Distribution of papA and papG Variants among Escherichia coli Genotypes: Association with Major Extraintestinal Pathogenic Lineages
Abstract
:1. Introduction
2. Results
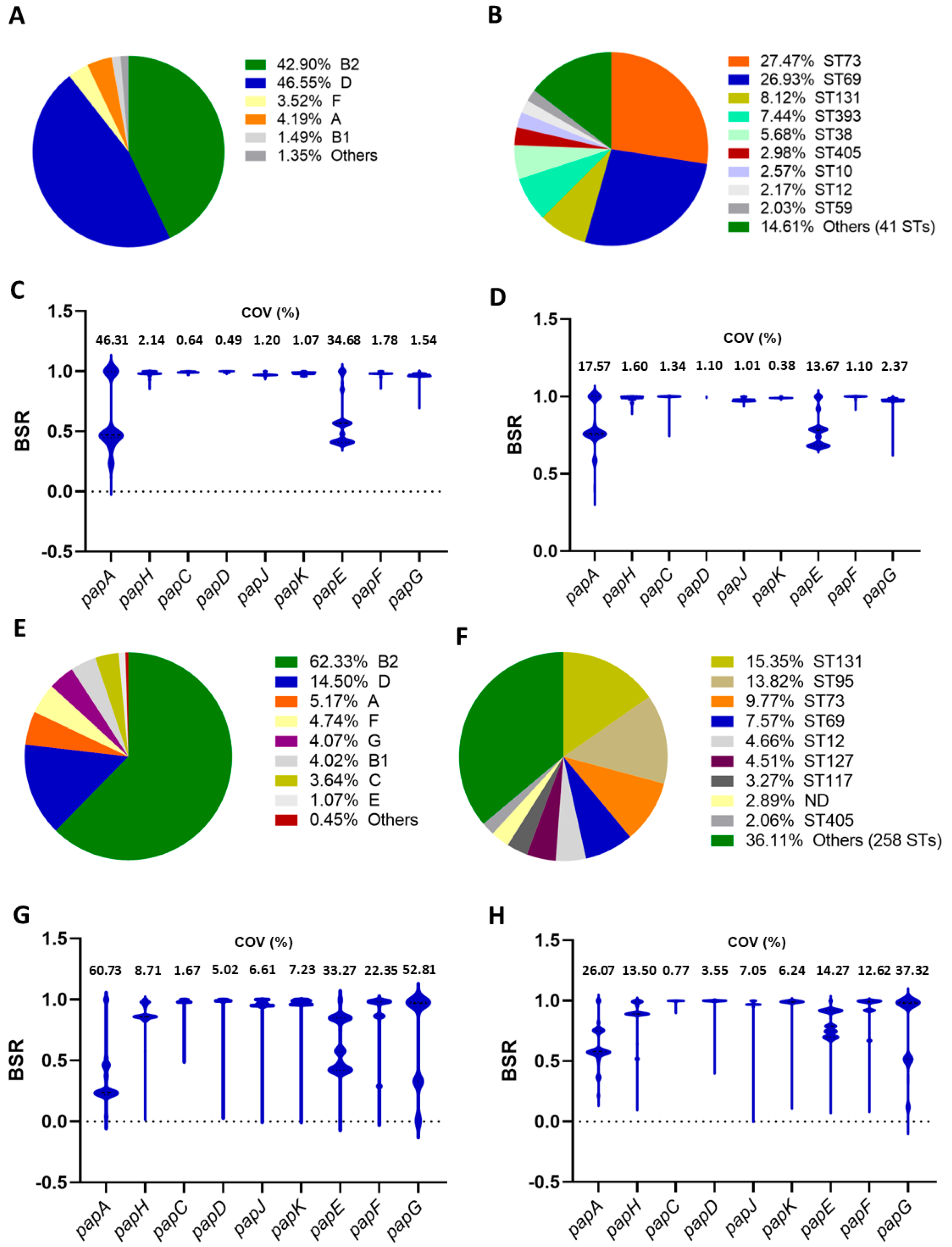
2.1. Screening of papA and papG Variants
2.2. Genomes Harboring Two or Three papA Variants
2.3. Presence of papA and papG Variants in E. coli Isolated from Humans and Animals
2.4. Screening of papA and papG Variants in a Collection of Chilean UPEC Strains
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GBD 2019 Antimicrobial Resistance Collaborators. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef] [PubMed]
- Mellata, M. Human and avian extraintestinal pathogenic Escherichia coli: Infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 2013, 10, 916–932. [Google Scholar] [CrossRef] [PubMed]
- Sora, V.M.; Meroni, G.; Martino, P.A.; Soggiu, A.; Bonizzi, L.; Zecconi, A. Extraintestinal pathogenic Escherichia coli: Virulence factors and antibiotic resistance. Pathogens 2021, 10, 1355. [Google Scholar] [CrossRef] [PubMed]
- Kathayat, D.; Lokesh, D.; Ranjit, S.; Rajashekara, G. Avian Pathogenic Escherichia coli (APEC): An overview of virulence and pathogenesis factors, zoonotic potential, and control strategies. Pathogens 2021, 10, 467. [Google Scholar] [CrossRef] [PubMed]
- Barros, M.M.; Castro, J.; Araújo, D.; Campos, A.M.; Oliveira, R.; Silva, S.; Outor-Monteiro, D.; Almeida, C. Swine colibacillosis: Global epidemiologic and antimicrobial scenario. Antibiotics 2023, 12, 682. [Google Scholar] [CrossRef]
- Goulart, D.B.; Mellata, M. Escherichia coli mastitis in dairy cattle: Etiology, diagnosis, and treatment challenges. Front. Microbiol. 2022, 13, 928346. [Google Scholar] [CrossRef] [PubMed]
- Yeruham, I.; Elad, D.; Avidar, Y.; Goshen, T. A herd level analysis of urinary tract infection in dairy cattle. Vet. J. 2006, 171, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.F.; Litster, A.L.; Platell, J.L.; Trott, D.J. Canine bacterial urinary tract infections: New developments in old pathogens. Vet. J. 2011, 190, 22–27. [Google Scholar] [CrossRef]
- Dorsch, R.; Teichmann-Knorrn, S.; Lund, H.S. Urinary tract infection and subclinical bacteriuria in cats: A clinical update. J. Feline Med. Surg. 2019, 21, 1023–1038. [Google Scholar] [CrossRef]
- Manges, A.R.; Johnson, J.R. Reservoirs of extraintestinal pathogenic Escherichia coli. Microbiol. Spectr. 2015, 3, UTI-0006-2012. [Google Scholar] [CrossRef]
