Beyond the Usual Suspects: Examining the Role of Understudied Histone Variants in Breast Cancer
Abstract
:1. Introduction
2. Canonical and Non-Canonical Histones
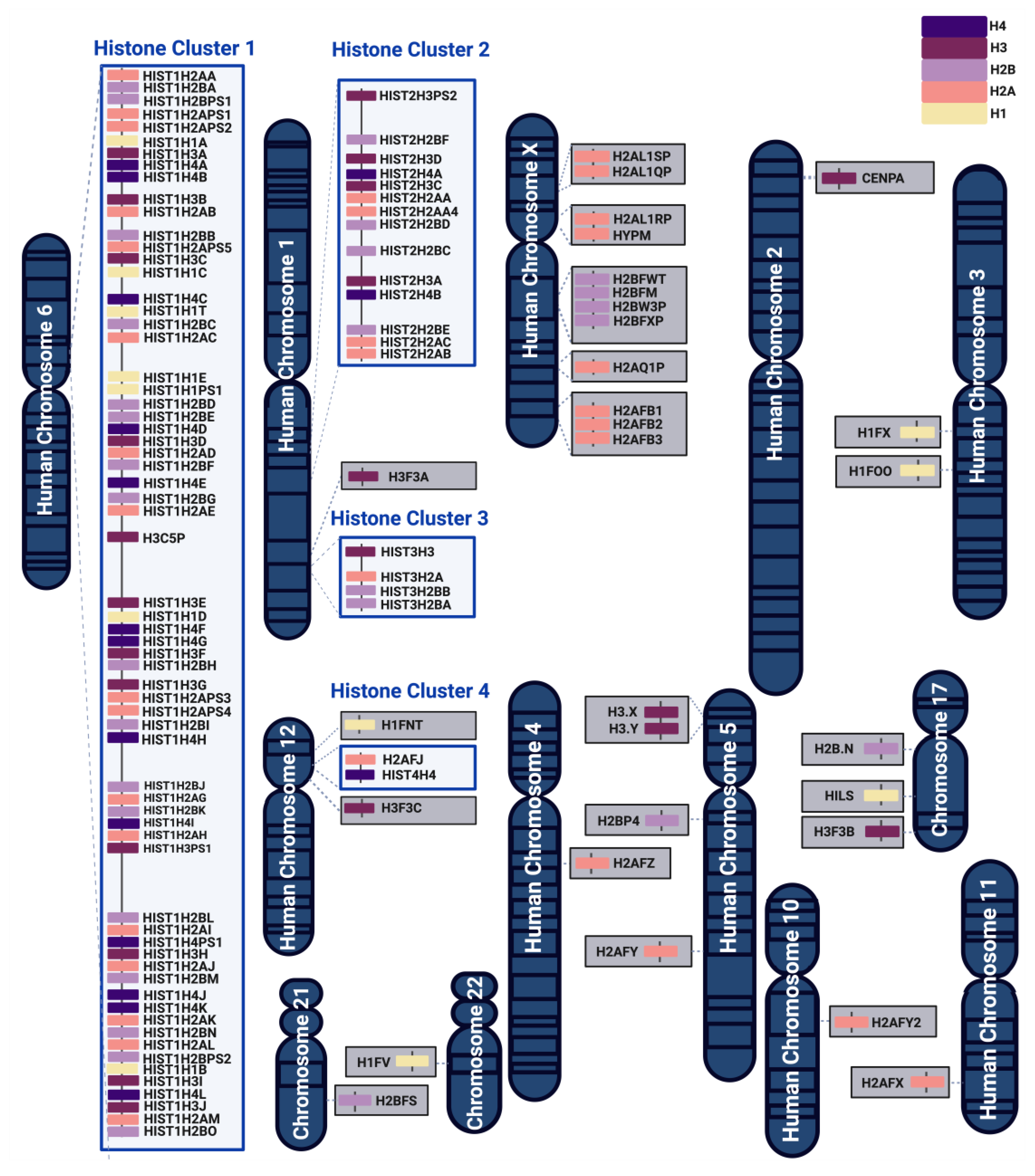
3. Histone Gene Organization and Expression
4. Histone Isoforms and Nucleosome Structure
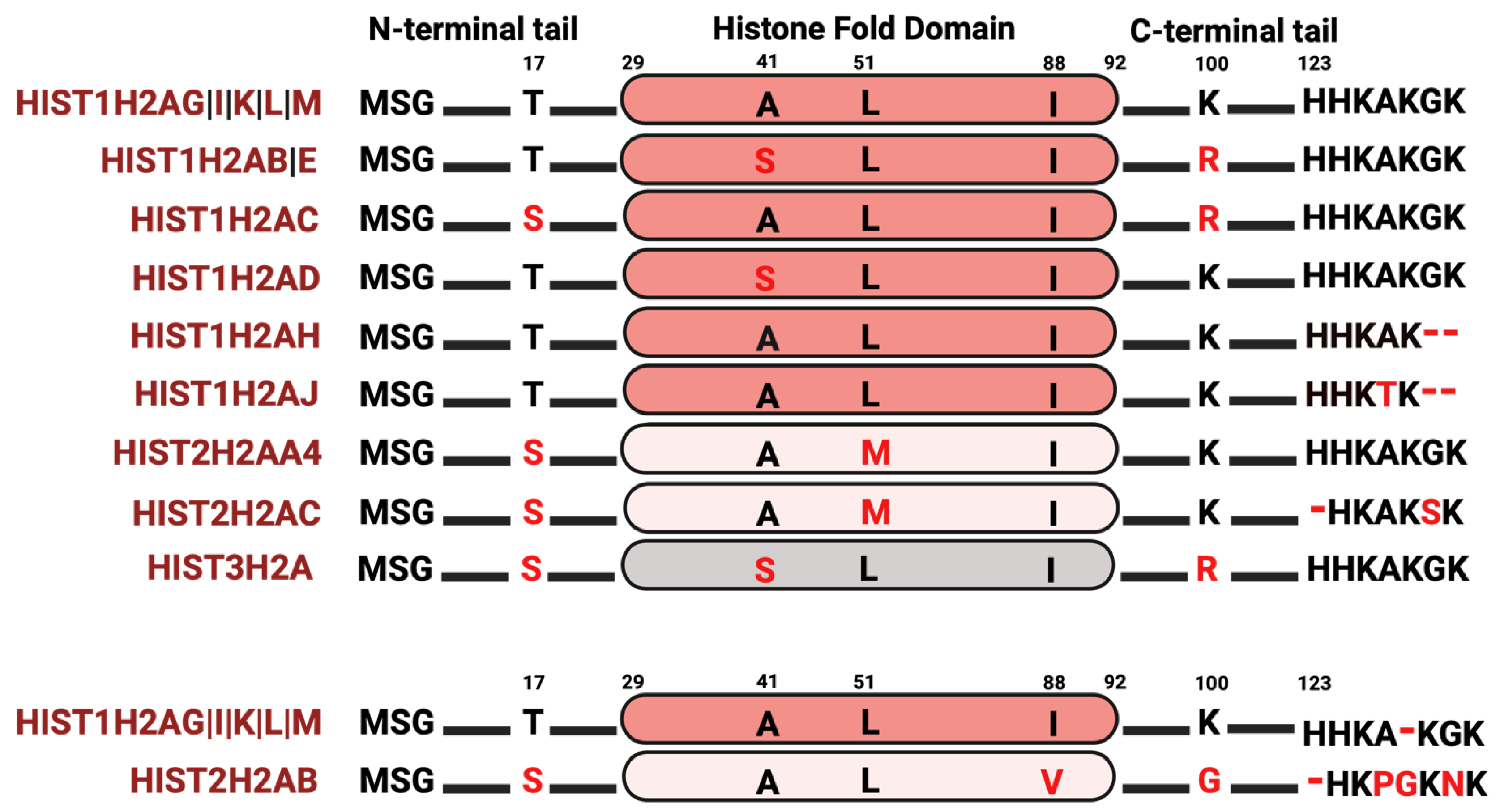
4.1. Histone H2A Isoforms
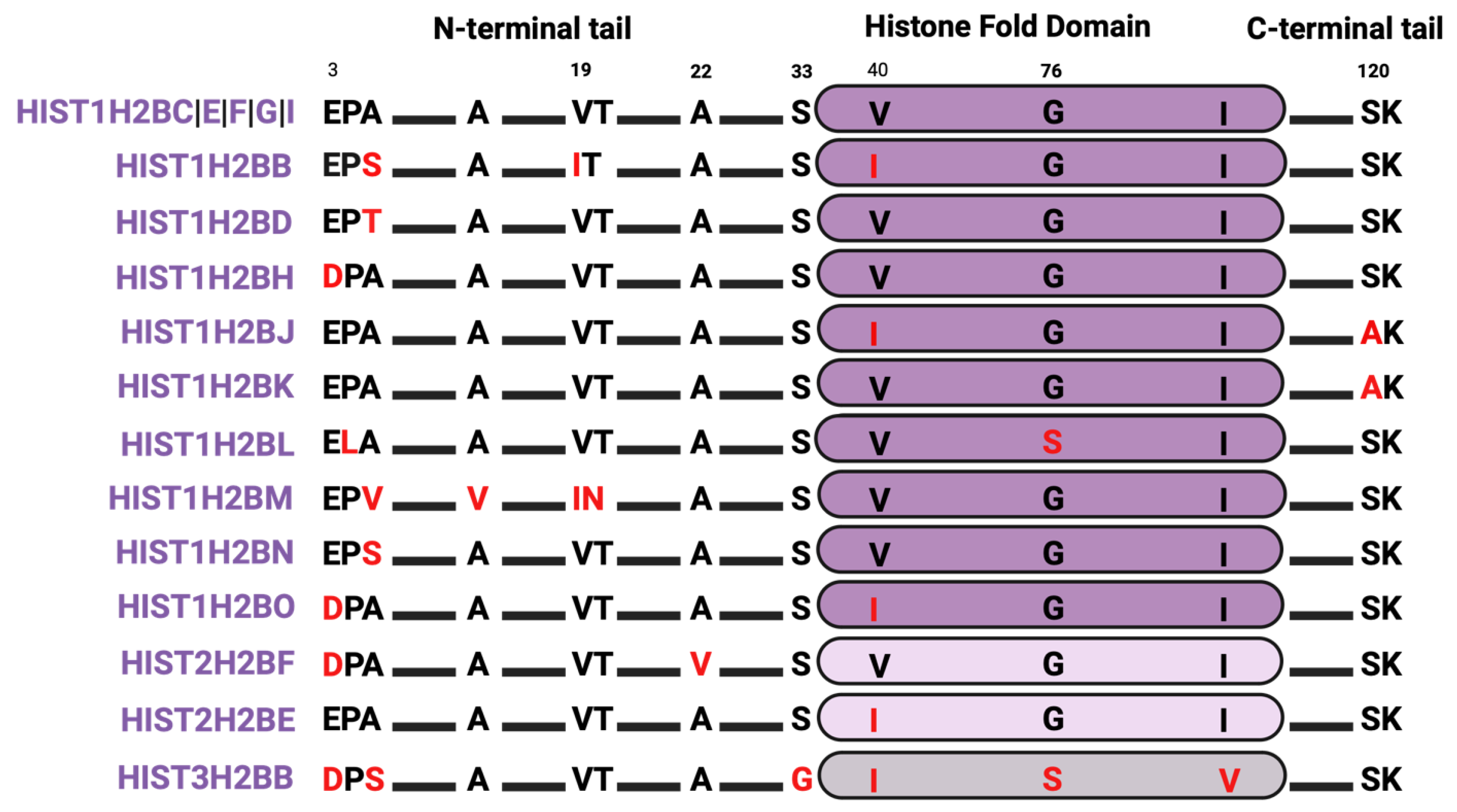
4.2. Histone H2B Isoforms
4.3. Histone H3 Isoforms
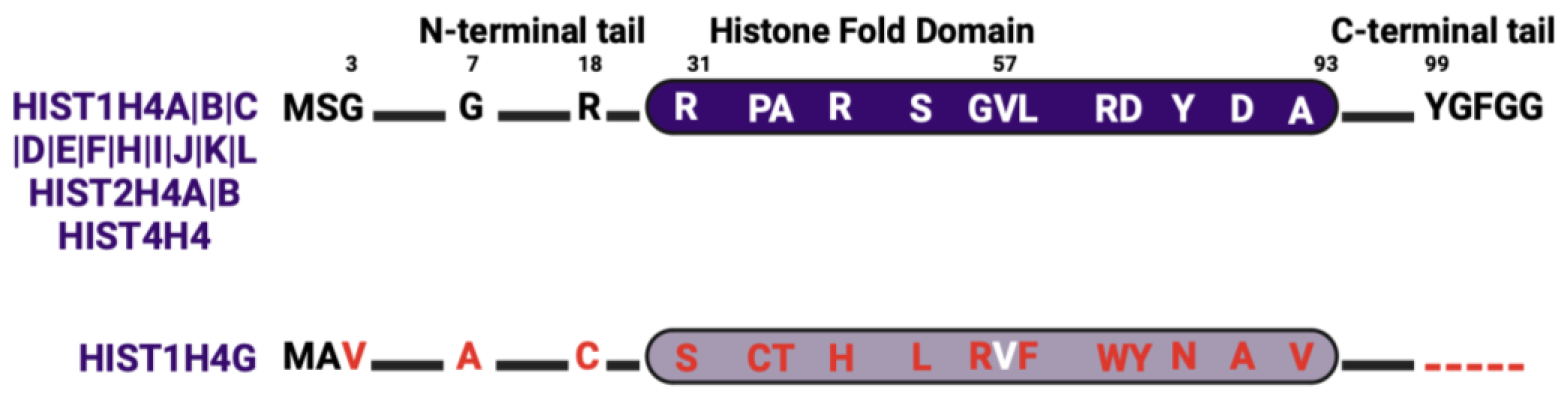
4.4. Histone H4 Isoforms
5. Dysregulated Histone Genes in Breast Cancer
5.1. Histone H2A Variants
5.2. Histone H2B Variants
5.3. Histone H3 Variants
5.4. Histone H4 Variants
6. Onco-Histones and Breast Cancer
7. Epigenetic Inhibitors and Clinical Trials
8. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Chromosome | Name | Gene Name | Protein Name |
|---|---|---|---|
| 1q21.2 | H2A clustered histone 18 | H2AC18 | HIST2H2AA |
| 1q21.2 | H2A clustered histone 19 | H2AC19 | HIST2H2AA4 |
| 1q21.2 | H2A clustered histone 20 | H2AC20 | HIST2H2AC |
| 1q21.2 | H2A clustered histone 21 | H2AC21 | HIST2H2AB |
| 1q42.13 | H2A clustered histone 25 | H2AC25 | HIST3H2A |
| 1q21.2 | H2B clustered histone 18 | H2BC18 | HIST2H2BF |
| 1q21.2 | H2B clustered histone 19, pseudogene | H2BC19P | HIST2H2BD |
| 1q21.2 | H2B clustered histone 20, pseudogene | H2BC20P | HIST2H2BC |
| 1q21.2 | H2B clustered histone 21 | H2BC21 | HIST2H2BE |
| 1q42.13 | H2B clustered histone 26 | H2BC26 | HIST3H2BB |
| 1q42.13 | H2B clustered histone 27, pseudogene | H2BC27P | HIST3H2BA |
| 1q21.1 | H3.7 histone | H3-7 | HIST2H3PS2 |
| 1q21.2 | H3 clustered histone 13 | H3C13 | HIST2H3D |
| 1q21.2 | H3 clustered histone 14 | H3C14 | HIST2H3C |
| 1q21.2 | H3 histone clustered 15 | H3C15 | HIST2H3A |
| 1q42.12 | H3.3 histone A | H3-3A | H3F3A |
| 1q42.13 | H3.4 histone, cluster member | H3-4 | HIST3H3 |
| 1q21.2 | H4 clustered histone 14 | H4C14 | HIST2H4A |
| 1q21.2 | H4 clustered histone 15 | H4C15 | HIST2H4B |
| 2p23.3 | Centromere protein A | CENPA | CENP-A |
| 3q21.3 | H1.10 linker histone | H1-10 | H1FX |
| 3q22.1 | H1.8 linker histone | H1-8 | H1FOO |
| 4q23 | H2A.Z variant histone 1 | H2AZ1 | H2AFZ |
| 5q31.1 | macroH2A.1 histone | MACROH2A1 | H2AFY |
| 5q13.2 | H2B.L histone variant 1, pseudogene | H2BL1P | H2BP4 |
| 5p15.1 | H3.Y histone 1 | H3Y1 | H3.Y |
| 5p15.1 | H3.Y histone 2 | H3Y2 | H3.X |
| 6p22.2 | H1.1 linker histone, cluster member | H1-1 | HIST1H1A |
| 6p22.2 | H1.2 linker histone, cluster member | H1-2 | HIST1H1C |
| 6p22.2 | H1.3 linker histone, cluster member | H1-3 | HIST1H1D |
| 6p22.2 | H1.4 linker histone, cluster member | H1-4 | HIST1H1E |
| 6p22.1 | H1.5 linker histone, cluster member | H1-5 | HIST1H1B |
| 6p22.2 | H1.6 linker histone, cluster member | H1-6 | HIST1H1T |
| 6p22.2 | H1.12 linker histone, cluster member, pseudogene | H1-12P | HIST1H1PS1 |
| 6p22.2 | H2A clustered histone 1 | H2AC1 | HISTH2AA |
| 6p22.2 | H2A clustered histone 2, pseudogene | H2AC2P | HIST1H2APS1 |
| 6p22.2 | H2A clustered histone 3, pseudogene | H2AC3P | HIST1H2APS2 |
| 6p22.2 | H2A clustered histone 4 | H2AC4 | HIST1H2AB |
| 6p22.2 | H2A clustered histone 5, pseudogene | H2AC5P | HIST1H2APS5 |
| 6p22.2 | H2A clustered histone 6 | H2AC6 | HIST1H2AC |
| 6p22.2 | H2A clustered histone 7 | H2AC7 | HIST1H2AD |
| 6p22.2 | H2A clustered histone 8 | H2AC8 | HIST1H2AE |
| 6p22.2 | H2A clustered histone 9, pseudogene | H2AC9P | HIST1H2APS3 |
| 6p22.2 | H2A clustered histone 10, pseudogene | H2AC10P | HIST1H2APS4 |
| 6p22.1 | H2A clustered histone 11 | H2AC11 | HIST1H2AG |
| 6p22.1 | H2A clustered histone 12 | H2AC12 | HIST1H2AH |
| 6p22.1 | H2A clustered histone 13 | H2AC13 | HIST1H2AI |
| 6p22.1 | H2A clustered histone 14 | H2AC14 | HIST1H2AJ |
| 6p22.1 | H2A clustered histone 15 | H2AC15 | HIST1H2AK |
| 6p22.1 | H2A clustered histone 16 | H2AC16 | HIST1H2AL |
| 6p22.1 | H2A clustered histone 17 | H2AC17 | HIST1H2AM |
| 6p22.2 | H2B clustered histone 1 | H2BC1 | HIST1H2BA |
| 6p22.2 | H2B clustered histone 2, pseudogene | H2BC2P | HIST1H2BPS1 |
| 6p22.2 | H2B clustered histone 3 | H2BC3 | HIST1H2BB |
| 6p22.2 | H2B clustered histone 4 | H2BC4 | HIST1H2BC |
| 6p22.2 | H2B clustered histone 5 | H2BC5 | HIST1H2BD |
| 6p22.2 | H2B clustered histone 6 | H2BC6 | HIST1H2BE |
| 6p22.2 | H2B clustered histone 7 | H2BC7 | HIST1H2BF |
