. Chem. 1993, 268, 7868–7873. [Google Scholar] [CrossRef]
- Kickhoefer, V.A.; Rajavel, K.S.; Scheffer, G.L.; Dalton, W.S.; Scheper, R.J.; Rome, L.H. Vaults are up-regulated in multidrug-resistant cancer cell lines. J. Biol. Chem. 1998, 273, 8971–8974. [Google Scholar] [CrossRef]
- van Zon, A.; Mossink, M.H.; Schoester, M.; Scheffer, G.L.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A. Multiple human vault RNAs. Expression and association with the vault complex. J. Biol. Chem. 2001, 276, 37715–37721. [Google Scholar] [CrossRef]
- Stephen, A.G.; Raval-Fernandes, S.; Huynh, T.; Torres, M.; Kickhoefer, V.A.; Rome, L.H. Assembly of vault-like particles in insect cells expressing only the major vault protein. J. Biol. Chem. 2001, 276, 23217–23220. [Google Scholar] [CrossRef]
- Mrazek, J.; Toso, D.; Ryazantsev, S.; Zhang, X.; Zhou, Z.H.; Fernandez, B.C.; Kickhoefer, V.A.; Rome, L.H. Polyribosomes are molecular 3D nanoprinters that orchestrate the assembly of vault particles. ACS Nano 2014, 8, 11552–11559. [Google Scholar] [CrossRef] [PubMed]
- Berger, W.; Steiner, E.; Grusch, M.; Elbling, L.; Micksche, M. Vaults and the major vault protein: Novel roles in signal pathway regulation and immunity. Cell. Mol. Life Sci. 2009, 66, 43–61. [Google Scholar] [CrossRef]