- Tourret, J.; Denamur, E. Population phylogenomics of extraintestinal pathogenic Escherichia coli. Microbiol. Spectr. 2016, 4, UTI-0010-2012. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Banting, G.; Neumann, N.F. A review of the taxonomy, genetics, and biology of the genus Escherichia and the type species Escherichia coli. Can. J. Microbiol. 2021, 67, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Croxen, M.A.; Finlay, B.B. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 2010, 8, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Secher, T.; Brehin, C.; Oswald, E. Early settlers: Which, E. coli strains do you not want at birth? Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, G123–G129. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Roach, D.J.; Kitzman, J.O.; Snyder, M.W.; Stackhouse, B.; Butler-Wu, S.M.; Lee, C.; Cookson, B.T.; Shendure, J. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res. 2015, 25, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Antão, E.M.; Wieler, L.H.; Ewers, C. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 2009, 1, 22. [Google Scholar] [CrossRef]
- Waksman, G. Structural and molecular biology of a protein-polymerizing nanomachine for pilus biogenesis. J. Mol. Biol. 2017, 429, 2654–2666. [Google Scholar] [CrossRef]
- Båga, M.; Norgren, M.; Normark, S. Biogenesis of E. coli Pap pili: papH, a minor pilin subunit involved in cell anchoring and length modulation. Cell 1987, 49, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Tennent, J.M.; Lindberg, F.; Normark, S. Integrity of Escherichia coli P pili during biogenesis: Properties and role of PapJ. Mol. Microbiol. 1990, 4, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Werneburg, G.T.; Thanassi, D.G. Pili assembled by the chaperone/usher pathway in Escherichia coli and Salmonella. EcoSal Plus 2018, 8, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Stell, A.L.; Scheutz, F.; O‘Bryan, T.T.; Russo, T.A.; Carlino, U.B.; Fasching, C.; Kavle, J.; Van Dijk, L.; Gaastra, W. Analysis of the F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect. Immun. 2000, 68, 1587–1599. [Google Scholar] [CrossRef] [PubMed]
- Manning, S.D.; Zhang, L.; Foxman, B.; Spindler, A.; Tallman, P.; Marrs, C.F. Prevalence of known P-fimbrial G alleles in Escherichia coli and identification of a new adhesin class. Clin. Diagn. Lab. Immunol. 2001, 8, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Malberg Tetzschner, A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef] [PubMed]
- Kudinha, T.; Kong, F. Distribution of papG alleles among uropathogenic Escherichia coli from reproductive age women. J. Biomed. Sci. 2022, 29, 66. [Google Scholar] [CrossRef] [PubMed]
- Salamzade, R.; McElheny, C.L.; Manson, A.L.; Earl, A.M.; Shaikh, N.; Doi, Y. Genomic epidemiology and antibiotic susceptibility profiling of uropathogenic Escherichia coli among children in the United States. mSphere 2023, 8, e0018423. [Google Scholar] [CrossRef] [PubMed]
- Welch, R.A.; Burland, V.; Plunkett, G., 3rd; Redford, P.; Roesch, P.; Rasko, D.; Buckles, E.L.; Liou, S.R.; Boutin, A.; Hackett, J.; et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 17020–17024. [Google Scholar] [CrossRef]