| 6p22.2 | H2B clustered histone 8 | H2BC8 | HIST1H2BG |
| 6p22.2 | H2B clustered histone 9 | H2BC9 | HIST1H2BH |
| 6p22.2 | H2B clustered histone 10 | H2BC10 | HIST1H2BI |
| 6p22.1 | H2B clustered histone 11 | H2BC11 | HIST1H2BJ |
| 6p22.1 | H2B clustered histone 12 | H2BC12 | HIST1H2BK |
| 6p22.1 | H2B clustered histone 13 | H2BC13 | HIST1H2BL |
| 6p22.1 | H2B clustered histone 14 | H2BC14 | HIST1H2BM |
| 6p22.1 | H2B clustered histone 15 | H2BC15 | HIST1H2BN |
| 6p22.1 | H2B clustered histone 16, pseudogene | H2BC16P | HIST1H2BPS2 |
| 6p22.1 | H2B clustered histone 17 | H2BC17 | HIST1H2BO |
| 6p22.2 | H3 clustered histone 1 | H3C1 | HIST1H3A |
| 6p22.2 | H3 clustered histone 2 | H3C2 | HIST1H3B |
| 6p22.2 | H3 clustered histone 3 | H3C3 | HIST1H3C |
| 6p22.2 | H3 clustered histone 4 | H3C4 | HIST1H3D |
| 6p22.2 | H3 clustered histone 5, pseudogene | H3C5P | |
| 6p22.2 | H3 clustered histone 6 | H3C6 | HIST1H3E |
| 6p22.2 | H3 clustered histone 7 | H3C7 | HIST1H3F |
| 6p22.2 | H3 clustered histone 8 | H3C8 | HIST1H3G |
| 6p22.2 | H3 clustered histone 9, pseudogene | H3C9P | HIST1H3PS1 |
| 6p22.1 | H3 clustered histone 10 | H3C10 | HIST1H3H |
| 6p22.1 | H3 clustered histone 11 | H3C11 | HIST1H3I |
| 6p22.1 | H3 clustered histone 12 | H3C12 | HIST1H3J |
| 6p22.2 | H4 clustered histone 1 | H4C1 | HIST1H4A |
| 6p22.2 | H3 clustered histone 2 | H4C2 | HIST1H4B |
| 6p22.2 | H3 clustered histone 3 | H4C3 | HIST1H4C |
| 6p22.2 | H3 clustered histone 4 | H4C4 | HIST1H4D |
| 6p22.2 | H3 clustered histone 5 | H4C5 | HIST1H4E |
| 6p22.2 | H3 clustered histone 6 | H4C6 | HIST1H4F |
| 6p22.2 | H3 clustered histone 7 | H4C7 | HIST1H4G |
| 6p22.2 | H3 clustered histone 8 | H4C8 | HIST1H4H |
| 6p22.1 | H3 clustered histone 9 | H4C9 | HIST1H4I |
| 6p22.1 | H3 clustered histone 10, pseudogene | H4C10P | HIST1H4PS1 |
| 6p22.1 | H3 clustered histone 11 | H4C11 | HIST1H4J |
| 6p22.1 | H3 clustered histone 12 | H4C12 | HIST1H4K |
| 6p22.1 | H3 clustered histone 13 | H4C13 | HIST1H4L |
| 7p13 | H2A.Z variant histone 2 | H2AZ2 | H2AFV |
| 7q36.1 | H2B.K variant histone 1 | H2BK1 | H2BE1 |
| 10q22.1 | macroH2A.2 histone | MACROH2A2 | H2AFY2 |
| 11q23.3 | H2A.X variant histone | H2AX | H2AFX |
| 12q13.11 | H1.7 linker histone | H1-7 | H1FNT |
| 12p12.3 | H2A.J histone | H2AJ | H2AFJ |
| 12p11.21 | H3.5 histone | H3-5 | H3F3C |
| 12p12.3 | H4 histone 16 | H4C16 | HIST4H4 |
| 17q21.33 | H1.9 linker histone, pseudogene | H1-9P | HILS1 |
| 17q11.2 | H2B.N variant histone | H2BN1 | H2B.N |
| 17q25.1 | H3.3 histone B | H3-3B | H3F3B |
| 21q22.3 | H2B clustered histone 12 like | H2BC12L | H2BFS |
| 22q13.1 | H1.0 linker histone | H1-0 | H1FV |
| Xq28 | H2A.B variant histone 1 | H2AB1 | H2AFB1 |
| Xq28 | H2A.B variant histone 2 | H2AB2 | H2AFB2 |
| Xq28 | H2A.B variant histone 3 | H2AB3 | H2AFB3 |
| Xp21.1 | H2A.L variant histone 1M, pseudogene | H2AL1MP | H2AL1SP |
| Xp21.1 | H2A.L variant histone 1Q | H2AL1Q | H2AL1QP |
| Xp11.4 | H2A.L variant histone 3 | H2AL3 | H2AL1RP |
| Xp11.4 | H2A.P histone | H2AP | HYPM (CXorf27) |
| Xq26.3 | H2A.Q variant histone 1, pseudogene | H2AQ1P | |
| Xq22.2 | H2B.W histone 1 | H2BW1 | H2BFWT |
| Xq22.2 | H2B.W histone 2 | H2BW2 | H2BFM |
| Xq22.2 | H2B.W histone 3, pseudogene | H2BW3P | |
| Xq22.2 | H2B.W histone 4, pseudogene | H2BW4P | H2BFXP |
References
- Chioda, M.; Eskeland, R.; Thompson, E.M. Histone Gene Complement, Variant Expression, and mRNA Processing in a Urochordate Oikopleura dioica that Undergoes Extensive Polyploidization. Mol. Biol. Evol. 2002, 19, 2247–2260. [Google Scholar] [CrossRef]
- Talbert, P.B.; Ahmad, K.; Almouzni, G.; Ausió, J.; Berger, F.; Bhalla, P.L.; Bonner, W.M.; Cande, W.Z.; Chadwick, B.P.; Chan, S.W.L.; et al. A unified phylogeny-based nomenclature for histone variants. Epigenet. Chromatin 2012, 5, 7. [Google Scholar] [CrossRef]
- Grau-Bové, X.; Navarrete, C.; Chiva, C.; Pribasnig, T.; Antó, M.; Torruella, G.; Galindo, L.J.; Lang, B.F.; Moreira, D.; López-Garcia, P.; et al. A phylogenetic and proteomic reconstruction of eukaryotic chromatin evolution. Nat. Ecol. Evol. 2022, 6, 1007–1023. [Google Scholar] [CrossRef]
- Zhou, B.R.; Feng, H.; Kale, S.; Fox, T.; Khant, H.; de Val, N.; Ghirlando, R.; Panchenko, A.R.; Bai, Y. Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms. Mol. Cell 2021, 81, 166–182.e6. [Google Scholar] [CrossRef]
