Reprogramming in Candida albicans Gene Expression Network under Butanol Stress Abrogates Hyphal Development
Abstract
1. Introduction
2. Results
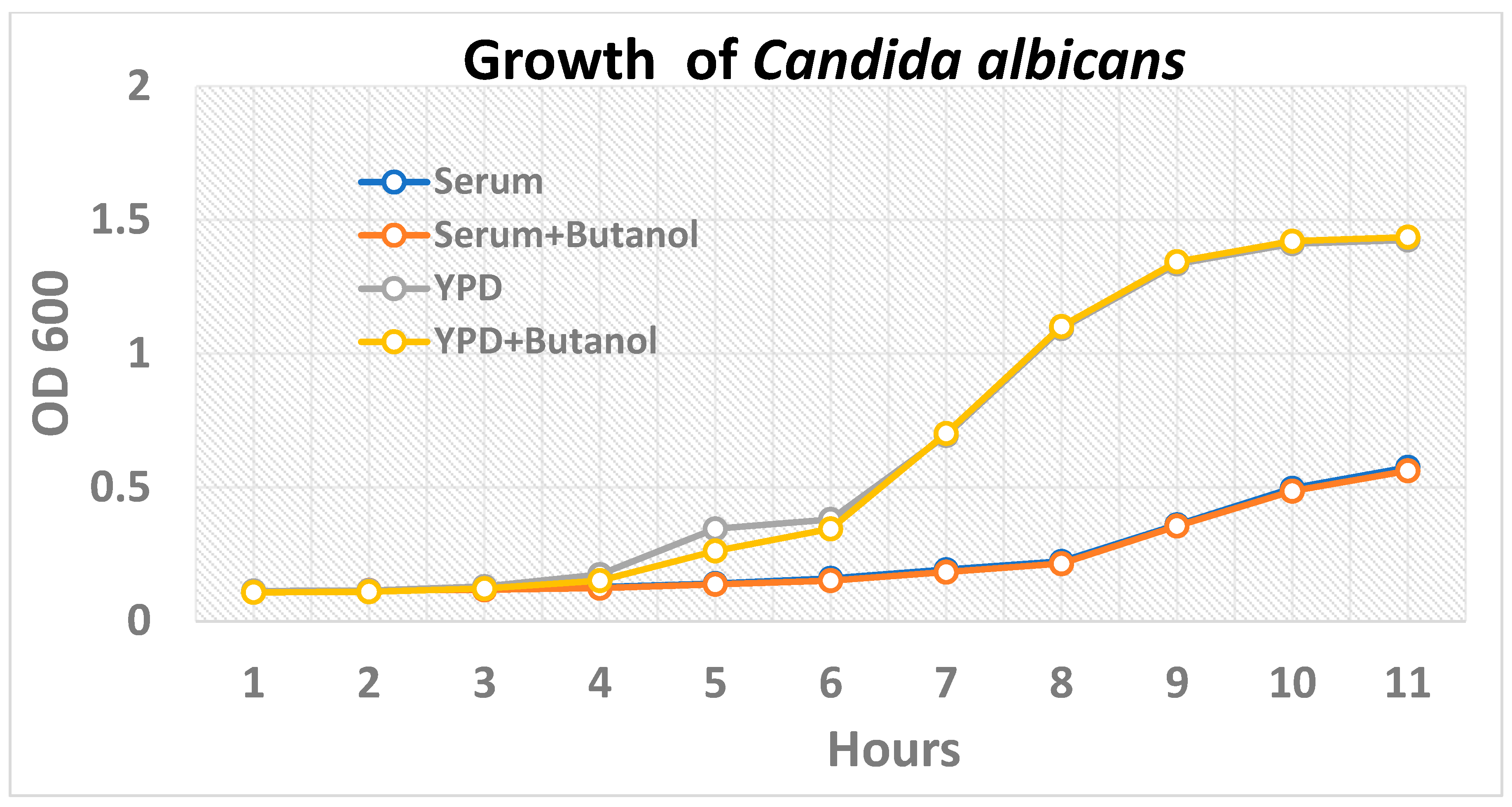
2.1. Inhibition of Hypha Formation by Butanol
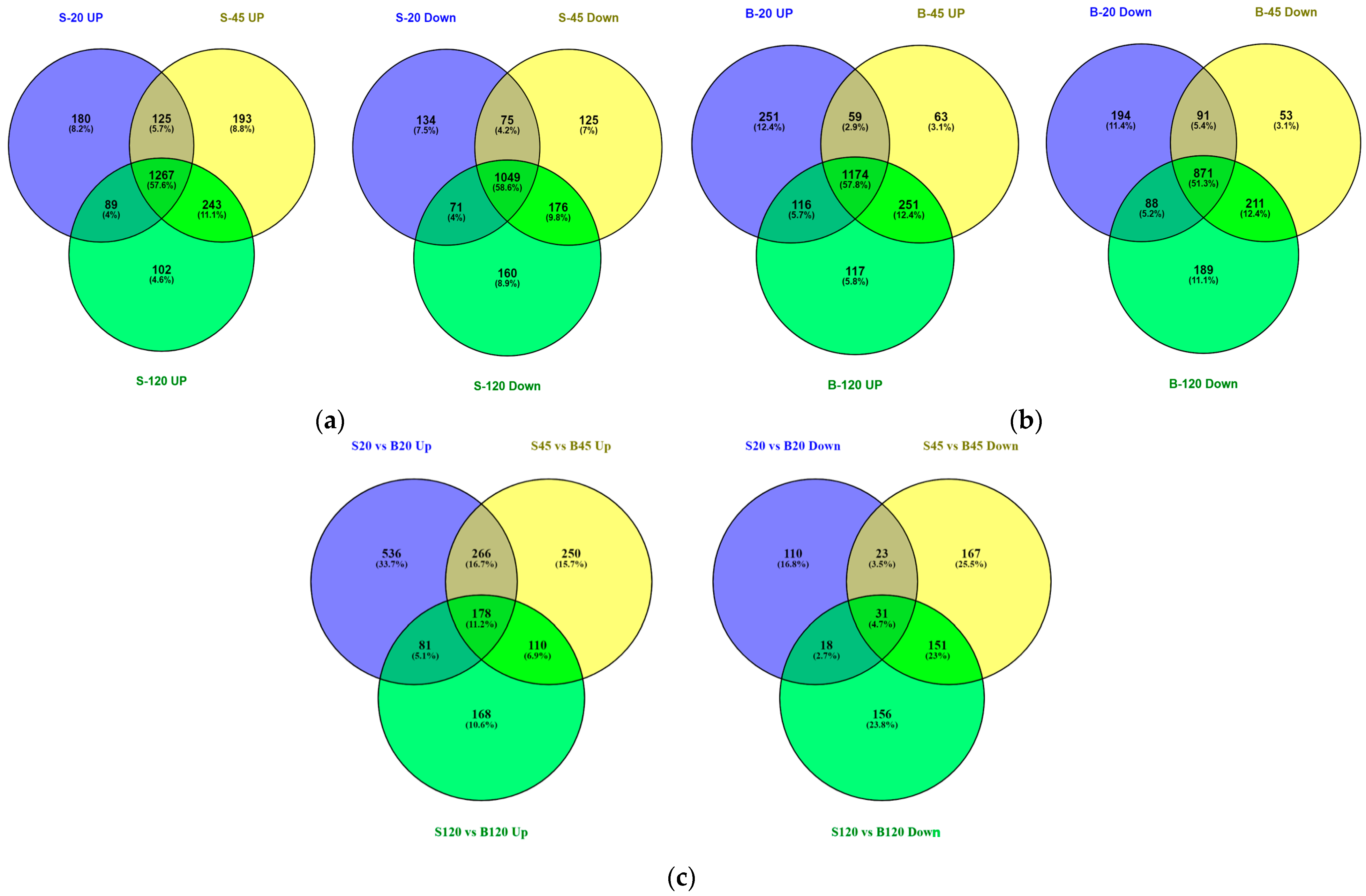
2.2. Differential Gene Expression Pattern of C. albicans in Serum and the Serum with Butanol
2.3. Altered Gene Expression Signatures of Many Hyphae-Associated Transcription Factors Repressors and Other Genes in Serum with Butanol Compared to the Serum
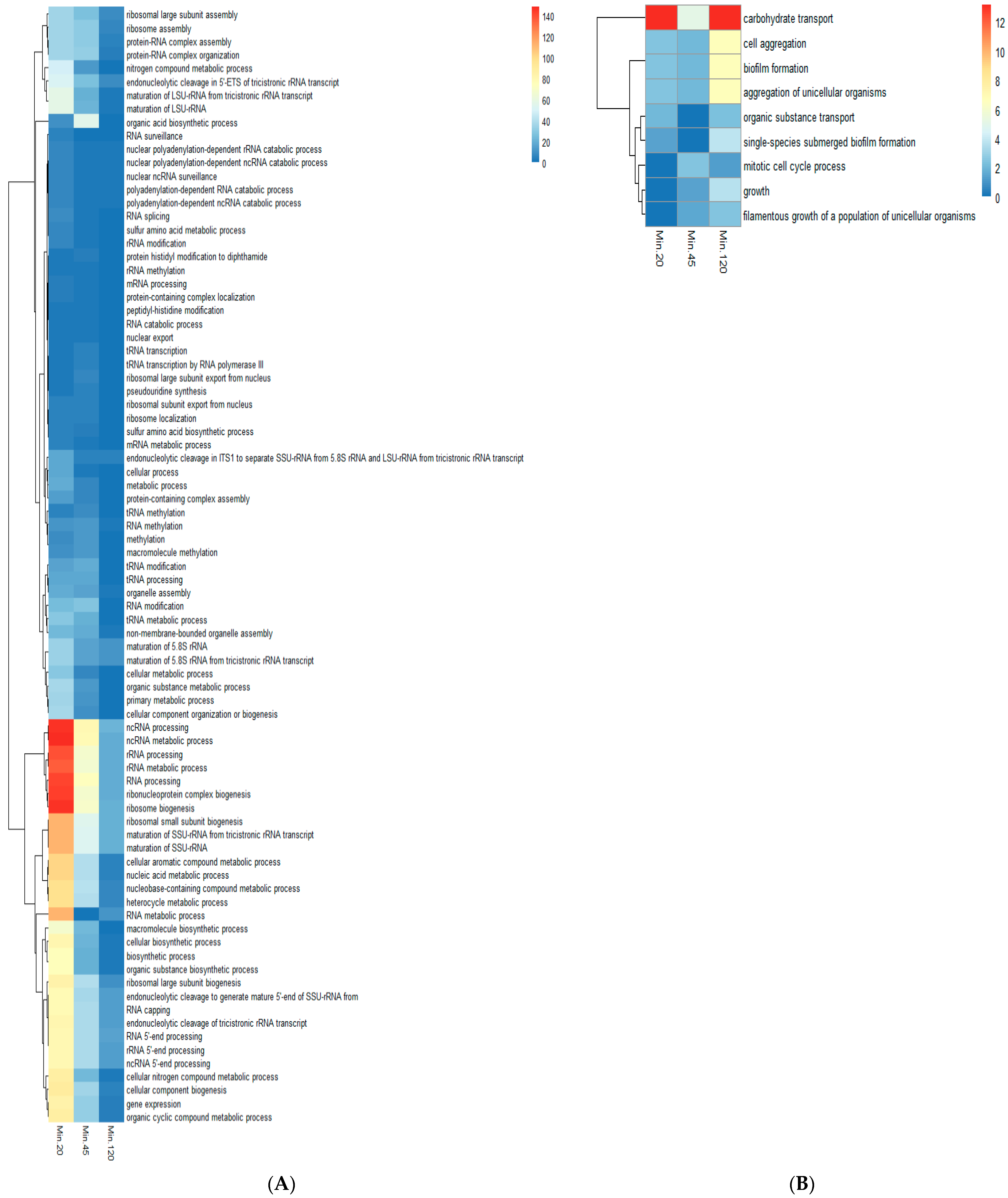
2.4. Gene Ontology (GO) Analysis of Comparative Gene Expression Data Obtained Using RPKM Values of Serum and Serum with Butanol Indicated Induction and Repression of Specific Processes in Presence of Butanol
2.5. Unique Stress Associated Transcripts in Serum with Butanol Compared to Serum
2.6. RNA Seq Gene-Expression Data and qRT-PCR Data Correlated with Each Other
3. Discussion
3.1. Hyphae Transcripts under the Influence of Butanol
3.2. Induction and Repression of Specific GO Processes in the Serum with Butanol
3.3. Stress Response of Candida albicans in the Serum with Butanol
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Selection of Strain and Preparation of Inoculum
4.3. Microscopy for the Determination of Inhibition of Hyphae Formation
4.3.1. Visualization of Candida albicans under Scanning Electron Microscope
4.3.2. Transmission Electron Microscopy for the Observation of Morphology of Candida albicans under the Influence of Butanol
4.4. Growth Kinetics of Candida albicans in Presence of Butanol
4.5. Extraction of Total RNA
4.6. RNA Library Preparation, Sequencing, and Data Analyses
4.7. Real-Time PCR Analysis of Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TF | Transcription Factor |
References
- Horn, D.L.; Neofytos, D.; Anaissie, E.J.; Fishman, J.A.; Steinbach, W.J.; Olyaei, A.J.; Marr, K.A.; Pfaller, M.A.; Chang, C.H.; Webster, K.M. Epidemiology and outcomes of candidemia in 2019 patients: Data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 2009, 48, 1695–1703. [Google Scholar] [CrossRef] [PubMed]
- Neofytos, D.; Horn, D.; Anaissie, E.; Steinbach, W.; Olyaei, A.; Fishman, J.; Pfaller, M.; Chang, C.; Webster, K.; Marr, K. Epidemiology and outcome of invasive fungal infection in adult hematopoietic stem cell transplant recipients: Analysis of Multicenter Prospective Antifungal Therapy (PATH) Alliance registry. Clin. Infect. Dis. 2009, 48, 265–273. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 2010, 36, 1–53. [Google Scholar] [CrossRef]
- Ciurea, C.N.; Kosovski, I.-B.; Mare, A.D.; Toma, F.; Pintea-Simon, I.A.; Man, A. Candida and Candidiasis—Opportunism versus Pathogenicity: A Review of the Virulence Traits. Microorganisms 2020, 8, 857. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Ghosh, A.; Batra, R.; Kaushal, A.; Roy, P.; Singh, H. Antifungal susceptibility pattern of non-albicans Candida species & distribution of species isolated from Candidaemia cases over a 5 year period. Indian J. Med. Res. 1996, 104, 171–176. [Google Scholar]
- Chakrabarti, A.; Mohan, B.; Shrivastava, S.K.; Marak, R.S.K. Change in distribution & antifungal susceptibility of Candida species isolated from candidaemia cases in a tertiary care centre during 1996–2000. Indian J. Med. Res. 2002, 116, 5. [Google Scholar]
- Verma, A.K.; Prasad, K.N.; Singh, M.; Dixit, A.K.; Ayyagari, A. Candidaemia in patients of a tertiary health care hospital from north India. Indian J. Med.Res. 2003, 117, 122–128. [Google Scholar] [PubMed]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.S.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef]