- Bélanger, L.; Garenaux, A.; Harel, J.; Boulianne, M.; Nadeau, E.; Dozois, C.M. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol. Med. Microbiol. 2011, 62, 1–10. [Google Scholar] [CrossRef]
- Uhlin, B.E.; Norgren, M.; Båga, M.; Normark, S. Adhesion to human cells by Escherichia coli lacking the major subunit of a digalactoside-specific pilus-adhesin. Proc. Natl. Acad. Sci. USA 1985, 82, 1800–1804. [Google Scholar] [CrossRef] [PubMed]
- Lane, M.C.; Mobley, H.L. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 2007, 72, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Melican, K.; Sandoval, R.M.; Kader, A.; Josefsson, L.; Tanner, G.A.; Molitoris, B.A.; Richter-Dahlfors, A. Uropathogenic Escherichia coli P and Type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 2011, 7, e1001298. [Google Scholar] [CrossRef] [PubMed]
- Nuccio, S.P.; Bäumler, A.J. Evolution of the chaperone/usher assembly pathway: Fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 2007, 71, 551–575. [Google Scholar] [CrossRef] [PubMed]
- Biggel, M.; Xavier, B.B.; Johnson, J.R.; Nielsen, K.L.; Frimodt-Møller, N.; Matheeussen, V.; Goossens, H.; Moons, P.; Van Puyvelde, S. Horizontally acquired papGII-containing pathogenicity islands underlie the emergence of invasive uropathogenic Escherichia coli lineages. Nat. Commun. 2020, 11, 5968. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Stell, A.L.; Delavari, P. Canine feces as a reservoir of extraintestinal pathogenic Escherichia coli. Infect. Immun. 2001, 69, 1306–1314. [Google Scholar] [CrossRef] [PubMed]
- Stromberg, Z.R.; Johnson, J.R.; Fairbrother, J.M.; Kilbourne, J.; Van Goor, A.; Curtiss, R., 3rd; Mellata, M. Evaluation of Escherichia coli isolates from healthy chickens to determine their potential risk to poultry and human health. PLoS ONE 2017, 12, e0180599. [Google Scholar] [CrossRef] [PubMed]
- Melendez, D.; Roberts, M.C.; Greninger, A.L.; Weissman, S.; No, D.; Rabinowitz, P.; Wasser, S. Whole-genome analysis of extraintestinal pathogenic Escherichia coli (ExPEC) MDR ST73 and ST127 isolated from endangered southern resident killer whales (Orcinus orca). J. Antimicrob. Chemother. 2019, 74, 2176–2180. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Sahl, J.W.; Caporaso, J.G.; Rasko, D.A.; Keim, P. The large-scale blast score ratio (LS-BSR) pipeline: A method to rapidly compare genetic content between bacterial genomes. PeerJ 2014, 2, e332. [Google Scholar] [CrossRef]
- Waters, N.R.; Abram, F.; Brennan, F.; Holmes, A.; Pritchard, L. Easy phylotyping of Escherichia coli via the EzClermont web app and command-line tool. Access Microbiol. 2020, 2, acmi000143. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. mlst. Github. Available online: https://github.com/tseemann/mlst (accessed on 8 January 2024).
