Residence of the Nucleotide Sugar Transporter Family Members SLC35F1 and SLC35F6 in the Endosomal/Lysosomal Pathway
Abstract
1. Introduction
2. Results
2.1. Human SLC35F1 (hSLC35F1) and SLC35F6 (hSLC35F6) Have Different Main Subcellular Localizations in HeLa Cells
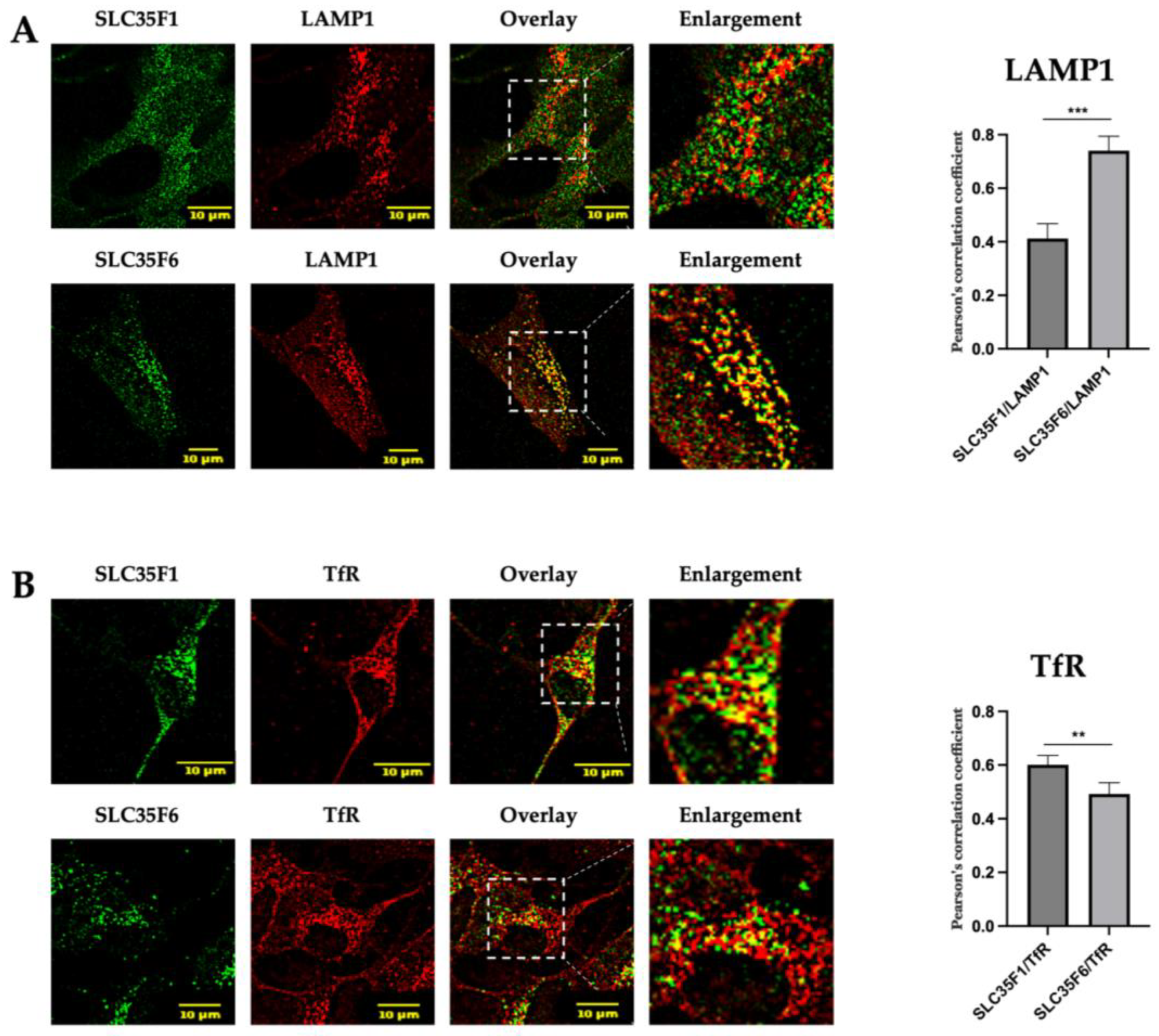
2.2. In SH-SY5Y Neuronal Cells, Endogenously Expressed SLC35F1 and SLC35F6 Are Located in Recycling Endosomes and Lysosomes, Respectively
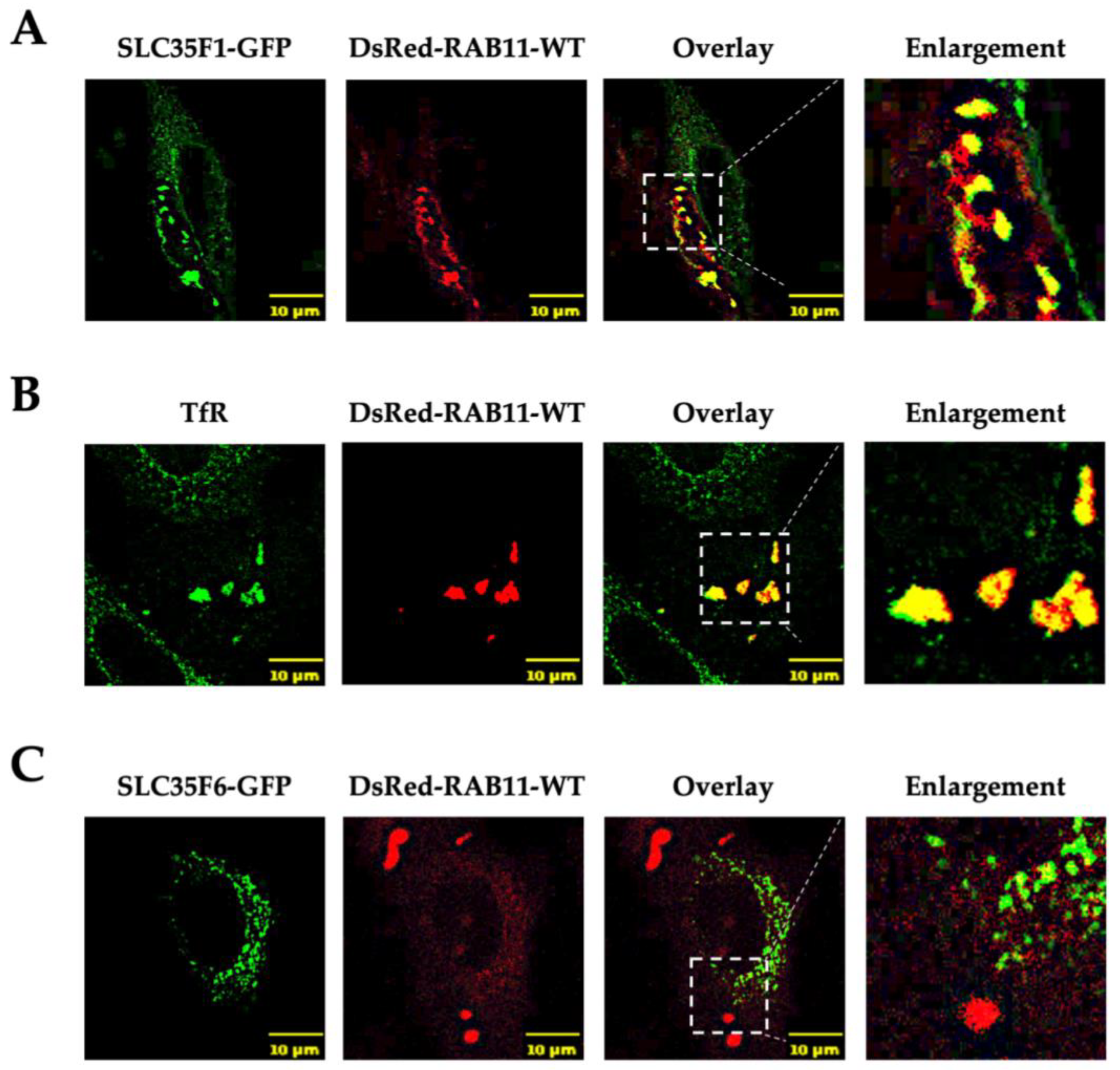
2.3. Overexpression of Rab11 Disrupts the Intracellular Distribution of SLC35F1 but Has Little Effect on SLC35F6 Localization
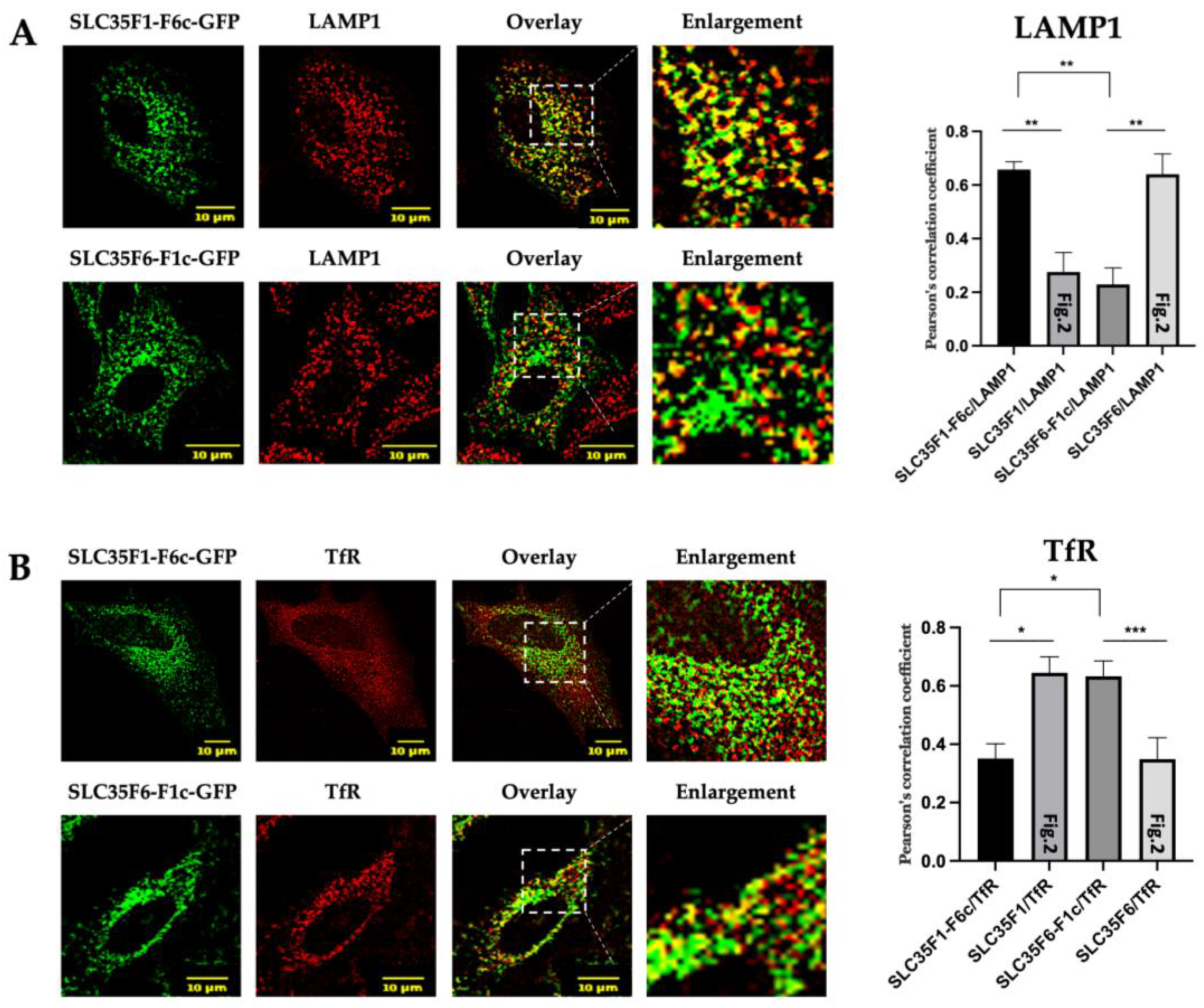
2.4. The C-Terminal Tails of SLC35F1 and SLC35F6 Contain the Molecular Information Responsible for Their Differential Trafficking
2.5. Two YXXΦ Sorting Motifs in SLC35F1 and a [D/E]XXXL[L/I] Motif in SLC35F6 C-Terminal Tails Are Critical for Their Respective Trafficking
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. DNA Constructs
4.3. Cell Culture and Transient Transfection
4.4. Western Blotting
4.5. Immunofluorescence
4.6. Cell Surface Biotinylation Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, Z. Roles of the Nucleotide Sugar Transporters (SLC35 Family) in Health and Disease. Mol. Asp. Med. 2013, 34, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Hadley, B.; Litfin, T.; Day, C.J.; Haselhorst, T.; Zhou, Y.; Tiralongo, J. Nucleotide Sugar Transporter SLC35 Family Structure and Function. Comput. Struct. Biotechnol. J. 2019, 17, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Orellana, A.; Moraga, C.; Araya, M.; Moreno, A. Overview of Nucleotide Sugar Transporter Gene Family Functions Across Multiple Species. J. Mol. Biol. 2016, 428, 3150–3165. [Google Scholar] [CrossRef] [PubMed]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global View of Human Protein Glycosylation Pathways and Functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef] [PubMed]