- Muñoz-Juan, A.; Carreño, A.; Mendoza, R.; Corchero, I.L. Latest advances in the development of eukaryotic vaults as targeted drug delivery systems. Pharmaceutics 2019, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Kickhoefer, V.A.; Han, M.; Raval-Fernandes, S.; Poderycki, M.J.; Moniz, R.J.; Vaccari, D.; Silvestry, M.; Stewart, P.L.; Kelly, K.A.; Rome, L.H. Targeting vault nanoparticles to specific cell surface receptors. ACS Nano 2009, 3, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Benner, N.L.; Zang, X.; Buehler, D.C.; Kickhoefer, V.A.; Rome, M.E.; Rome, L.H.; Wender, P.A. Vault nanoparticles: Chemical modifications for imaging and enhanced delivery. ACS Nano 2017, 11, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Braisted, A.C.; Wells, J.A. Minimizing a binding domain from protein A. Proc. Natl. Acad. Sci. USA 1996, 11, 5688–5692. [Google Scholar] [CrossRef] [PubMed]
- Tomaino, G.; Pantaleoni, C.; Ami, D.; Pellecchia, F.; Dutriaux, A.; Barbieri, L.; Garbujo, S.; Natalello, A.; Tortora, P.; Frascotti, G. Addressing critical issues related to storage and stability of the vault nanoparticle expressed and purified from Komagataella phaffi. Int. J. Mol. Sci. 2023, 24, 4214. [Google Scholar] [CrossRef]
- Wang, M.; Kickhoefer, V.A.; Rome, L.H.; Foellmer, O.K.; Mahendra, S. Synthesis and assembly of human vault particles in yeast. Biotechnol. Bioeng. 2018, 115, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, E.; Avvakumova, S.; La Rocca, A.; Pozzi, M.; Messali, S.; Magnaghi, P.; Colombo, M.; Prosperi, D.; Tortora, P. A fast and straightforward procedure for vault nanoparticle purification and the characterization of its endocytic uptake. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2254–2260. [Google Scholar] [CrossRef]