- Phillips, E.O.N.; Gunjan, A. Histone variants: The unsung guardians of the genome. DNA Repair. 2022, 112, 103301. [Google Scholar] [CrossRef]
- Williamson, E.A.; Wray, J.W.; Bansal, P.; Hromas, R. Overview for the histone codes for DNA repair. Prog. Mol. Biol. Transl. Sci. 2012, 110, 207–227. [Google Scholar]
- Bhattacharya, S.; Reddy, D.; Jani, V.; Gadewal, N.; Shah, S.; Reddy, R.; Bose, K.; Sonavane, U.; Joshi, R.; Smoot, D.; et al. Histone isoform H2A1H promotes attainment of distinct physiological states by altering chromatin dynamics. Epigenet. Chromatin 2017, 10, 48. [Google Scholar] [CrossRef]
- Singh, R.; Mortazavi, A.; Telu, K.H.; Nagarajan, P.; Lucas, D.M.; Thomas-Ahner, J.M.; Clinton, S.K.; Byrd, J.C.; Freitas, M.A.; Parthun, M.R. Increasing the complexity of chromatin: Functionally distinct roles for replication-dependent histone H2A isoforms in cell proliferation and carcinogenesis. Nucleic Acids Res. 2013, 41, 9284–9295. [Google Scholar] [CrossRef]
- Henikoff, S.; Smith, M.M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 2015, 7, a019364. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Ramachandran, S.; Henikoff, S. Replicating Nucleosomes. Sci. Adv. 2015, 1, e1500587. [Google Scholar] [CrossRef]
- Zhang, W.; Feng, J.; Li, Q. The replisome guides nucleosome assembly during DNA replication. Cell Biosci. 2020, 10, 37. [Google Scholar] [CrossRef]
- Korolev, N.; Vorontsova, O.V.; Nordenskiöld, L. Physicochemical analysis of electrostatic foundation for DNA–protein interactions in chromatin transformations. Progress. Biophys. Mol. Biol. 2007, 95, 23–49. [Google Scholar] [CrossRef]
- Rudnizky, S.; Khamis, H.; Ginosar, Y.; Goren, E.; Melamed, P.; Kaplan, A. Extended and dynamic linker histone-DNA Interactions control chromatosome compaction. Mol. Cell 2021, 81, 3410–3421.e4. [Google Scholar] [CrossRef]
- Burgess, R.J.; Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 2013, 20, 14–22. [Google Scholar] [CrossRef]
- He, H.; Lee, M.-C.; Zheng, L.-L.; Zheng, L.; Luo, Y. Integration of the metabolic/redox state, histone gene switching, DNA replication and S-phase progression by moonlighting metabolic enzymes. Biosci. Rep. 2013, 33, e00018. [Google Scholar] [CrossRef]
- Paik, J.; Giovinazzi, S.; Gunjan, A. Coordination of DNA Replication and Histone Synthesis during S Phase. In The Initiation of DNA Replication in Eukaryotes; Kaplan, D.L., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 213–237. [Google Scholar]
- Mei, Q.; Huang, J.; Chen, W.; Tang, J.; Xu, C.; Yu, Q.; Cheng, Y.; Ma, L.; Yu, X.; Li, S. Regulation of DNA replication-coupled histone gene expression. Oncotarget 2017, 8, 95005. [Google Scholar] [CrossRef]
- Nelson, D.M.; Ye, X.; Hall, C.; Santos, H.; Ma, T.; Kao, G.D.; Yen, T.J.; Harper, J.W.; Adams, P.D. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell Biol. 2002, 22, 7459–7472. [Google Scholar] [CrossRef]
- Marzluff, W.F.; Wagner, E.J.; Duronio, R.J. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 2008, 9, 843–854. [Google Scholar] [CrossRef]
- Duronio, R.J.; Marzluff, W.F. Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol. 2017, 14, 726–738. [Google Scholar] [CrossRef]
- Kemp, J.P., Jr.; Yang, X.C.; Dominski, Z.; Marzluff, W.F.; Duronio, R.J. Superresolution light microscopy of the Drosophila histone locus body reveals a core-shell organization associated with expression of replication-dependent histone genes. Mol. Biol. Cell 2021, 32, 942–955. [Google Scholar] [CrossRef]
- Romeo, V.; Schümperli, D. Cycling in the nucleus: Regulation of RNA 3′ processing and nuclear organization of replication-dependent histone genes. Curr. Opin. Cell Biol. 2016, 40, 23–31. [Google Scholar] [CrossRef]
- Koreski, K.P.; Rieder, L.E.; McLain, L.M.; Chaubal, A.; Marzluff, W.F.; Duronio, R.J. Drosophila histone locus body assembly and function involves multiple interactions. Mol. Biol. Cell 2020, 31, 1525–1537. [Google Scholar] [CrossRef]
- Zhang, J.; Tan, D.; DeRose, E.F.; Perera, L.; Dominski, Z.; Marzluff, W.F.; Tong, L.; Hall, T.M. Molecular mechanisms for the regulation of histone mRNA stem-loop-binding protein by phosphorylation. Proc. Natl. Acad. Sci. USA 2014, 111, E2937–E2946. [Google Scholar] [CrossRef]