- Mikkola, S. Nucleotide Sugars in Chemistry and Biology. Molecules 2020, 25, 5755. [Google Scholar] [CrossRef] [PubMed]
- Farenholtz, J.; Artelt, N.; Blumenthal, A.; Endlich, K.; Kroemer, H.K.; Endlich, N.; von Bohlen und Halbach, O. Expression of Slc35f1 in the Murine Brain. Cell Tissue Res. 2019, 377, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Boonen, M.; Hamer, I.; Boussac, M.; Delsaute, A.-F.; Flamion, B.; Garin, J.; Jadot, M. Intracellular Localization of P40, a Protein Identified in a Preparation of Lysosomal Membranes. Biochem. J. 2006, 395, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Wu, Z.; Sun, R.; Yu, H.; Zeng, J.; Rao, Y.; Li, Y. Localization, Proteomics, and Metabolite Profiling Reveal a Putative Vesicular Transporter for UDP-Glucose. eLife 2021, 10, e65417. [Google Scholar] [CrossRef] [PubMed]
- Chintala, S.; Tan, J.; Gautam, R.; Rusiniak, M.E.; Guo, X.; Li, W.; Gahl, W.A.; Huizing, M.; Spritz, R.A.; Hutton, S.; et al. The Slc35d3 Gene, Encoding an Orphan Nucleotide Sugar Transporter, Regulates Platelet-Dense Granules. Blood 2007, 109, 1533–1540. [Google Scholar] [CrossRef]
- Meng, R.; Wang, Y.; Yao, Y.; Zhang, Z.; Harper, D.C.; Heijnen, H.F.G.; Sitaram, A.; Li, W.; Raposo, G.; Weiss, M.J.; et al. SLC35D3 Delivery from Megakaryocyte Early Endosomes Is Required for Platelet Dense Granule Biogenesis and Is Differentially Defective in Hermansky-Pudlak Syndrome Models. Blood 2012, 120, 404–414. [Google Scholar] [CrossRef]
- Lee, M.M.; Nasirikenari, M.; Manhardt, C.T.; Ashline, D.J.; Hanneman, A.J.; Reinhold, V.N.; Lau, J.T.Y. Platelets Support Extracellular Sialylation by Supplying the Sugar Donor Substrate. J. Biol. Chem. 2014, 289, 8742–8748. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Traub, L.M. Signals for Sorting of Transmembrane Proteins to Endosomes and Lysosomes. Annu. Rev. Biochem. 2003, 72, 395–447. [Google Scholar] [CrossRef] [PubMed]