- Nikolaou, E.; Agrafioti, I.; Stumpf, M.; Quinn, J.; Stansfield, I.; Brown, A.J.P. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol. Biol. 2009, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.P.; Haynes, K.; Gow, N.A.R.; Quinn, J. Stress responses in Candida. In Candida and Candidiasis, 2nd ed.; Calderone, R.A., Clancy, C.J., Eds.; ASM Press: Washington, DC, USA, 2012; pp. 225–242. [Google Scholar]
- Odds, F.C.; Calderone, R.A.; Hube, B.; Nombela, C. Virulence in Candida species: Views and suggestions from a peer-group workshop. ASM News 2003, 69, 54–55. [Google Scholar]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Green, C.B.; Oh, S.-H.; Zhao, X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—A sticky pursuit. Med. Mycol. 2008, 46, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Dalle, F.; Wächtler, B.; L’Ollivier, C.; Holland, G.; Bannert, N.; Wilson, D.; Labruère, C.; Bonnin, A.; Hube, B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell. Microbiol. 2010, 12, 248–271. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.C.; Bender, J.A.; Fink, G.R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 2004, 3, 1076–1087. [Google Scholar] [CrossRef]
- Banerjee, M.; Thompson, D.S.; Lazzell, A.; Carlisle, P.L.; Pierce, C.; Monteagudo, C.; Lopez-Ribot, J.L.; Kadosh, D. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell. 2008, 19, 1354–1365. [Google Scholar] [CrossRef]
- Zeidler, U.; Lettner, T.; Lassnig, C.; Müller, M.; Lajko, R.; Hintner, H.; Breitenbach, M.; Bito, A. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 2009, 9, 126–142. [Google Scholar] [CrossRef]
- Carlisle, P.L.; Banerjee, M.; Lazzell, A.; Monteagudo, C.; López-Ribot, J.L.; Kadosh, D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. USA 2009, 106, 599–604. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, Y.; Wang, Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004, 23, 1845–1856. [Google Scholar] [CrossRef]
- Zheng, X.D.; Lee, R.T.H.; Wang, Y.M.; Lin, Q.S.; Wang, Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007, 26, 3760–3769. [Google Scholar] [CrossRef]
- Wang, A.; Raniga, P.P.; Lane, S.; Lu, Y.; Liu, H. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol. Cell. Biol. 2009, 29, 4406–4416. [Google Scholar] [CrossRef]
- Bishop, A.; Lane, R.; Beniston, R.; Chapa-y-Lazo, B.; Smythe, C.; Sudbery, P. Hyphal growth in Candida albicans requires the phosphorylation of Sec2 by the Cdc28-Ccn1/Hgc1 kinase. EMBO J. 2010, 29, 2930–2942. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Escribano, P.; González-Novo, A.; Suárez, M.B.; Li, C.R.; Wang, Y.; de Aldana, C.R.V.; Correa-Bordes, J. CDK-dependent phosphorylation of Mob2 is essential for hyphal development in Candida albicans. Mol. Biol. Cell. 2011, 22, 2458–2469. [Google Scholar] [CrossRef] [PubMed]
- González-Novo, A.; Correa-Bordes, J.; Labrador, L.; Sánchez, M.; Vazquez de Aldana, C.R.; Jiménez, J. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol. Biol. Cell. 2008, 19, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Lima, D.; Sudbery, P.E. In Candida albicans, phosphorylation of Exo84 by Cdk1-Hgc1 is necessary for efficient hyphal extension. Mol. Biol. Cell. 2014, 25, 1097–1110. [Google Scholar] [CrossRef]
- Sinha, I.; Wang, Y.M.; Philp, R.; Li, C.R.; Yap, W.H.; Wang, Y. Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev. Cell 2007, 13, 421–432. [Google Scholar] [CrossRef]
- Taschdjian, C.L.; Burchall, J.J.; Kozinn, P.J. Rapid identification of Candida albicans by filamentation on serum and serum substitutes. AMA J. Dis. Child. 1960, 99, 212–215. [Google Scholar] [CrossRef]
- Simonetti, N.; Strippoli, V.; Cassone, A. Yeast-mycelialconversioninduced by N-acetyl-D-glucosamine in Candida albicans. Nature 1974, 250, 344–346. [Google Scholar] [CrossRef]