- Nagel, G.; Tschiche, H.R.; Wedepohl, S.; Calderón, M. Modular approach for theranostic polymer conjugates with activatable fluorescence: Impact of linker design on the stimuli-induced release of doxorubicin. J. Control. Release 2018, 285, 200–211. [Google Scholar] [CrossRef]
- Louage, B.; De Wever, O.; Hennink, W.E.; De Geest, B.G. Developments and future clinical outlook of taxane nanomedicines. J. Control. Release 2017, 253, 137–152. [Google Scholar] [CrossRef]
- Mousavizadeha, A.; Jabbarib, A.; Akramic, M.; Bardaniab, H. Cell targeting peptides as smart ligands for targeting of therapeutic or diagnostic agents: A systematic review. Colloids Surf. B Biointerfaces 2017, 158, 507–517. [Google Scholar] [CrossRef]
- Hudis, C.A. Trastuzumab—Mechanism of action and use in clinical practice. N. Engl. J. Med. 2007, 357, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Kasi, P.M.; Afable, M.G.; Herting, C.; Lukanowski, M.; Jin, Z. Anti-EGFR antibodies in the management of advanced colorectal cancer. Oncologist 2023, 28, 1034–1048. [Google Scholar] [CrossRef]
- Conrad, M.; Fechner, P.; Proll, G.; Gauglitz, G. Comparison of methods for quantitative biomolecular interaction analysis. Anal. Bioanal. Chem. 2022, 414, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Zhang, X.; Mrazek, J.; Kickhoefer, V.A.; Lai, M.; Ng, H.L.; Yang, O.O.; Rome, L.H.; Zhou, Z.H. Solution