- Staudt, C.; Puissant, E.; Boonen, M. Subcellular Trafficking of Mammalian Lysosomal Proteins: An Extended View. Int. J. Mol. Sci. 2016, 18, 47. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.S. Forty Years of Clathrin-coated Vesicles. Traffic 2015, 16, 1210–1238. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Guo, X. Adaptor Protein Complexes and Intracellular Transport. Biosci. Rep. 2014, 34, e00123. [Google Scholar] [CrossRef]
- Sanger, A.; Hirst, J.; Davies, A.K.; Robinson, M.S. Adaptor Protein Complexes and Disease at a Glance. J. Cell Sci. 2019, 132, jcs222992. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Carosi, J.M.; Yang, Z.; Ariotti, N.; Kerr, M.C.; Parton, R.G.; Sargeant, T.J.; Teasdale, R.D. Retromer Has a Selective Function in Cargo Sorting via Endosome Transport Carriers. J. Cell Biol. 2019, 218, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Weeratunga, S.; Paul, B.; Collins, B.M. Recognising the Signals for Endosomal Trafficking. Curr. Opin. Cell Biol. 2020, 65, 17–27. [Google Scholar] [CrossRef]
- Cullen, P.J.; Steinberg, F. To Degrade or Not to Degrade: Mechanisms and Significance of Endocytic Recycling. Nat. Rev. Mol. Cell Biol. 2018, 19, 679–696. [Google Scholar] [CrossRef]
- Fölsch, H. Role of the Epithelial Cell-Specific Clathrin Adaptor Complex AP-1B in Cell Polarity. Cell. Logist. 2015, 5, e1074331. [Google Scholar] [CrossRef]
- Bonifacino, J.S. Adaptor Proteins Involved in Polarized Sorting. J. Cell Biol. 2014, 204, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Boonen, M.; Rezende de Castro, R.; Cuvelier, G.; Hamer, I.; Jadot, M. A Dileucine Signal Situated in the C-Terminal Tail of the Lysosomal Membrane Protein P40 Is Responsible for Its Targeting to Lysosomes. Biochem. J. 2008, 414, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Mayle, K.M.; Le, A.M.; Kamei, D.T. The Intracellular Trafficking Pathway of Transferrin. Biochim. Biophys. Acta BBA-Gen. Subj. 2012, 1820, 264–281. [Google Scholar] [CrossRef] [PubMed]