- Buffo, J.; Herman, M.A.; Soll, D.R. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia 1984, 85, 21–30. [Google Scholar] [CrossRef]
- Lu, Y.; Su, C.; Wang, A.; Liu, H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 2011, 9, e1001105. [Google Scholar] [CrossRef]
- Klengel, T.; Liang, W.J.; Chaloupka, J.; Ruoff, C.; Schröppel, K.; Naglik, J.R.; Eckert, S.E.; Mogensen, E.G.; Haynes, K.; Tuite, M.F.; et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 2005, 15, 2021–2026. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.H., Jr.; Giusani, A.D.; Chen, X.; Kumamoto, C.A. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 1999, 34, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Rocha, C.R.; Schroppel, K.; Harcus, D.; Marcil, A.; Dignard, D.; Taylor, B.N.; Thomas, D.Y.; Whiteway, M.; Leberer, E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell. 2001, 12, 3631–3643. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Sundstrom, P. CAP1, an adenylatecyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 2001, 183, 3211–3223. [Google Scholar] [CrossRef]
- Zou, H.; Fang, H.M.; Zhu, Y.; Wang, Y. Candida albicans Cyr1, Cap1 and G-actin form a sensor/effector apparatus for activating cAMP synthesis in hyphal growth. Mol. Microbiol. 2010, 75, 579–591. [Google Scholar] [CrossRef]
- Hogan, D.A.; Sundstrom, P. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 2009, 4, 1263–1270. [Google Scholar] [CrossRef]
- Kumamoto, C.A.; Vinces, M.D. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 2005, 7, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Whiteway, M.; Bachewich, C. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 2007, 61, 529–553. [Google Scholar] [CrossRef]
- Brown, A.J.P. Integration of metabolism with virulence in Candida albicans. In Fungal Genomics (The Mycota); Brown, A.J.P., Esser, K., Eds.; Springer: Berlin, Germany; London, UK, 2005; pp. 85–203. [Google Scholar]
- Brown, A.J.; Budge, S.; Kaloriti, D.; Tillmann, A.; Jacobsen, M.D.; Yin, Z.; Ene, I.V.; Bohovych, I.; Sandai, D.; Kastora, S.; et al. Stress adaptation in a pathogenic fungus. J. Exp. Biol. 2014, 217, 144–155. [Google Scholar] [CrossRef]
- Enjalbert, B.; MacCallum, D.M.; Odds, F.C.; Brown, A.J.P. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect. Immun. 2007, 75, 2143–2151. [Google Scholar] [CrossRef]
- Miramón, P.; Dunker, C.; Windecker, H.; Bohovych, I.M.; Brown, A.J.P.; Kurzai, O.; Hube, B. Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS ONE 2012, 7, e52850. [Google Scholar] [CrossRef]
- Cottier, F.; Tan, A.S.; Chen, J.; Lum, J.; Zolezzi, F.; Poidinger, M.; Pavelka, N. The transcriptional stress response of Candida albicans to weak organic acids. G3 2015, 5, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; Smith, D.A.; Cornell, M.J.; Alam, I.; Nicholls, S.; Brown, A.J.P.; Quinn, J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 2006, 17, 1018–1032. [Google Scholar] [CrossRef] [PubMed]
- Kaloriti, D.; Tillmann, A.; Cook, E.; Jacobsen, M.D.; You, T.; Lenardon, M.D.; Ames, L.; Barahona, M.; Chandrasekaran, K.; Coghill, G.; et al. Combinatorial stresses kill pathogenic Candida species. Med. Mycol. 2012, 50, 699–709. [Google Scholar] [CrossRef]
- Chauhan, N.M.; Raut, J.S.; Karuppayil, S.M. A morphogenetic regulatory role for ethyl alcohol in Candida albicans. Mycoses 2011, 54, e697–e703. [Google Scholar] [CrossRef]
- Chauhan, N.M.; Shinde, R.B.; Karuppayil, S.M. Effect of alcohols on filamentation, growth, viability and biofilm development in Candida albicans. Braz. J. Microbiol. 2013, 44, 1315–1320. [Google Scholar] [CrossRef][Green Version]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. 2002, 66, 300–372. [Google Scholar] [CrossRef]