- Gales, A.C.; Castanheira, M.; Jones, R.N.; Sader, H.S. Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: Results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008–2010). Diagn. Microbiol. Infect. Dis. 2012, 73, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology 2012, 158, 1005–1015. [Google Scholar] [CrossRef]
| papA Variant | N° of Positive Genomes | Distribution among Phylogroups (Phylogroup: n° Genomes) | Significant Association with Sequence Types: n° Genomes (n° Genomes per Phylogroup) |
|---|---|---|---|
| F7-1 | 62 |
|
|
| F7-2 | 235 |
|
|
| F8 | 59 |
|
|
| F9 | 85 |
|
|
| F10 | 511 |
|
|
| F11 | 893 |
|
|
| F12 | 287 |
|
|
| F13 | 231 |
|
|
| F14 | 137 |
|
|
| F15 | 15 |
|
|
| F16 | 371 |
|
|
| F48 | 379 |
|
|
| papG Variant | N° of Positive Genomes | Distribution among Phylogroups (Phylogroup: n° Genomes) | Significant Association with Sequence Types (ST: n° Genomes per Phylogroup) | Significant Association with papA Variants (n° Genomes per ST) |
|---|---|---|---|---|
| papGI | 26 |
|
|
|
| papGII | 2569 |
|
|
|
| papGIII | 942 |
|
|
|
| papA Variants | N° of Genomes | Phylogroups (Phylogroup: n° Genomes) | Sequence Types (ST: n° Genomes) | papG Variants (Variant: n° Genomes) |
|---|---|---|---|---|
| F7-1/F7-2 | 3 | B2: 3 | ST73: 3 | papGII: 3 |
| F7-1/F10 | 2 | B2: 2 | ST144: 2 | papGII: 2 |
| F7-1/F12 | 1 | B2: 1 | ST73: 1 | papGIII: 1 |
| F7-1/F14 | 8 | B2: 8 | ST73: 8 | papGII/papGIII: 8 |
| F7-1/F16 | 1 | B2: 1 | ST12: 1 | papGII: 1 |
| F7-2/F10 | 4 | B2: 3 | ST6355: 3 | papGII: 2 |
| D: 4 | ST8767: 1 | papGII: 1 | ||
| F7-2/F13 | 11 | B2: 11 | ST73: 10 | papGII: 2 papGII/papGIII: 8 |
| ST131: 1 | papGII: 1 | |||
| F7-2/F48 | 8 | B2: 8 | ST127: 7 | papGII: 8 |
| ST8312: 1 | papGIII: 1 | |||
| F8/F9 | 15 | F: 14 | ST59: 13 | papGII: 13 |
| ST6199: 1 | papGII: 1 | |||
| G: 1 | ST59: 1 | papGII: 1 | ||
| F8/F14 | 1 | F: 1 | ST59: 1 | papGII: 1 |
| F9/F11 | 1 | B2: 1 | ST12: 1 | papGII: 1 |
| F9/F12 | 1 | B2: 1 | ST141: 1 | ND: 1 |
| F9/F13 | 5 | B2: 5 | ST12: 5 | papGII: 1 papGII/papGIII: 4 |
| F9/F15 | 1 | B2: 1 | ST8118: 1 | papGII: 1 |
| F11/F13 | 2 | B2: 2 | ST12: 2 | papGII/papGIII: 2 |
| F10/F11 | 1 | B2:1 | ST12: 1 | papGII: 1 |
| F10/F12 | 33 | B2: 32 | ST12: 25 | papGIII: 24 ND: 1 |
| ST625: 1 | papGI/papGIII: 1 | |||
| ST961: 3 | papGIII: 2 ND: 1 | |||
| ST2604: 3 | papGIII: 3 | |||
| Cryptic: 1 | ST961: 1 | papGIII: 1 | ||
| F10/F13 | 1 | B2: 1 | ST131: 1 | papGIII: 1 |
| F10/F14 | 1 | B2: 1 | ST12: 1 | papGIII: 1 |
| F10/F16 | 15 | B2: 15 | ST12: 15 | papGII: 15 |
| F11/F13 | 2 | B2: 2 | ST12: 2 | papGI/papGIII: 2 |