- Wang, Z.F.; Whitfield, M.L.; Ingledue, T.C., 3rd; Dominski, Z.; Marzluff, W.F. The protein that binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes. Dev. 1996, 10, 3028–3040. [Google Scholar] [CrossRef]
- Allard, P.; Yang, Q.; Marzluff, W.F.; Clarke, H.J. The stem-loop binding protein regulates translation of histone mRNA during mammalian oogenesis. Dev. Biol. 2005, 286, 195–206. [Google Scholar] [CrossRef]
- Sullivan, E.; Santiago, C.; Parker, E.D.; Dominski, Z.; Yang, X.; Lanzotti, D.J.; Ingledue, T.C.; Marzluff, W.F.; Duronio, R.J. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes. Dev. 2001, 15, 173–187. [Google Scholar] [CrossRef]
- Lanzotti, D.J.; Kupsco, J.M.; Yang, X.C.; Dominski, Z.; Marzluff, W.F.; Duronio, R.J. Drosophila stem-loop binding protein intracellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Mol. Biol. Cell 2004, 15, 1112–1123. [Google Scholar] [CrossRef]
- Terme, J.-M.; Sesé, B.; Millán-Ariño, L.; Mayor, R.; Belmonte, J.C.I.; Barrero, M.J.; Jordan, A. Histone H1 Variants Are Differentially Expressed and Incorporated into Chromatin during Differentiation and Reprogramming to Pluripotency*. J. Biol. Chem. 2011, 286, 35347–35357. [Google Scholar] [CrossRef]
- Dzhondzhurov, L.; Yancheva, N.; Ivanova, E. Histones of terminally differentiated cells undergo continuous turnover. Biochemistry 1983, 22, 4095–4102. [Google Scholar] [CrossRef]
- Armstrong, C.; Spencer, S.L. Replication-dependent histone biosynthesis is coupled to cell-cycle commitment. Proc. Natl. Acad. Sci. USA 2021, 118, e2100178118. [Google Scholar] [CrossRef]
- Amatori, S.; Tavolaro, S.; Gambardella, S.; Fanelli, M. The dark side of histones: Genomic organization and role of oncohistones in cancer. Clin. Epigenet. 2021, 13, 71. [Google Scholar] [CrossRef]
- Kurumizaka, H.; Kujirai, T.; Takizawa, Y. Contributions of Histone Variants in Nucleosome Structure and Function. J. Mol. Biol. 2021, 433, 166678. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants at a glance. J. Cell Sci. 2021, 134, jcs244749. [Google Scholar] [CrossRef]
- Ferrand, J.; Rondinelli, B.; Polo, S.E. Histone Variants: Guardians of Genome Integrity. Cells 2020, 9, 2424. [Google Scholar] [CrossRef]
- Alonso-Curbelo, D.; Ho, Y.J.; Burdziak, C.; Maag, J.L.V.; Morris, J.P.t.; Chandwani, R.; Chen, H.A.; Tsanov, K.M.; Barriga, F.M.; Luan, W.; et al. A gene-environment-induced epigenetic program initiates tumorigenesis. Nature 2021, 590, 642–648. [Google Scholar] [CrossRef]
- Martire, S.; Banaszynski, L.A. The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell Biol. 2020, 21, 522–541. [Google Scholar] [CrossRef]
- Ballouz, S.; Pena, M.T.; Knight, F.M.; Adams, L.B.; Gillis, J.A. The transcriptional legacy of developmental stochasticity. bioRxiv 2019. [Google Scholar] [CrossRef]
- Hsu, C.J.; Meers, O.; Buschbeck, M.; Heidel, F.H. The Role of MacroH2A Histone Variants in Cancer. Cancers 2021, 13, 3003. [Google Scholar] [CrossRef]
- Kirkiz, E.; Meers, O.; Grebien, F.; Buschbeck, M. Histone Variants and Their Chaperones in Hematological Malignancies. HemaSphere 2023, 7, e927. [Google Scholar] [CrossRef]
- Johal, K.S.; Cheema, M.S.; Stefanelli, G. Histone Variants and Their Chaperones: An Emerging Epigenetic Mechanism in Neurodevelopment and Neurodevelopmental Disorders. JIN 2023, 22, 108. [Google Scholar] [CrossRef] [PubMed]
- Siddaway, R.; Milos, S.; Coyaud, É.; Yun, H.Y.; Morcos, S.M.; Pajovic, S.; Campos, E.I.; Raught, B.; Hawkins, C. The in vivo Interaction Landscape of Histones H3.1 and H3.3. Mol. Cell Proteom. 2022, 21, 100411. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhao, J.; Wang, Y.; Wang, M.; Long, H.; Liang, D.; Huang, L.; Wen, Z.; Li, W.; Li, X.; et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes. Dev. 2013, 27, 2109–2124. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes. Dev. 2007, 21, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Tvardovskiy, A.; Schwämmle, V.; Kempf, S.J.; Rogowska-Wrzesinska, A.; Jensen, O.N. Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res. 2017, 45, 9272–9289. [Google Scholar] [CrossRef]
- Loyola, A.; Almouzni, G. Marking histone H3 variants: How, when and why? Trends Biochem. Sci. 2007, 32, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Loyola, A.; Bonaldi, T.; Roche, D.; Imhof, A.; Almouzni, G. PTMs on H3 Variants before Chromatin Assembly Potentiate Their Final Epigenetic State. Mol. Cell 2006, 24, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wei, T.; Panchenko, A.R. Histone variant H2A.Z modulates nucleosome dynamics to promote DNA accessibility. Nat. Commun. 2023, 14, 769. [Google Scholar] [CrossRef]