- Enjalbert, B.; Nantel, A.; Whiteway, M. Stress-induced gene expression in Candida albicans: Absence of a general stress response. Mol. Biol. Cell 2003, 14, 1460–1467. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.A.; Nicholls, S.; Morgan, B.A.; Brown, A.J.; Quinn, J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida Albicans. Mol. Biol. Cell 2004, 15, 4179–4190. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, M. Effects of methanol, ethanol, propanol, and butanol on bacterial attachment to surfaces. J. Gen. Microbiol. 1983, 129, 633–641. [Google Scholar] [CrossRef]
- Leberer, E.; Harcus, D.; Broadbent, I.D.; Clark, K.L.; Dignard, D.; Ziegelbauer, K.; Schmit, A.; Gow, N.A.R.; Brown, A.J.P.; Thomas, D.Y. Homologs of the Ste20p and Ste7p protein kinases are involved in hyphal formation of Candida albicans. Proc. Natl. Acad. Sci. USA 1996, 93, 13217–13222. [Google Scholar] [CrossRef] [PubMed]
- Leng, P. Gene Regulation During Morphogenesis in Candida albicans. Ph.D. Thesis, University of Aberdeen, Aberdeen, UK, 1999. [Google Scholar]
- Song, W.; Wang, H.; Chen, J. Candida albicans Sfl2, a temperature-induced transcriptional regulator, is required for virulence in a murine gastrointestinal infection model. FEMS Yeast Res. 2011, 11, 209–222. [Google Scholar] [CrossRef]
- Chibana, H.; Uno, J.; Cho, T.; Mikami, Y. Mutation in IRO1 tightly linked with URA3 gene reduces virulence of Candida albicans. Microbiol. Immunol. 2005, 49, 937–939. [Google Scholar] [CrossRef]
- Schweizer, A.; Rupp, S.; Taylor, B.N.; Röllinghoff, M.; Schröppel, K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 2000, 38, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Murad, A.M.A.; Leng, P.; Straffon, M.; Wishart, J.; Macaskill, S.; MacCallum, D.; Schnell, N.; Talibi, D.; Marechal, D.; Tekaia, F.; et al. NRG1 represses yeast–hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001, 20, 4742–4752. [Google Scholar] [CrossRef] [PubMed]
- Braun, B.R.; Kadosh, D.; Johnson, A.D. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001, 20, 4753–4761. [Google Scholar] [CrossRef] [PubMed]
- Talibi, D.; Raymond, M. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J. Bacteriol. 1999, 181, 231–240. [Google Scholar] [CrossRef]
- Uhl, M.A.; Biery, M.; Craig, N.; Johnson, A.D. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 2003, 22, 2668–2678. [Google Scholar] [CrossRef]
- Bauer, J.; Wendland, J. Candida albicans Sfl1 suppresses flocculation and filamentation. Eukar. Cell 2007, 6, 1736–1744. [Google Scholar] [CrossRef] [PubMed]
- Askew, C.; Sellam, A.; Epp, E.; Mallick, J.; Hogues, H.; Mullick, A.; Nantel, A.; Whiteway, M. The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol. Microbiol. 2011, 79, 940–953. [Google Scholar] [CrossRef]
- Cleary, I.A.; Reinhard, S.M.; Miller, C.L.; Murdoch, C.; Thornhill, M.H.; Lazzell, A.L.; Monteagudo, C.; Thomas, D.P.; Saville, S.P. Candida albicans adhesin Als3p is dispensable for virulence in the mouse model of disseminated candidiasis. Microbiology 2011, 157, 1806–1815. [Google Scholar] [CrossRef] [PubMed]
- Yau, K.P.S.; Weerasinghe, H.; Olivier, F.A.B.; Lo, T.L.; Powell, D.R.; Koch, B.; Beilharz, T.H.; Traven, A. The proteasome regulator Rpn4 controls antifungal drug tolerance by coupling protein homeostasis with metabolic responses to drug stress. PLoS Pathog. 2023, 19, e1011338. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.G.; Yang, Y.L.; Shih, H.I.; Su, C.L.; Lo, H.J. CaNdt80 is involved in drug resistance in Candida albicans by regulating CDR1. Antimicrob. Agents Chemother. 2004, 48, 4505–4512. [Google Scholar] [CrossRef]