| F11/F14 | 2 | B2: 1 | ST12: 1 | papGII/papGIII: 1 |
| C: 1 | ST88: 1 | papGIII: 1 | ||
| F11/F16 | 25 | B2: 25 | ST12: 25 | papGII: 24 ND: 1 |
| F11/F48 | 2 | A: 1 | ST10: 1 | papGII: 1 |
| B2: 1 | ST131: 1 | papGIII: 1 | ||
| F12/F14 | 2 | B2: 2 | ST12: 1 | papGIII: 1 |
| ST372: 1 | papGII/papGIII: 1 | |||
| F12/F48 | 6 | B2: 6 | ST144: 6 | papGII: 6 |
| F13/F14 | 5 | B2: 5 | ST12: 3 | papGI/papGIII: 1 papGII/papGIII: 1 ND: 1 |
| ST599: 2 | papGI/papGIII: 2 | |||
| F13/F48 | 2 | B2: 2 | ST555: 2 | papGII/papGIII: 2 |
| F14/F16 | 4 | B2: 3 | ST12: 3 | papGII: 3 |
| D: 1 | ST69: 1 | papGII: 1 | ||
| F15/F16 | 2 | B2: 2 | ST827: 2 | papGIII: 2 |
| F7-1/F9/F15 | 1 | B2: 1 | ST703: 1 | ND: 1 |
| F7-1/F15/F16 | 1 | B2: 1 | ST73: 1 | papGIII: 1 |
| F9/F10/F16 | 1 | B2: 1 | ST12: 1 | papGII: 1 |
| F9/F11/F16 | 1 | B2: 1 | ST12: 1 | papGII: 1 |
| F10/F11/F16 | 3 | B2: 3 | ST12: 3 | papGII: 3 |
| F10/F14/F16 | 1 | B2: 1 | ST12: 1 | ND |
| F13/F14/F16 | 2 | B2: 2 | ST12: 1 | ND |
| ND: 1 | papGII: 1 |
| Strain (NCBI Assembly Code) | Phylogroup/ST | 1st papA/papG Pair (pap Locus Coordinates) | 2nd papA/papG Pair (pap Locus Coordinates) | 3rd papA/papG Pair (pap Locus Coordinates) |
|---|---|---|---|---|
| E. coli GN02350 (GCF_026651165.1) | B2/ST12 | F13/papGII (c1,566,503–-1,558,580) | F9/papGIII (c2,122,156–2,114,207) | - |
| E. coli C 691-04A GCF_025946565.1 | B2/ST73 | F7-2/papGIII (899,304–907,248) | F13/papGIII (c4,560,907–4,552,986) | - |
| E. coli CFT073 GCF_014262945.1 | B2/ST73 | F7-2/papGII (c3,448,359–3,440,421) | F7-1/papGII (c4,959,718–4,951,783) | - |
| E. coli BH100N substr. MG2017 GCF_002900305.1 | B2/ST127 | F48/papGIII (c3,287,322–3,279,405) | F7-2/papGIII (c4,881,632–4,873,708) | - |
| E. coli BH100 substr. MG2014 GCF_002763515.2 | B2/ST127 | F48/papGIII (c3,214,562–3,206,645) | F7-2/papGIII (c4,824,548–4,816,627) FS | - |
| E. coli K-15KW01 GCF_001683435.1 | B2/ST127 | F7-2/papGIII (c1,676,021–1,669,148) | F48/papGIII (c3,310,504–3,302,587) | - |
| E. coli EC5654 GCF_022919035.1 | B2/ST12 | F16/papGII (878,908–886,816) | F10/papGII (4,458,416–4,466,350) | F11/papGII (c4,638,209–4,630,290) |
| E. coli strain UPEC132 GCF_007833875.1 | B2/ND | F14/papGII (79,054–86,999) | F16/papGII (934,430–942,336) | F13/papGII (c4,629,322–4,621,398) |
| Sequence Type | papA/papG Variant | N° Genomes | N° Genomes with Host Information | N° Genomes per Host |
|---|---|---|---|---|
| ST131 | All | 633 | 531 | Human: 505 |
| Dog: 16 | ||||
| Others: 10 (6 types) | ||||
| F14 | 10 | 9 | Human: 8 | |
| Porcine: 1 | ||||
| F48 | 43 | 39 | Human: 38 | |
| Dog: 1 | ||||