structures of engineered vault particles. Structure 2018, 26, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Deisenhofer, J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-Å resolution. Biochemistry 1981, 20, 2361–2370. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-H.; Huan, L.M.; Chu, X.-S.; Sun, Y.; Shi, Q.-H. A comparative investigation of random and oriented immobilization of protein A ligands on the binding of immunoglobulin G. Biochem. Eng. J. 2018, 139, 15–24. [Google Scholar] [CrossRef]
- Galovic, M.; Xu, D.; Areces, L.B.; van der Kammen, R.; Innocenti, M. Interplay between N-WASP and CK2 optimizes clathrin-mediated endocytosis of EGFR. J. Cell Sci. 2011, 124, 2001–2012. [Google Scholar] [CrossRef] [PubMed]
- Iravani, S.; Varma, R.S. Vault, viral, and virus-like nanoparticles for targeted cancer therapy. Mater. Adv. 2023, 4, 2909–2917. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, M.; Shah, K.; Kalra, S.S.; Rome, L.H.; Mahendra, S. Decolorization and detoxification of synthetic dye compounds by laccase immobilized in vault nanoparticles. Bioresour. Technol. 2022, 351, 127040. [Google Scholar] [CrossRef]
- Yu, K.; Yau, Y.H.; Sinha, A.; Tan, T.; Kickhoefer, V.A.; Rome, L.H.; Lee, H.; Shochat, S.G.; Lim, S. Modulation of the vault protein-protein interaction for tuning of molecular release. Sci. Rep. 2017, 7, 14816. [Google Scholar] [CrossRef]