- Gammella, E.; Buratti, P.; Cairo, G.; Recalcati, S. The Transferrin Receptor: The Cellular Iron Gate. Metallomics 2017, 9, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Welz, T.; Wellbourne-Wood, J.; Kerkhoff, E. Orchestration of Cell Surface Proteins by Rab11. Trends Cell Biol. 2014, 24, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Baetz, N.W.; Goldenring, J.R. Rab11-Family Interacting Proteins Define Spatially and Temporally Distinct Regions within the Dynamic Rab11a-Dependent Recycling System. Mol. Biol. Cell 2013, 24, 643–658. [Google Scholar] [CrossRef]
- Takahashi, S.; Kubo, K.; Waguri, S.; Yabashi, A.; Shin, H.-W.; Katoh, Y.; Nakayama, K. Rab11 Regulates Exocytosis of Recycling Vesicles at the Plasma Membrane. J. Cell Sci. 2012, 125, 4049–4057. [Google Scholar] [CrossRef] [PubMed]
- Hölttä-Vuori, M.; Tanhuanpää, K.; Möbius, W.; Somerharju, P.; Ikonen, E. Modulation of Cellular Cholesterol Transport and Homeostasis by Rab11. Mol. Biol. Cell 2002, 13, 3107–3122. [Google Scholar] [CrossRef]
- Wilcke, M.; Johannes, L.; Galli, T.; Mayau, V.; Goud, B.; Salamero, J. Rab11 Regulates the Compartmentalization of Early Endosomes Required for Efficient Transport from Early Endosomes to the Trans-Golgi Network. J. Cell Biol. 2000, 151, 1207–1220. [Google Scholar] [CrossRef]
- Vergarajauregui, S.; Puertollano, R. Two Di-Leucine Motifs Regulate Trafficking of Mucolipin-1 to Lysosomes. Traffic 2006, 7, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Kyttälä, A.; Yliannala, K.; Schu, P.; Jalanko, A.; Luzio, J.P. AP-1 and AP-3 Facilitate Lysosomal Targeting of Batten Disease Protein CLN3 via Its Dileucine Motif. J. Biol. Chem. 2005, 280, 10277–10283. [Google Scholar] [CrossRef] [PubMed]
- Mayor, S.; Presley, J.F.; Maxfield, F.R. Sorting of Membrane Components from Endosomes and Subsequent Recycling to the Cell Surface Occurs by a Bulk Flow Process. J. Cell Biol. 1993, 121, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.D.; Donaldson, J.G. Pathways and Mechanisms of Endocytic Recycling. Nat. Rev. Mol. Cell Biol. 2009, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- White, S.; Hatton, S.R.; Siddiqui, M.A.; Parker, C.D.; Trowbridge, I.S.; Collawn, J.F. Analysis of the Structural Requirements for Lysosomal Membrane Targeting Using Transferrin Receptor Chimeras. J. Biol. Chem. 1998, 273, 14355–14362. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Li, J.; Bos, E.; Porcionatto, M.; Premont, R.T.; Bourgoin, S.; Peters, P.J.; Hsu, V.W. ACAP1 Promotes Endocytic Recycling by Recognizing Recycling Sorting Signals. Dev. Cell 2004, 7, 771–776. [Google Scholar] [CrossRef]
- Bai, M.; Pang, X.; Lou, J.; Zhou, Q.; Zhang, K.; Ma, J.; Li, J.; Sun, F.; Hsu, V.W. Mechanistic Insights into Regulated Cargo Binding by ACAP1 Protein. J. Biol. Chem. 2012, 287, 28675–28685. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Peters, P.J.; Bai, M.; Dai, J.; Bos, E.; Kirchhausen, T.; Kandror, K.V.; Hsu, V.W. An ACAP1-Containing Clathrin Coat Complex for Endocytic Recycling. J. Cell Biol. 2007, 178, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Gallon, M.; Clairfeuille, T.; Steinberg, F.; Mas, C.; Ghai, R.; Sessions, R.B.; Teasdale, R.D.; Collins, B.M.; Cullen, P.J. A Unique PDZ Domain and Arrestin-like Fold Interaction Reveals Mechanistic Details of Endocytic Recycling by SNX27-Retromer. Proc. Natl. Acad. Sci. USA 2014, 111, E3604–E3613. [Google Scholar] [CrossRef]
- Bartuzi, P.; Billadeau, D.D.; Favier, R.; Rong, S.; Dekker, D.; Fedoseienko, A.; Fieten, H.; Wijers, M.; Levels, J.H.; Huijkman, N.; et al. CCC- and WASH-Mediated Endosomal Sorting of LDLR Is Required for Normal Clearance of Circulating LDL. Nat. Commun. 2016, 7, 10961. [Google Scholar] [CrossRef]
- Wang, J.; Fedoseienko, A.; Chen, B.; Burstein, E.; Jia, D.; Billadeau, D.D. Endosomal Receptor Trafficking: Retromer and Beyond. Traffic 2018, 19, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Neiss, W.F. A Coat of Glycoconjugates on the Inner Surface of the Lysosomal Membrane in the Rat Kidney. Histochem. Cell Biol. 1984, 80, 603–608. [Google Scholar] [CrossRef]