- Li, R.; Kumar, R.; Tati, S.; Puri, S.; Edgerton, M. Candida albicans flu1-mediated efflux of salivary histatin 5 reduces its cytosolic concentration and fungicidal activity. Antimicrob. Agents Chemother. 2013, 57, 1832–1839. [Google Scholar] [CrossRef]
- Nobile, C.J.; Fox, E.P.; Nett, J.E.; Sorrells, T.R.; Mitrovich, Q.M.; Hernday, A.D.; Tuch, B.B.; Andes, D.R.; Johnson, A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 2012, 148, 126–138. [Google Scholar] [CrossRef]
- García-Sánchez, S.; Aubert, S.; Iraqui, I.; Janbon, G.; Ghigo, J.M.; d’Enfert, C. Candida albicans biofilms: A developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell 2004, 3, 536–545. [Google Scholar] [CrossRef]
- Tournu, H.; Tripathi, G.; Bertram, G.; Macaskill, S.; Mavor, A.; Walker, L.; Odds, F.C.; Gow, N.A.; Brown, A.J. Global role of the protein kinase Gcn2 in the human pathogen Candida albicans. Eukaryot. Cell 2005, 4, 1687–1696. [Google Scholar] [CrossRef]
- Singh, R.P.; Prasad, H.K.; Sinha, I.; Agarwal, N.; Natarajan, K. Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J. Biol. Chem. 2011, 286, 25154–25170. [Google Scholar] [CrossRef]
- Fradin, C.; De Groot, P.; MacCallum, D.; Schaller, M.; Klis, F.; Odds, F.C.; Hube, B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 2005, 56, 397–415. [Google Scholar] [CrossRef]
- Navarathna, D.H.M.L.P.; Das, A.; Morschhäuser, J.; Nickerson, K.W.; Roberts, D.D. Dur3 is the major urea transporter in Candida albicans and is co-regulated with the urea amidolyase Dur1, 2. Microbiology 2011, 157 Pt 1, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Vylkova, S.; Carman, A.J.; Danhof, H.A.; Collette, J.R.; Zhou, H.; Lorenz, M.C. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio 2011, 2, e00055-11. [Google Scholar] [CrossRef]
- Reuss, O.; Morschhäuser, J. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol. Microbiol. 2006, 60, 795–812. [Google Scholar] [CrossRef]
- Theiss, S.; Ishdorj, G.; Brenot, A.; Kretschmar, M.; Lan, C.Y.; Nichterlein, T.; Hacker, J.; Nigam, S.; Agabian, N.; Köhler, G.A. Inactivation of the phospholipase B gene PLB5 in wild-type Candida albicans reduces cell-associated phospholipase A2 activity and attenuates virulence. IJMM 2006, 296, 405–420. [Google Scholar] [CrossRef]
- Niewerth, M.; Kunze, D.; Seibold, M.; Schaller, M.; Korting, H.C.; Hube, B. Ciclopiroxolamine treatment affects the expression pattern of Candida albicans genes encoding virulence factors, iron metabolism proteins, and drug resistance factors. Antimicrob. Agents Chemother. 2003, 47, 1805–1817. [Google Scholar] [CrossRef]
- Bowles, L.K.; Ellefson, W.L. Effects of butanol on Clostridium acetobutylicum. Appl. Environ. Microbiol. 1985, 50, 1165–1170. [Google Scholar] [CrossRef]
- Heipieper, H.J.; Neumann, G.; Cornelissen, S.; Meinhardt, F. Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 2007, 74, 961–973. [Google Scholar] [CrossRef]
- González-Ramos, D.; van den Broek, M.; van Maris, A.J.; Pronk, J.T.; Daran, J.-M.G. Genome-scale analyses of butanol tolerance in Saccharomyces cerevisiae reveal an essential role of protein degradation. Biotechnol. Biofuels 2013, 6, 48. [Google Scholar] [CrossRef]
- Setiawan, I.; Blanchard, G.J. Structural disruption of phospholipid bilayers over a range of length scales by n-butanol. J. Phys. Chem. B 2014, 118, 3085–3093. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Mohamed, S.; Chandra, J.; Kuhn, D.; Liu, S.; Antar, O.S.; Munyon, R.; Mitchell, A.P.; Andes, D.; Chance, M.R.; et al. Alcohol dehydrogenase restricts the ability of the pathogen Candida albicans to form a biofilm on catheter surfaces through an ethanol-based mechanism. Infect. Immun. 2006, 74, 3804–3816. [Google Scholar] [CrossRef]