- Fernández, R.; Carreño, A.; Mendoza, R.; Benito, A.; Neus Ferrer-Miralles, N.; Céspedes, M.V.; Corchero, J.L. Escherichia coli as a new platform for the fast production of vault-like nanoparticles: An optimized protocol. Int. J. Mol. Sci. 2022, 23, 15543. [Google Scholar] [CrossRef] [PubMed]
- Martín, F.; Carreño, A.; Mendoza, R.; Caruana, P.; Rodriguez, F.; Bravo, M.; Benito, A.; Ferrer-Miralles, N.; Céspedes, M.V.; Corchero, J.L. All-in-one biofabrication and loading of recombinant vaults in human cells. Biofabrication 2022, 14, 025018. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kickhoefer, V.A.; Ng, B.C.; Gopal, A.; Bentolila, L.A.; John, S.; Tolbert, S.H.; Rome, L.H. Vaults are dynamically unconstrained cytoplasmic nanoparticles capable of half vault exchange. ACS Nano 2010, 4, 7229–7240. [Google Scholar] [CrossRef]
- Guerra, P.; González-Alamos, M.; Llauró, A.; Casañas, A.; Querol-Audí, J.; de Pablo, P.J.; Verdaguer, N. Symmetry disruption commits vault particles to disassembly. Sci. Adv. 2022, 8, eabj7795. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.T.; Beatty, W.; Moyo, B.; Alves-Bezerra, M.; Hurley, A.; Lagor, W.; Bao, G.; Ponnazhagan, S.; McNally, R.; Rome, L.; et al. Encapsulation of AAVs into protein vault nanoparticles as a novel solution to gene therapy’s neutralizing antibody problem. bioRxiv 2023. [Google Scholar] [CrossRef]