| papGII | 584 | 487 | Human: 464 | |
| Dog: 14 | ||||
| Others: 9 (6 types) | ||||
| papGIII | 41 | 37 | Human: 34 | |
| Dog: 2 | ||||
| Porcine: 1 | ||||
| ST95 | All | 570 | 513 | Human: 427 |
| Poultry: 69 | ||||
| Others: 16 (9 types) | ||||
| F11 | 477 | 434 | Human: 350 | |
| Poultry: 69 | ||||
| Others: 15 (8 types) | ||||
| papGII | 480 | 436 | Human: 351 | |
| Poultry: 69 | ||||
| Others: 16 (9 types) | ||||
| papGIII | 85 | 72 | Human: 71 | |
| Non-human primate: 1 | ||||
| ST73 | All | 403 | 305 | Human: 278 |
| Dog: 8 | ||||
| Feline: 3 | ||||
| Others: 16 (10 types) | ||||
| F7-1 | 29 | 25 | Human: 24 | |
| Dog: 1 | ||||
| F7-2 | 135 | 62 | Human: 61 | |
| Porcine: 1 | ||||
| F13 | 35 | 31 | Human: 28 | |
| Non-human primate: 3 | ||||
| F14 | 67 | 65 | Human: 61 | |
| Orca: 4 | ||||
| F48 | 15 | 13 | Human: 9 | |
| Dog: 3 | ||||
| Common polecat: 1 | ||||
| papGII | 295 | 208 | Human: 199 | |
| Orca: 4 | ||||
| Others: 5 (4 types) | ||||
| papGIII | 138 | 124 | Human: 110 | |
| Dog: 7 | ||||
| Others: 7 (5 types) | ||||
| ST69 | All | 312 | 261 | Human: 241 |
| Poultry: 9 | ||||
| Others: 6 (4 types) | ||||
| F9 | 19 | 12 | Human: 12 | |
| F16 | 182 | 149 | Human: 147 | |
| Others: 2 (2 types) | ||||
| papGII | 246 | 205 | Human: 191 | |
| Poultry: 8 | ||||
| Others: 6 (4 types) | ||||
| ST12 | All | 192 | 173 | Human: 140 |
| Dog: 16 | ||||
| Others: 17 (13 types) | ||||
| F10 | 107 | 92 | Human: 69 | |
| Dog: 9 | ||||
| Others: 14 (12 types) | ||||
| F12 | 31 | 29 | Human: 20 | |
| Dog: 4 | ||||
| Others: 5 (4 types) | ||||
| F13 | 24 | 23 | Human: 19 | |
| Dog: 4 | ||||
| F14 | 14 | 13 | Human: 11 | |
| Others: 2 (2 types) | ||||
| F16 | 49 | 45 | Human: 45 | |
| papGI | 14 | 13 | Human: 6 | |
| Dog: 4 | ||||
| Others: 3 (3 types) | ||||
| papGIII | 122 | 108 | Human: 77 | |
| Dog: 16 | ||||
| Others: 15 (11 types) | ||||
| ST127 | All | 186 | 168 | Human: 144 |
| Dog: 7 | ||||
| Others: 17 (13 types) | ||||
| F48 | 170 | 153 | Human: 136 | |
| Dog: 4 | ||||
| Others: 13 (9 types) | ||||
| papGIII | 153 | 136 | Human: 118 | |
| Dog: 5 | ||||
| Others: 13 (10 types) | ||||
| ST117 | All | 135 | 116 | Poultry: 69 |
| Bovine: 21 | ||||
| Human: 9 | ||||
| Others: 18 (6 types) | ||||
| F11 | 66 | 61 | Poultry: 41 | |
| Bovine: 8 | ||||
| Human: 5 | ||||
| Others: 7 (3 types) | ||||
| ST393 | All | 57 | 53 | Human: 53 |
| F16 | 57 | 53 | Human: 53 | |
| papGII | 57 | 53 | Human: 53 |
| Strain | Diagnosis | Year of Isolation | Phylogroup | Sequence Type | papA Variant | papG Variant |
|---|---|---|---|---|---|---|