- Bruinenberg, P.M.; van Dijken, J.P.; Scheffers, W.A. An enzymic analysis of NADPH production and consumption in Candida utilis. J. Gen. Microbiol. 1983, 129, 965–971. [Google Scholar] [CrossRef]
- Piekarska, K.; Hardy, G.; Mol, E.; van den Burg, J.; Strijbis, K.; van Roermund, C.; van den Berg, M.; Distel, B. The activity of the glyoxylate cycle in peroxisomes of Candida albicans depends on a functional beta-oxidation pathway: Evidence for reduced metabolite transport across the peroxisomal membrane. Microbiology 2008, 154, 3061–3072. [Google Scholar] [CrossRef] [PubMed]
- Hromatka, B.S.; Noble, S.M.; Johnson, A.D. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell 2005, 16, 4814–4826. [Google Scholar] [CrossRef]
- Liu, T.T.; Lee, R.E.; Barker, K.S.; Lee, R.E.; Wei, L.; Homayouni, R.; Rogers, P.D. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 2005, 49, 2226–2236. [Google Scholar] [CrossRef]
- Lan, C.Y.; Rodarte, G.; Murillo, L.A.; Jones, T.; Davis, R.W.; Dungan, J.; Newport, G.; Agabian, N. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 2004, 53, 1451–1469. [Google Scholar] [CrossRef] [PubMed]
- Synnott, J.M.; Guida, A.; Mulhern-Haughey, S.; Higgins, D.G.; Butler, G. Regulation of the hypoxic response in Candida albicans. Eukaryot. Cell 2010, 9, 1734–1746. [Google Scholar] [CrossRef]
- Fan, J.; Chaturvedi, V.; Shen, S.H. Identification and phylogenetic analysis of a glucose transporter gene family from the human pathogenic yeast Candida albicans. J. Mol. Evol. 2002, 55, 336–346. [Google Scholar] [CrossRef]
- Copping, V.M.; Barelle, C.J.; Hube, B.; Gow, N.A.; Brown, A.J.; Odds, F.C. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Agents Chemother. 2005, 55, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Kuloyo, O.; Fourie, R.; Cason, E.; Albertyn, J.; Pohl, C.H. Transcriptome Analyses of Candida albicans Biofilms, Exposed to Arachidonic Acid and Fluconazole, Indicates Potential Drug Targets. G3 2020, 10, 3099–3108. [Google Scholar] [CrossRef]
- Kos-Braun, I.C.; Koš, M. Post-transcriptional Regulation of Ribosome Biogenesis in Yeast. Microb. Cell 2017, 4, 179–181. [Google Scholar] [CrossRef]
- Fleischmann, J.; Rocha, M.A. Decrease in Ribosomal RNA in Candida albicans Induced by Serum Exposure. PLoS ONE 2015, 10, e0124430. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, L.; Caplice, N.; Coleman, D.C.; Sullivan, D.J.; Moran, G.P. Differential filamentation of Candida albicans and Candida dubliniensis is governed by nutrient regulation of UME6 expression. Eukaryot. Cell. 2010, 9, 1383–1397. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.T.; Specian, A.F.; Andrade, C.G.; França, E.J.; Furlaneto-Maia, L.; Furlaneto, M.C. Interaction of Candida parapsilosis isolates with human hair and nail surfaces revealed by scanning electron microscopy analysis. Micron 2010, 41, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Wright, R. Transmission electron microscopy of yeast. Microsc. Res. Tech. 2000, 51, 496–510. [Google Scholar] [CrossRef] [PubMed]
- IlluminaTruSeq® RNA Sample Preparation v2 Guide. 2014. Available online: https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_truseq/truseqrna/truseq-rna-sample-prep-v2-guide-15026495-f.pdf (accessed on 11 March 2019).
- CLC Genomics Workbench Manuals. 2015. Available online: https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/852/index.php?manual=Sequence_alignment.html (accessed on 5 April 2018).
- Inglis, D.O.; Arnaud, M.B.; Binkley, J.; Shah, P.; Skrzypek, M.S.; Wymore, F.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida genome database incorporates multiple Candida species: Multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 2012, 40, D667–D674. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