- Colombo, M.; Fiandra, L.; Alessio, G.; Mazzucchelli, S.; Nebuloni, M.; De Palma, C.; Kantner, K.; Pelaz, B.; Rotem, R.; Corsi, F.; et al. Tumour homing and therapeutic effect of colloidal nanoparticles depend on the number of attached antibodies. Nat. Commun. 2016, 7, 13818. [Google Scholar] [CrossRef] [PubMed]
- Subik, K.; Lee, J.F.; Baxter, L.; Strzepek, T.; Costello, D.; Crowley, P.; Xing, L.; Hung, M.C.; Bonfiglio, T.; Hicks, D.G.; et al. The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer 2010, 4, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Brockhoff, G.; Heckel, B.; Schmidt-Bruecken, E.; Plander, M.; Hofstaedter, F.; Vollmann, A.; Diermeier, S. Differential impact of Cetuximab, Pertuzumab and Trastuzumab on BT474 and SK-BR-3 breast cancer cell proliferation. Cell Prolif. 2007, 40, 488–507. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Kickhoefer, V.A.; Nemerow, G.R.; Rome, L.H. Targeted vault nanoparticles engineered with an endosomolytic peptide deliver biomolecules to the cytoplasm. ACS Nano 2011, 5, 6128–6137. [Google Scholar] [CrossRef]
- Gleeson, M.A.G.; White, C.E.; Meininger, D.P.; Komives, E.A. Generation of protease-deficient strains and their use in heterologous protein expression. In Pichia Protocols; Higgins, D.R., Cregg, J.M., Eds.; Humana Press: Totowa, NJ, USA, 1998; Volume 103, pp. 81–94. [Google Scholar]
- Medzihradszky, K.F. In-solution digestion of proteins for mass spectrometry. Methods Enzymol. 2005, 405, 50–65. [Google Scholar]
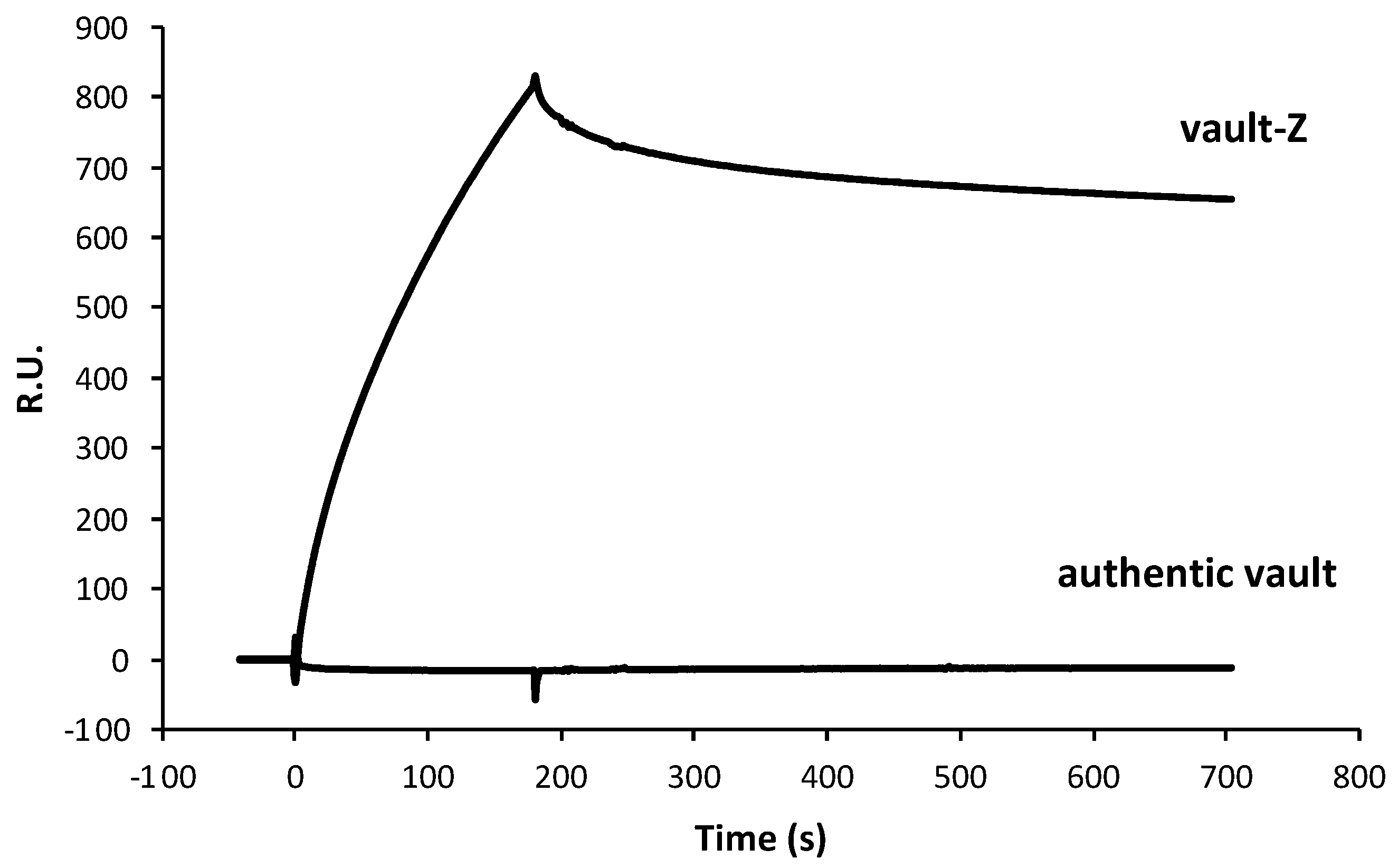