- Kundra, R.; Kornfeld, S. Asparagine-Linked Oligosaccharides Protect Lamp-1 and Lamp-2 from Intracellular Proteolysis. J. Biol. Chem. 1999, 274, 31039–31046. [Google Scholar] [CrossRef]
- Rudnik, S.; Damme, M. The Lysosomal Membrane—Export of Metabolites and Beyond. FEBS J. 2021, 288, 4168–4182. [Google Scholar] [CrossRef]
- Sleat, D.E.; Della Valle, M.C.; Zheng, H.; Moore, D.F.; Lobel, P. The Mannose 6-Phosphate Glycoprotein Proteome. J. Proteome Res. 2008, 7, 3010–3021. [Google Scholar] [CrossRef]
- Sleat, D.E.; Sun, P.; Wiseman, J.A.; Huang, L.; El-Banna, M.; Zheng, H.; Moore, D.F.; Lobel, P. Extending the Mannose 6-Phosphate Glycoproteome by High Resolution/Accuracy Mass Spectrometry Analysis of Control and Acid Phosphatase 5-Deficient Mice. Mol. Cell. Proteom. 2013, 12, 1806–1817. [Google Scholar] [CrossRef] [PubMed]
- Abu-Remaileh, M.; Wyant, G.A.; Kim, C.; Laqtom, N.N.; Abbasi, M.; Chan, S.H.; Freinkman, E.; Sabatini, D.M. Lysosomal Metabolomics Reveals V-ATPase- and mTOR-Dependent Regulation of Amino Acid Efflux from Lysosomes. Science 2017, 358, 807–813. [Google Scholar] [CrossRef]
- Damme, M.; Morelle, W.; Schmidt, B.; Andersson, C.; Fogh, J.; Michalski, J.-C.; Lübke, T. Impaired Lysosomal Trimming of N-Linked Oligosaccharides Leads to Hyperglycosylation of Native Lysosomal Proteins in Mice with α-Mannosidosis. Mol. Cell. Biol. 2010, 30, 273–283. [Google Scholar] [CrossRef]
- Nasirikenari, M.; Veillon, L.; Collins, C.C.; Azadi, P.; Lau, J.T.Y. Remodeling of Marrow Hematopoietic Stem and Progenitor Cells by Non-Self ST6Gal-1 Sialyltransferase. J. Biol. Chem. 2014, 289, 7178–7189. [Google Scholar] [CrossRef]
- Lee-Sundlov, M.M.; Ashline, D.J.; Hanneman, A.J.; Grozovsky, R.; Reinhold, V.N.; Hoffmeister, K.M.; Lau, J.T. Circulating Blood and Platelets Supply Glycosyltransferases That Enable Extrinsic Extracellular Glycosylation. Glycobiology 2017, 27, 188–198. [Google Scholar] [CrossRef]