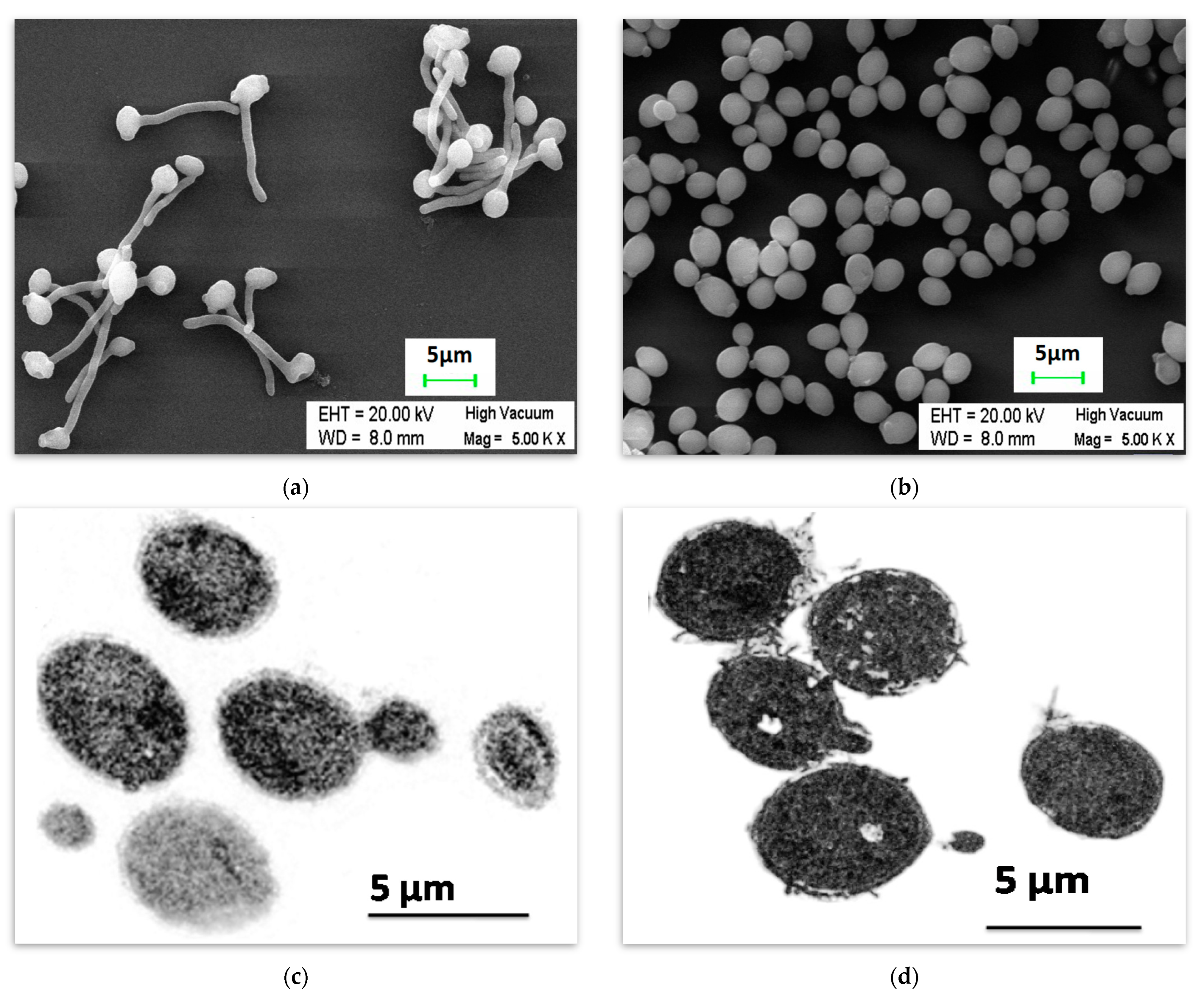
| Descriptions | Fold Change in Serum with Butanol Medium Compared to the Serum Alone Medium | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene Name/ID | 20 min | 45 min | 120 min | Gene Name/ID | 20 min | 45 min | 120 min | |
| Hypha-Specific TFs/ Repressor | UME6 | −677.49 | −264.33 | −75.17 | NRG1 | 2.45 | 8.44 | 11.72 |
| CSR1 | −5.79 | NS | −2.52 | FGR17 | NS | 3.7 | 4.06 | |
| IRO1 | −4.55 | −8.58 | −6.38 | FCR1 | NS | 3.56 | 8.41 | |
| SFL2 | −2.45 | −4.19 | NS | SFL1 | NS | 2.93 | 2.84 | |
| CPH1 | NS | −5.8 | −4.54 | TEC1 | −2.98 | −3.62 | NS | |
| Hypha-Specific Genes | ECE1 | NS | −1800.63 | −1732.65 | SHE3 | NS | −4.4 | −4.03 |
| HWP1 | −1265.6 | −2361.3 | −528.71 | RSR1 | NS | −4.65 | −6.12 | |
| SAP5 | NS | −511.46 | −425.61 | CLA4 | NS | −6.32 | −6.43 | |
| CDC11 | NS | −2.62 | −2.83 | C1_06250W_A | NS | −7.71 | −3.07 | |
| HYR1 | NS | −38.73 | −22.14 | DCK1 | NS | −2.97 | −4.02 | |
| CDC10 | NS | −2.5 | −2.28 | SOD5 | NS | −51.19 | −52.42 | |
| IHD1 | −42.14 | −63.83 | −145.49 | GPR1 | −7.84 | −3.23 | −3.78 | |
| C6_02330W_A | −4.86 | −17.36 | −31.74 | SAP6 | NS | −418.36 | −222.69 | |
| RBT1 | −27.61 | −57.1 | NS | C7_03480W_A | −2.29 | −6.01 | −7.15 | |
| RAX2 | NS | −2.15 | −3.04 | TUB2 | NS | −3.28 | −3.92 | |
| HGC1 | NS | −10.23 | −10 | PHR1 | −2.05 | −2.46 | −3.12 | |
| PGA54 | NS | −7.49 | −4.25 | CHS2 | −5.34 | NS | −2.25 | |
| RAS1 | −3.77 | −4.87 | −3.4 | AXL2 | NS | −9.14 | −7.88 | |
| PGA56 | NS | −2.88 | −3.73 | C4_01860C_A | −2.7 | −3.44 | −2.87 | |
| PST1 | NS | −3.93 | −2.36 | PTP3 | −4.01 | NS | −3.5 | |
| RBT4 | NS | −3.76 | −4.77 | SUN41 | NS | −4.49 | −3.57 | |
| PHO13 | NS | −3.50 | −7.07 | SHE3 | NS | −4.40 | −4.03 | |
| SNG4 | NS | −3.37 | −2.49 | MYO2 | NS | −2.60 | −2.39 | |
| IHD2 | NS | −3.14 | −3.6 | CAS4 | NS | −2.49 | −2.14 | |
| RHO3 | NS | −2.19 | −2.4 | MRP8 | NS | −2.47 | −2.12 | |
| KIP4 | NS | −2.31 | −5.52 | |||||
| Adhesion-Specific TFs/Repressor | UME6 | −677.49 | −264.33 | −75.17 | RFX2 | NS | −15.85 | −26.78 |
| AHR1 | −3.73 | −5.14 | −5.68 | |||||
| Adhesion-Specific Genes | C1_13100W_A | −5.1 | −7.06 | −12.61 | MSB2 | NS | −3.14 | −2.69 |
| ALS3 | NS | −105.50 | −96.27 | EAP1 | NS | −9.27 | −4.88 | |
| ECM33 | NS | −2.37 | −3.38 | PGA59 | −2.25 | −2.23 | NS | |
| FAV2 | NS | −20.73 | −18.53 | |||||
| Descriptions | Fold Changein Serum with Butanol Medium Compared to the Serum Alone Medium | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene Name/ID | 20 min | 45 min | 120 min | Gene Name/ID | 20 min | 45 min | 120 min | |
| Other TFs/Repressor | RLM1 | −3.49 | −3.33 | −4.27 | RPN4 | 2.38 | 6.15 | |
| SUC1 | NS | −2.44 | −2.59 | CTA4 | 2.35 | 3.78 | 2.02 | |
| AHR1 | −3.73 | −5.14 | −5.68 | ZCF5 | NS | 4.49 | 6.02 | |
| C2_00640W_A | NS | −2.15 | −3.11 | UGA3 | NS | 3.97 | 3.15 | |
| ZCF25 | 5.8 | NS | 13.8 | LYS144 | NS | 3.03 | 2.34 | |
| WOR2 | 5.73 | 10.45 | NS | C4_03160C_A | NS | 2.44 | 3.8 | |
| CTA8 | 3.25 | 3.04 | NS | NDT80 | NS | 2.19 | 2.38 | |
| C3_04860W_A | 3.11 | 4.72 | 3.81 | C2_01870C_A | 2.92 | 2.06 | 2.58 | |
| Fungal-Specific Genes | ATO1 | NS | −35.11 | −23.01 | FLU1 | 14.33 | 7.25 | NS |
| PLB3 | NS | −2.51 | −2.12 | MET16 | 5.56 | 2.77 | 2.33 | |
| PLB5 | NS | −4.74 | −6.03 | HOM3 | 5.1 | NS | 5.56 | |
| OPT6 | NS | −3.92 | −5.75 | C1_13130C_A | 3.08 | 4.18 | NS | |
| LSP1 | NS | −2.20 | −2.13 | LEU42 | 2.77 | 2.38 | 3.44 | |
| CR_03710C_A | NS | 4.67 | 2.38 | DUR4 | 2.25 | 2.84 | NS | |
| RIT1 | 2.66 | 2.50 | NS | C3_03070W_A | 2.05 | 2.54 | NS | |