- Rangasamy, D. Histone variant H2A.Z can serve as a new target for breast cancer therapy. Curr. Med. Chem. 2010, 17, 3155–3161. [Google Scholar] [CrossRef]
- Buschbeck, M.; Hake, S.B. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 2017, 18, 299–314. [Google Scholar] [CrossRef]
- Xia, W.; Jiao, J. Histone variant H3.3 orchestrates neural stem cell differentiation in the developing brain. Cell Death Differ. 2017, 24, 1548–1563. [Google Scholar] [CrossRef]
- Maze, I.; Noh, K.-M.; Soshnev, A.A.; Allis, C.D. Every amino acid matters: Essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 2014, 15, 259–271. [Google Scholar] [CrossRef]
- Gomes, A.P.; Ilter, D.; Low, V.; Rosenzweig, A.; Shen, Z.J.; Schild, T.; Rivas, M.A.; Er, E.E.; McNally, D.R.; Mutvei, A.P.; et al. Dynamic Incorporation of Histone H3 Variants into Chromatin Is Essential for Acquisition of Aggressive Traits and Metastatic Colonization. Cancer Cell 2019, 36, 402–417.e13. [Google Scholar] [CrossRef]
- Taguchi, H.; Xie, Y.; Horikoshi, N.; Maehara, K.; Harada, A.; Nogami, J.; Sato, K.; Arimura, Y.; Osakabe, A.; Kujirai, T.; et al. Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8. Biochemistry 2017, 56, 2184–2196. [Google Scholar] [CrossRef]
- Susano Pinto, D.M.; Flaus, A. The human canonical core histone catalogue. bioRxiv 2019. [Google Scholar] [CrossRef]
- Kurat, C.F.; Recht, J.; Radovani, E.; Durbic, T.; Andrews, B.; Fillingham, J. Regulation of histone gene transcription in yeast. Cell Mol. Life Sci. 2014, 71, 599–613. [Google Scholar] [CrossRef]
- Mariño-Ramírez, L.; Jordan, I.K.; Landsman, D. Multiple independent evolutionary solutions to core histone gene regulation. Genome Biol. 2006, 7, R122. [Google Scholar] [CrossRef]
- Singh, R.; Bassett, E.; Chakravarti, A.; Parthun, M.R. Replication-dependent histone isoforms: A new source of complexity in chromatin structure and function. Nucleic Acids Res. 2018, 46, 8665–8678. [Google Scholar] [CrossRef]
- Molden, R.C.; Bhanu, N.V.; LeRoy, G.; Arnaudo, A.M.; Garcia, B.A. Multi-faceted quantitative proteomics analysis of histone H2B isoforms and their modifications. Epigenet. Chromatin 2015, 8, 15. [Google Scholar] [CrossRef]
- Shah, S.; Verma, T.; Rashid, M.; Gadewal, N.; Gupta, S. Histone H2A isoforms: Potential implications in epigenome plasticity and diseases in eukaryotes. J. Biosci. 2020, 45, 4. [Google Scholar] [CrossRef]
- Heintz, N.; Zernik, M.; Roeder, R.G. The structure of the human histone genes: Clustered but not tandemly repeated. Cell 1981, 24, 661–668. [Google Scholar] [CrossRef]
- Marzluff, W.F.; Gongidi, P.; Woods, K.R.; Jin, J.; Maltais, L.J. The human and mouse replication-dependent histone genes. Genomics 2002, 80, 487–498. [Google Scholar] [CrossRef]
- Luense, L.J.; Wang, X.; Schon, S.B.; Weller, A.H.; Lin Shiao, E.; Bryant, J.M.; Bartolomei, M.S.; Coutifaris, C.; Garcia, B.A.; Berger, S.L. Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenet. Chromatin 2016, 9, 24. [Google Scholar] [CrossRef]
- Lyons, S.M.; Cunningham, C.H.; Welch, J.D.; Groh, B.; Guo, A.Y.; Wei, B.; Whitfield, M.L.; Xiong, Y.; Marzluff, W.F. A subset of replication-dependent histone mRNAs are expressed as polyadenylated RNAs in terminally differentiated tissues. Nucleic Acids Res. 2016, 44, 9190–9205. [Google Scholar] [CrossRef]
- Bonner, W.M.; Mannironi, C.; Orr, A.; Pilch, D.R.; Hatch, C.L. Histone H2A.X gene transcription is regulated differently than transcription of other replication-linked histone genes. Mol. Cell Biol. 1993, 13, 984–992. [Google Scholar]
- Mannironi, C.; Bonner, W.M.; Hatch, C.L. H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3′ processing signals. Nucleic Acids Res. 1989, 17, 9113–9126. [Google Scholar] [CrossRef]
- Marzluff, W.F.; Koreski, K.P. Birth and Death of Histone mRNAs. Trends Genet. 2017, 33, 745–759. [Google Scholar] [CrossRef]
- Panyim, S.; Chalkley, R. A new histone found only in mammalian tissues with little cell division. Biochem. Biophys. Res. Commun. 1969, 37, 1042–1049. [Google Scholar] [CrossRef]