- Wandall, H.H.; Rumjantseva, V.; Sørensen, A.L.T.; Patel-Hett, S.; Josefsson, E.C.; Bennett, E.P.; Italiano, J.E., Jr.; Clausen, H.; Hartwig, J.H.; Hoffmeister, K.M. The Origin and Function of Platelet Glycosyltransferases. Blood 2012, 120, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.B.; Oswald, D.M.; Joshi, S.; Whiteheart, S.W.; Orlando, R.; Cobb, B.A. B-Cell–Independent Sialylation of IgG. Proc. Natl. Acad. Sci. USA 2016, 113, 7207–7212. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.; Dominguez, M.; Puri, V.; Sharma, D.K.; Narita, K.; Wheatley, C.L.; Marks, D.L.; Pagano, R.E. Rab Proteins Mediate Golgi Transport of Caveola-Internalized Glycosphingolipids and Correct Lipid Trafficking in Niemann-Pick C Cells. J. Clin. Investig. 2002, 109, 1541–1550. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van den Bossche, F.; Tevel, V.; Gilis, F.; Gaussin, J.-F.; Boonen, M.; Jadot, M. Residence of the Nucleotide Sugar Transporter Family Members SLC35F1 and SLC35F6 in the Endosomal/Lysosomal Pathway. Int. J. Mol. Sci. 2024, 25, 6718. https://doi.org/10.3390/ijms25126718
Van den Bossche F, Tevel V, Gilis F, Gaussin J-F, Boonen M, Jadot M. Residence of the Nucleotide Sugar Transporter Family Members SLC35F1 and SLC35F6 in the Endosomal/Lysosomal Pathway. International Journal of Molecular Sciences. 2024; 25(12):6718. https://doi.org/10.3390/ijms25126718
Chicago/Turabian StyleVan den Bossche, François, Virginie Tevel, Florentine Gilis, Jean-François Gaussin, Marielle Boonen, and Michel Jadot. 2024. "Residence of the Nucleotide Sugar Transporter Family Members SLC35F1 and SLC35F6 in the Endosomal/Lysosomal Pathway" International Journal of Molecular Sciences 25, no. 12: 6718. https://doi.org/10.3390/ijms25126718
APA StyleVan den Bossche, F., Tevel, V., Gilis, F., Gaussin, J.-F., Boonen, M., & Jadot, M. (2024). Residence of the Nucleotide Sugar Transporter Family Members SLC35F1 and SLC35F6 in the Endosomal/Lysosomal Pathway. International Journal of Molecular Sciences, 25(12), 6718. https://doi.org/10.3390/ijms25126718