| Heat-shock Genes | C7_01360C_A | 4.72 | 4.25 | NS | SSB1 | 2.01 | 2.09 | NS |
| CTA8 | 3.25 | 3.04 | NS | |||||
| Serum vs. Serum with Butanol | Expression in Serum vs. Control (0 min) and Serum with Butanol vs. Control (0 min) | Expression in Serum with Butanol vs. Control (0 min) | |||||
|---|---|---|---|---|---|---|---|
| Genes as observed in Osmotic Stress + Oxidative Stress [45,49] | Upregulated | Downregulated | Upregulated in Both | Downregulated in Both | No Change | Upregulated | Downregulated |
| IFR2 | RHD3 | C1_02040C_A | C2_07640W_A | AGP2 | CR_01090W_A | ||
| CR_03710C_A | CRG1 | GLC3 | C7_01430C_A | C3_06490W_A | |||
| C4_06710W_A | C1_07980C_A | ARE2 | MSN4 | ||||
| TPO3 | CDR11 | PST2 | C6_03370W_A | ||||
| RAD16 | CBP1 | HRT2 | LPI9 | ||||
| C7_01940C_A | GSY1 | STV1 | C1_06660W_A | ||||
| RHR2 | C1_06670W_A | C2_06600W_A | |||||
| C1_07990C_A | C2_04280W_A | ||||||
| C6_03320W_A | TPS3 | ||||||
| CR_02460W_A | C1_14250C_A | ||||||
| C3_06860C_A | MHP1 | ||||||
| C6_02420W_A | PRB1 | ||||||
| C2_07630C_A | C6_03780C_A | ||||||
| AHP1 | |||||||
| MLS1 | |||||||
| Genes as observed in heavy metal Stress + Oxidative Stress [45,50] | OYE32 | HSP21 | ZRT2 | CAP1 | C3_03230C_A | ||
| GST2 | OYE23 | ZRT2 | |||||
| OPT8 | TPS2 | C1_11530C_A | |||||
| GCS1 | HSP78 | ||||||
| CR_07480W_A | |||||||
| LEU42 | |||||||
| ALP1 | |||||||
| NRG1 | |||||||
| Genes as observed in heavy metal Stress + Osmotic Stress [45,49] | SOU1 | RCT1 | SRG1 | C3_01540W_A | SLK19 | ||
| C2_03020C_A | HMX1 | ALD5 | |||||
| HSP70 | ASR1 | ||||||
| HSP12 | C1_00310W_A | ||||||
| DLD1 | C1_10460W_A | ||||||
| PIR1 | C4_02160C_A | ||||||
| GAD1 | CIT1 | ||||||
| Genes as observed in heat shock Stress [45,50,51] | MRF1 | C7_00350C_A | PHO15 | TPS2 | HGT6 | ||
| RPN4 | C1_11270W_A | GLX3 | C1_11670W_A | GLK1 | |||
| C6_02480W_A | DAP1 | ||||||
| CAT1 | GRP2 | ||||||
| CDR4 | GPD2 | ||||||
| HGT8 | ZWF1 | ||||||
| NPR1 | ECM4 | ||||||
| RHR2 | |||||||
| AHP1 | |||||||
| HSP12 | |||||||
| Gene | 120 min (qRT-PCR) ± SEM * | 120 min (RNA Seq) | 120 min p Value (RNA Seq) |
|---|---|---|---|
| ACT1 | 1.8 ± 0.08 | 17.0 | 4.3 × 10−36 |
| ALS3 | 125 ± 3 | 215.3 | 1.3 × 10−130 |
| ECE1 | 26,851 ± 3749 | 25,907.7 | 1.0 × 10−8 |
| HWP1 | 1540 ± 288 | 7404.9 | 2.0 × 10−23 |
| RME1 | −98 ± 5 | −270.0 | 6.6 × 10−59 |
| SAP5 | 1669 ± 305 | 1156.0 | 7.7 × 10−5 |
| UME6 | 22 ± 2 | 87.0 | 6.1 × 10−7 |
| YWP1 | −129 ± 23 | −88.9 | 1.9 × 10−34 |
| C4_05010W_A | −1.6 ± 0.2 | −2.1 | 0.015604 |
| C5_03440W_A | −20 ± 2 | −28.4 | 1.9 × 10−12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anand, R.; Kashif, M.; Pandit, A.; Babu, R.; Singh, A.P. Reprogramming in Candida albicans Gene Expression Network under Butanol Stress Abrogates Hyphal Development. Int. J. Mol. Sci. 2023, 24, 17227. https://doi.org/10.3390/ijms242417227
Anand R, Kashif M, Pandit A, Babu R, Singh AP. Reprogramming in Candida albicans Gene Expression Network under Butanol Stress Abrogates Hyphal Development. International Journal of Molecular Sciences. 2023; 24(24):17227. https://doi.org/10.3390/ijms242417227
Chicago/Turabian StyleAnand, Rajesh, Mohammad Kashif, Awadhesh Pandit, Ram Babu, and Agam P. Singh. 2023. "Reprogramming in Candida albicans Gene Expression Network under Butanol Stress Abrogates Hyphal Development" International Journal of Molecular Sciences 24, no. 24: 17227. https://doi.org/10.3390/ijms242417227
APA StyleAnand, R., Kashif, M., Pandit, A., Babu, R., & Singh, A. P. (2023). Reprogramming in Candida albicans Gene Expression Network under Butanol Stress Abrogates Hyphal Development. International Journal of Molecular Sciences, 24(24), 17227. https://doi.org/10.3390/ijms242417227