- Kari, V.; Karpiuk, O.; Tieg, B.; Kriegs, M.; Dikomey, E.; Krebber, H.; Begus-Nahrmann, Y.; Johnsen, S.A. A subset of histone H2B genes produces polyadenylated mRNAs under a variety of cellular conditions. PLoS ONE 2013, 8, e63745. [Google Scholar] [CrossRef]
- Sullivan, K.D.; Mullen, T.E.; Marzluff, W.F.; Wagner, E.J. Knockdown of SLBP results in nuclear retention of histone mRNA. RNA 2009, 15, 459–472. [Google Scholar] [CrossRef]
- Monteiro, F.L.; Vitorino, R.; Wang, J.; Cardoso, H.; Laranjeira, H.; Simões, J.; Caldas, M.; Henrique, R.; Amado, F.; Williams, C.; et al. The histone H2A isoform Hist2h2ac is a novel regulator of proliferation and epithelial–mesenchymal transition in mammary epithelial and in breast cancer cells. Cancer Lett. 2017, 396, 42–52. [Google Scholar] [CrossRef]
- Tyagi, M.; Khade, B.; Khan, S.A.; Ingle, A.; Gupta, S. Expression of histone variant, H2A.1 is associated with the undifferentiated state of hepatocyte. Exp. Biol. Med. 2014, 239, 1335–1339. [Google Scholar] [CrossRef]
- Singh, R.; Harshman, S.W.; Ruppert, A.S.; Mortazavi, A.; Lucas, D.M.; Thomas-Ahner, J.M.; Clinton, S.K.; Byrd, J.C.; Freitas, M.A.; Parthun, M.R. Proteomic profiling identifies specific histone species associated with leukemic and cancer cells. Clin. Proteom. 2015, 12, 22. [Google Scholar] [CrossRef]
- Shah, S.G.; Rashid, M.; Natu, A.; Gupta, S. Differential expression of H2A isoforms contribute to tissue and lineage specificity with HIST2H2AC as a potential cancer biomarker. bioRxiv 2021. [Google Scholar] [CrossRef]
- Bowerman, S.; Wereszczynski, J. Effects of MacroH2A and H2A.Z on Nucleosome Dynamics as Elucidated by Molecular Dynamics Simulations. Biophys. J. 2016, 110, 327–337. [Google Scholar] [CrossRef]
- Kozlowski, M.; Corujo, D.; Hothorn, M.; Guberovic, I.; Mandemaker, I.K.; Blessing, C.; Sporn, J.; Gutierrez-Triana, A.; Smith, R.; Portmann, T.; et al. MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms. EMBO Rep. 2018, 19, e44445. [Google Scholar] [CrossRef]
- Pinto, D.M.; Flaus, A. Structure and function of histone H2AX. Subcell. Biochem. 2010, 50, 55–78. [Google Scholar]
- Foster, E.R.; Downs, J.A. Histone H2A phosphorylation in DNA double-strand break repair. FEBS J. 2005, 272, 3231–3240. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, C.K.; Lee, S.B.; Lee, K.-H.; Cho, S.-W.; Ahn, J.-Y. Akt attenuates apoptotic death through phosphorylation of H2A under hydrogen peroxide-induced oxidative stress in PC12 cells and hippocampal neurons. Sci. Rep. 2016, 6, 21857. [Google Scholar] [CrossRef]
- Abuetabh, Y.; Wu, H.H.; Chai, C.; Al Yousef, H.; Persad, S.; Sergi, C.M.; Leng, R. DNA damage response revisited: The p53 family and its regulators provide endless cancer therapy opportunities. Exp. Mol. Med. 2022, 54, 1658–1669. [Google Scholar] [CrossRef]
- Walser, F.; Mulder, M.P.C.; Bragantini, B.; Burger, S.; Gubser, T.; Gatti, M.; Botuyan, M.V.; Villa, A.; Altmeyer, M.; Neri, D.; et al. Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response. Mol. Cell 2020, 80, 423–436.e9. [Google Scholar] [CrossRef]
- Banerjee, T.; Chakravarti, D. A peek into the complex realm of histone phosphorylation. Mol. Cell Biol. 2011, 31, 4858–4873. [Google Scholar] [CrossRef]
- Chen, C.; Ha, B.H.; Thévenin, A.F.; Lou, H.J.; Zhang, R.; Yip, K.Y.; Peterson, J.R.; Gerstein, M.; Kim, P.M.; Filippakopoulos, P.; et al. Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity. Mol. Cell 2014, 53, 140–147. [Google Scholar] [CrossRef]
- Bönisch, C.; Hake, S.B. Histone H2A variants in nucleosomes and chromatin: More or less stable? Nucleic Acids Res. 2012, 40, 10719–10741. [Google Scholar] [CrossRef]
- Li, S.; Peng, Y.; Landsman, D.; Panchenko, A.R. DNA methylation cues in nucleosome geometry, stability and unwrapping. Nucleic Acids Res. 2022, 50, 1864–1874. [Google Scholar] [CrossRef] [PubMed]
- Huertas, J.; Schöler, H.R.; Cojocaru, V. Histone tails cooperate to control the breathing of genomic nucleosomes. PLoS Comput. Biol. 2021, 17, e1009013. [Google Scholar] [CrossRef]
- Chakraborty, K.; Loverde, S.M. Asymmetric breathing motions of nucleosomal DNA and the role of histone tails. J. Chem. Phys. 2017, 147, 065101. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Sakuraba, S.; Ishida, H. Free energy profiles for unwrapping the outer superhelical turn of nucleosomal DNA. PLoS Comput. Biol. 2018, 14, e1006024. [Google Scholar] [CrossRef]