| 23-UCH | Urosepsis | 2011 | B2 | ST14 | F8 | papGII |
| 29-UCH | Urosepsis | 2011 | B2 | ST12 | F13 | papGII/papGIII |
| 81-UCH | Urosepsis | 2009 | B2 | ST12 | F13 | papGII/papGIII |
| 92-UCH | Urosepsis | 2009 | D | ST69 | F7-2 | papGII |
| 104-UCH | Urosepsis | 2009 | D | ST69 | F16 | papGII |
| 112-UCH | Urosepsis | 2009 | D | ST69 | F16 | papGII |
| 150-UCH | Urosepsis | 2008 | B2 | ST131 | F48 | papGII |
| 151-UCH | Urosepsis | 2008 | B2 | ST73 | F7-1/F48 | papGII |
| 175-UCH | Urosepsis | 2008 | B2 | ST12 | F13 | papGII/papGIII |
| 176-UCH | Urosepsis | 2008 | B2 | ST73 | F7-2 | papGII |
| 177-UCH | Urosepsis | 2008 | B2 | ST12 | F9/F13 | papGII/papGIII |
| 197-UCH | UTI | 2017 | B2 | ST73 | F7-1 | papGII |
| 199-UCH | UTI | 2017 | B2 | ST73 | F14 | papGII/papGIII |
| 207-UCH | UTI | 2017 | B2 | ST12 | F11/F16 | papGII |
| 208-UCH | UTI | 2017 | B2 | ST73 | F13 | papGII |
| 235-UCH | UTI | 2017 | B2 | ST131 | ND | papGII |
| 253-UCH | UTI | 2017 | B2 | ST14 | F8 | papGII |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Yáñez, V.; Suazo, P.; Hormazábal, C.; Ibaceta, V.; Arenas-Salinas, M.; Vidal, R.M.; Silva-Ojeda, F.; Arellano, C.; Muñoz, I.; Del Canto, F. Distribution of papA and papG Variants among Escherichia coli Genotypes: Association with Major Extraintestinal Pathogenic Lineages. Int. J. Mol. Sci. 2024, 25, 6657. https://doi.org/10.3390/ijms25126657
Fernández-Yáñez V, Suazo P, Hormazábal C, Ibaceta V, Arenas-Salinas M, Vidal RM, Silva-Ojeda F, Arellano C, Muñoz I, Del Canto F. Distribution of papA and papG Variants among Escherichia coli Genotypes: Association with Major Extraintestinal Pathogenic Lineages. International Journal of Molecular Sciences. 2024; 25(12):6657. https://doi.org/10.3390/ijms25126657
Chicago/Turabian StyleFernández-Yáñez, Valentina, Patricio Suazo, Claudia Hormazábal, Valentina Ibaceta, Mauricio Arenas-Salinas, Roberto M. Vidal, Francisco Silva-Ojeda, Carolina Arellano, Ignacio Muñoz, and Felipe Del Canto. 2024. "Distribution of papA and papG Variants among Escherichia coli Genotypes: Association with Major Extraintestinal Pathogenic Lineages" International Journal of Molecular Sciences 25, no. 12: 6657. https://doi.org/10.3390/ijms25126657
APA StyleFernández-Yáñez, V., Suazo, P., Hormazábal, C., Ibaceta, V., Arenas-Salinas, M., Vidal, R. M., Silva-Ojeda, F., Arellano, C., Muñoz, I., & Del Canto, F. (2024). Distribution of papA and papG Variants among Escherichia coli Genotypes: Association with Major Extraintestinal Pathogenic Lineages. International Journal of Molecular Sciences, 25(12), 6657. https://doi.org/10.3390/ijms25126657