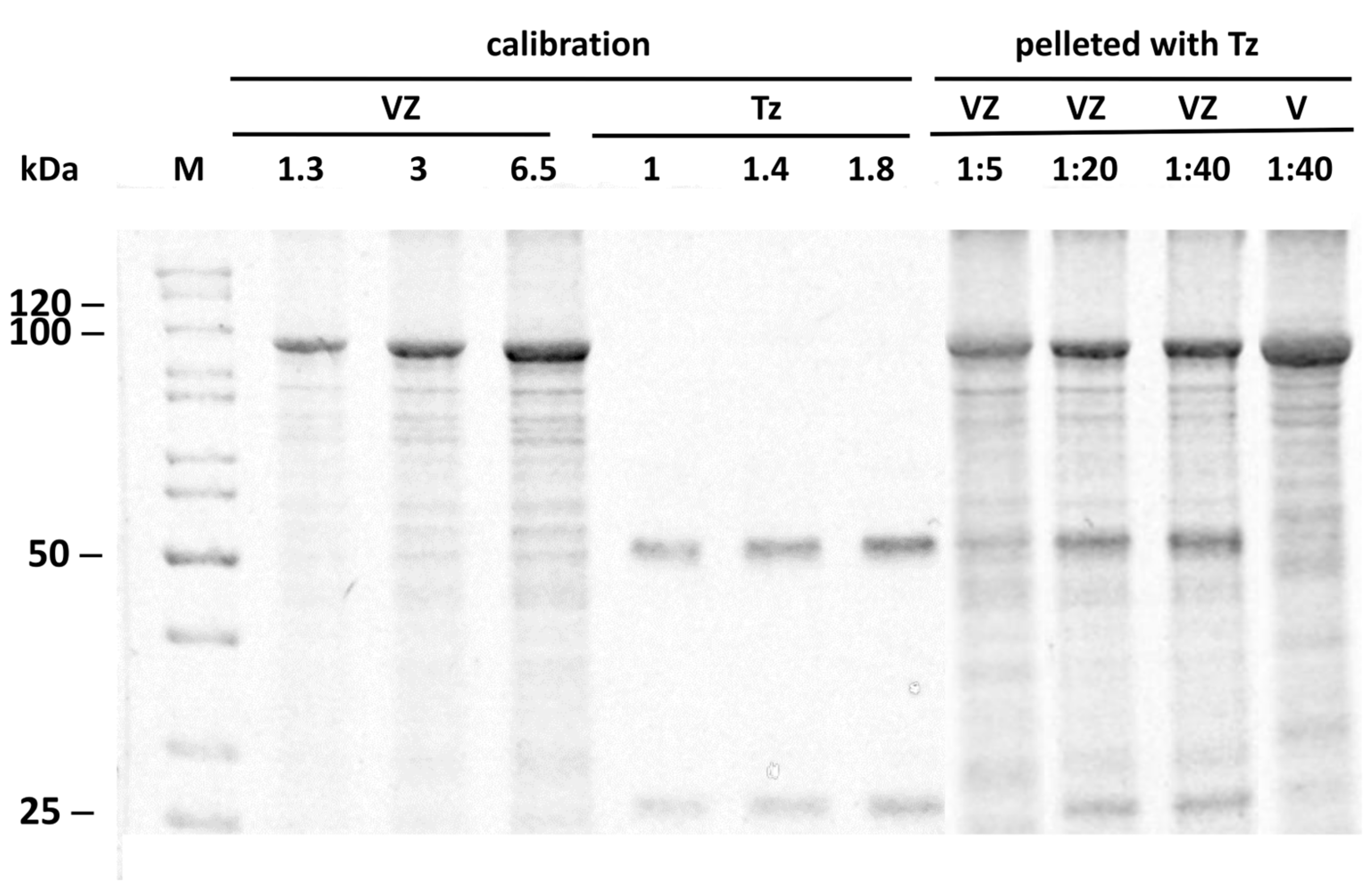
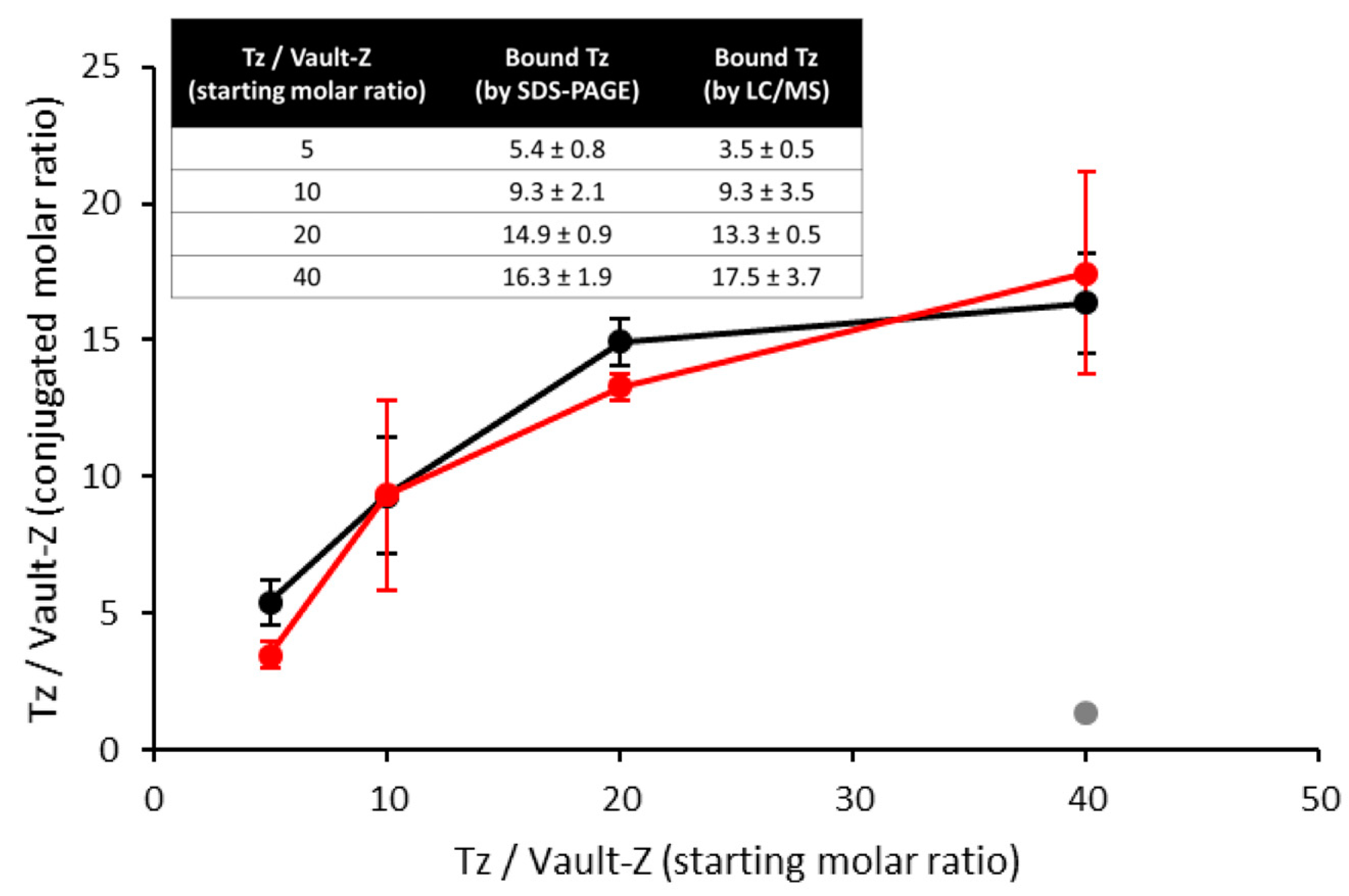

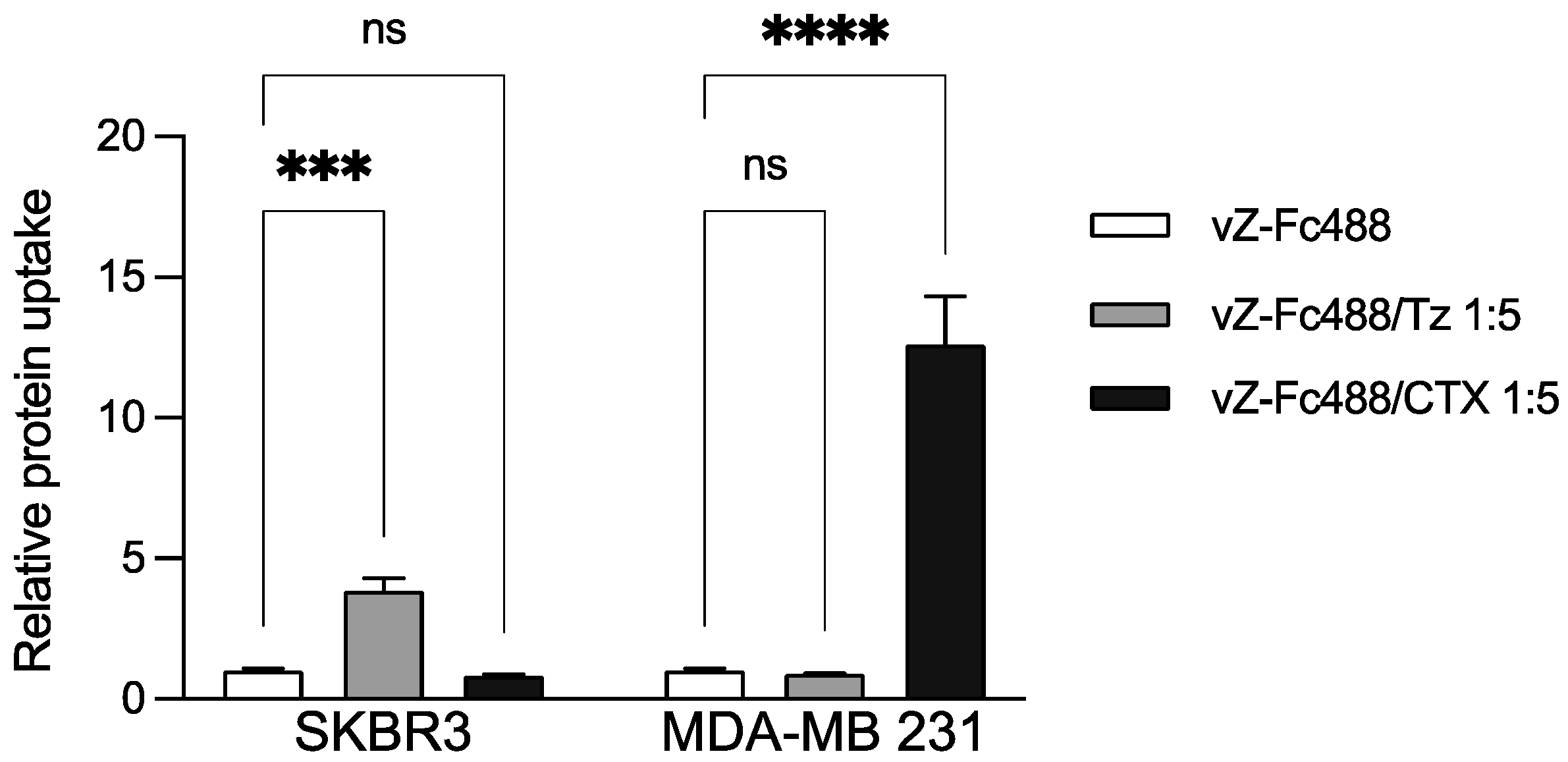
| Protein | ka (104 1/Ms) | kd (10−4 1/s) | KD (nM) |
|---|---|---|---|
| Vault-Z | 10.5 ± 5.0 | 2.59 ± 0.2 | 2.53 ± 0.4 |
| Z-domain | 44.6 ± 13.6 | 171 ± 84 | 38.3 ± 23.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomaino, G.; Pantaleoni, C.; D’Urzo, A.; Santambrogio, C.; Testa, F.; Ciprandi, M.; Cotugno, D.; Frascotti, G.; Vanoni, M.; Tortora, P. An Efficient Method for Vault Nanoparticle Conjugation with Finely Adjustable Amounts of Antibodies and Small Molecules. Int. J. Mol. Sci. 2024, 25, 6629. https://doi.org/10.3390/ijms25126629
Tomaino G, Pantaleoni C, D’Urzo A, Santambrogio C, Testa F, Ciprandi M, Cotugno D, Frascotti G, Vanoni M, Tortora P. An Efficient Method for Vault Nanoparticle Conjugation with Finely Adjustable Amounts of Antibodies and Small Molecules. International Journal of Molecular Sciences. 2024; 25(12):6629. https://doi.org/10.3390/ijms25126629
Chicago/Turabian StyleTomaino, Giulia, Camilla Pantaleoni, Annalisa D’Urzo, Carlo Santambrogio, Filippo Testa, Matilde Ciprandi, Davide Cotugno, Gianni Frascotti, Marco Vanoni, and Paolo Tortora. 2024. "An Efficient Method for Vault Nanoparticle Conjugation with Finely Adjustable Amounts of Antibodies and Small Molecules" International Journal of Molecular Sciences 25, no. 12: 6629. https://doi.org/10.3390/ijms25126629