- Zhang, K.; Tang, H. Analysis of core histones by liquid chromatography–mass spectrometry and peptide mapping. J. Chromatogr. B 2003, 783, 173–179. [Google Scholar] [CrossRef]
- Tessarz, P.; Santos-Rosa, H.; Robson, S.C.; Sylvestersen, K.B.; Nelson, C.J.; Nielsen, M.L.; Kouzarides, T. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 2014, 505, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Mobley, R.J.; Abell, A.N. Controlling Epithelial to Mesenchymal Transition through Acetylation of Histone H2BK5. J. Nat. Sci. 2017, 3, e432. [Google Scholar]
- Huang, D.; Camacho, C.V.; Setlem, R.; Ryu, K.W.; Parameswaran, B.; Gupta, R.K.; Kraus, W.L. Functional Interplay between Histone H2B ADP-Ribosylation and Phosphorylation Controls Adipogenesis. Mol. Cell 2020, 79, 934–949.e14. [Google Scholar] [CrossRef] [PubMed]
- Bungard, D.; Fuerth, B.J.; Zeng, P.-Y.; Faubert, B.; Maas, N.L.; Viollet, B.; Carling, D.; Thompson, C.B.; Jones, R.G.; Berger, S.L. Signaling Kinase AMPK Activates Stress-Promoted Transcription via Histone H2B Phosphorylation. Science 2010, 329, 1201–1205. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.A.; Um, S.H.; Lee, J.; Yoo, J.H.; Bang, S.Y.; Park, E.K.; Lee, M.G.; Nam, K.H.; Jeon, Y.J.; Park, J.W.; et al. S6K1 Phosphorylation of H2B Mediates EZH2 Trimethylation of H3: A Determinant of Early Adipogenesis. Mol. Cell 2016, 62, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Tachiwana, H.; Osakabe, A.; Shiga, T.; Miya, Y.; Kimura, H.; Kagawa, W.; Kurumizaka, H. Structures of human nucleosomes containing major histone H3 variants. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Hake, S.B.; Allis, C.D. Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis”. Proc. Natl. Acad. Sci. USA 2006, 103, 6428–6435. [Google Scholar] [CrossRef] [PubMed]
- Hake, S.B.; Garcia, B.A.; Duncan, E.M.; Kauer, M.; Dellaire, G.; Shabanowitz, J.; Bazett-Jones, D.P.; Allis, C.D.; Hunt, D.F. Expression Patterns and Post-translational Modifications Associated with Mammalian Histone H3 Variants*. J. Biol. Chem. 2006, 281, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Tachiwana, H.; Kagawa, W.; Osakabe, A.; Kawaguchi, K.; Shiga, T.; Hayashi-Takanaka, Y.; Kimura, H.; Kurumizaka, H. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc. Natl. Acad. Sci. USA 2010, 107, 10454–10459. [Google Scholar] [CrossRef] [PubMed]
- Espiritu, D.; Gribkova, A.K.; Gupta, S.; Shaytan, A.K.; Panchenko, A.R. Molecular Mechanisms of Oncogenesis through the Lens of Nucleosomes and Histones. J. Phys. Chem. B 2021, 125, 3963–3976. [Google Scholar] [CrossRef]
- Dorigo, B.; Schalch, T.; Bystricky, K.; Richmond, T.J. Chromatin fiber folding: Requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003, 327, 85–96. [Google Scholar] [CrossRef]
- Ishida, H.; Kono, H. H4 Tails Potentially Produce the Diversity in the Orientation of Two Nucleosomes. Biophys. J. 2017, 113, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Kan, P.Y.; Caterino, T.L.; Hayes, J.J. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol. Cell Biol. 2009, 29, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Bendandi, A.; Patelli, A.S.; Diaspro, A.; Rocchia, W. The role of histone tails in nucleosome stability: An electrostatic perspective. Comput. Struct. Biotechnol. J. 2020, 18, 2799–2809. [Google Scholar] [CrossRef] [PubMed]
- Chakkalakal, S.A.; Heilig, J.; Baumann, U.; Paulsson, M.; Zaucke, F. Impact of Arginine to Cysteine Mutations in Collagen II on Protein Secretion and Cell Survival. Int. J. Mol. Sci. 2018, 19, 541. [Google Scholar] [CrossRef] [PubMed]
- Flaus, A.; Rencurel, C.; Ferreira, H.; Wiechens, N.; Owen-Hughes, T. Sin mutations alter inherent nucleosome mobility. EMBO J. 2004, 23, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Chavez, M.S.; Scorgie, J.K.; Dennehey, B.K.; Noone, S.; Tyler, J.K.; Churchill, M.E. The conformational flexibility of the C-terminus of histone H4 promotes histone octamer and nucleosome stability and yeast viability. Epigenet. Chromatin 2012, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, E.; Chmúrčiaková, N.; Cmarko, D. Human rDNA and Cancer. Cells 2021, 10, 3452. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Sun, X.; Shi, W.; Yanru, A.; Leung, S.T.C.; Ding, D.; Cheema, M.S.; MacPherson, N.; Nelson, C.J.; Ausio, J.; et al. A novel histone H4 variant H4G regulates rDNA transcription in breast cancer. Nucleic Acids Res. 2019, 47, 8399–8409. [Google Scholar] [CrossRef] [PubMed]
- Su, C.H.; Tzeng, T.Y.; Cheng, C.; Hsu, M.T. An H2A histone isotype regulates estrogen receptor target genes by mediating enhancer-promoter-3′-UTR interactions in breast cancer cells. Nucleic Acids Res. 2014, 42, 3073–3088. [Google Scholar] [CrossRef]
- Toda, H.; Seki, N.; Kurozumi, S.; Shinden, Y.; Yamada, Y.; Nohata, N.; Moriya, S.; Idichi, T.; Maemura, K.; Fujii, T.; et al. RNA-sequence-based microRNA expression signature in breast cancer: Tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol. Oncol. 2020, 14, 426–446. [Google Scholar] [CrossRef]
- Mamoor, S. Differential Expression of Histone Cluster 1, H2ag in Triple Negative Breast Cancer. 2021. Available online: https://osf.io/t8swk (accessed on 5 May 2024).
- Komatsu, M.; Yoshimaru, T.; Matsuo, T.; Kiyotani, K.; Miyoshi, Y.; Tanahashi, T.; Rokutan, K.; Yamaguchi, R.; Saito, A.; Imoto, S.; et al. Molecular features of triple negative breast cancer cells by genome-wide gene expression profiling analysis. Int. J. Oncol. 2013, 42, 478–506. [Google Scholar] [CrossRef]
- Stander, B.A.; Marais, S.; Vorster, C.J.; Joubert, A.M. In vitro effects of 2-methoxyestradiol on morphology, cell cycle progression, cell death and gene expression changes in the tumorigenic MCF-7 breast epithelial cell line. J. Steroid Biochem. Mol. Biol. 2010, 119, 149–160. [Google Scholar] [CrossRef]
- Malvia, S.; Bagadi, S.A.R.; Pradhan, D.; Chintamani, C.; Bhatnagar, A.; Arora, D.; Sarin, R.; Saxena, S. Study of Gene Expression Profiles of Breast Cancers in Indian Women. Sci. Rep. 2019, 9, 10018. [Google Scholar] [CrossRef]
- Mamoor, S. Differential Expression of Histone Cluster 3, H2a in Triple Negative Breast Cancer. 2021. Available online: https://osf.io/89ceb (accessed on 6 May 2024).
- Mamoor, S. HIST3H2A is Differentially Expressed in the Brain Metastases of Patients with Metastatic Breast Cancer. 2020. Available online: https://osf.io/nb4q9 (accessed on 6 May 2024).
- Nayak, S.R.; Harrington, E.; Boone, D.; Hartmaier, R.; Chen, J.; Pathiraja, T.N.; Cooper, K.L.; Fine, J.L.; Sanfilippo, J.; Davidson, N.E.; et al. A Role for Histone H2B Variants in Endocrine-Resistant Breast Cancer. Horm. Cancer 2015, 6, 214–224. [Google Scholar] [CrossRef]
- Manna, P.R.; Molehin, D.; Ahmed, A.U. Chapter Eleven—Dysregulation of Aromatase in Breast, Endometrial, and Ovarian Cancers: An Overview of Therapeutic Strategies. In Progress in Molecular Biology and Translational Science; Pruitt, K., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 144, pp. 487–537. [Google Scholar]
- Liu, Y.-R.; Jiang, Y.-Z.; Xu, X.-E.; Hu, X.; Yu, K.-D.; Shao, Z.-M. Comprehensive Transcriptome Profiling Reveals Multigene Signatures in Triple-Negative Breast Cancer. Clin. Cancer Res. 2016, 22, 1653–1662. [Google Scholar] [CrossRef]
- Di Benedetto, M.; Toullec, A.; Buteau-Lozano, H.; Abdelkarim, M.; Vacher, S.; Velasco, G.; Christofari, M.; Pocard, M.; Bieche, I.; Perrot-Applanat, M. MDA-MB-231 breast cancer cells overexpressing single VEGF isoforms display distinct colonisation characteristics. Br. J. Cancer 2015, 113, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Mamoor, S. Differential Expression of Histone Cluster1, H2bo in Triple Negatibe Breast Cancer. 2021. Available online: https://osf.io/aphrb (accessed on 6 May 2024).
- He, Y.; Cao, Y.; Wang, X.; Jisiguleng, W.; Tao, M.; Liu, J.; Wang, F.; Chao, L.; Wang, W.; Li, P.; et al. Identification of Hub Genes to Regulate Breast Cancer Spinal Metastases by Bioinformatics Analyses. Comput. Math. Methods Med. 2021, 2021, 5548918. [Google Scholar] [CrossRef] [PubMed]
- Mamoor, S. HIST1H2BO Is Differentially Expressed in the Brain Metastases of Patients with Metastatic Breast Cancer. 2020. Available online: https://osf.io/h8uaq (accessed on 8 May 2024).
- Xie, W.; Zhang, J.; Zhong, P.; Qin, S.; Zhang, H.; Fan, X.; Yin, Y.; Liang, R.; Han, Y.; Liao, Y.; et al. Expression and potential prognostic value of histone family gene signature in breast cancer. Exp. Ther. Med. 2019, 18, 4893–4903. [Google Scholar] [CrossRef] [PubMed]
- Weber, L.; Maßberg, D.; Becker, C.; Altmüller, J.; Ubrig, B.; Bonatz, G.; Wölk, G.; Philippou, S.; Tannapfel, A.; Hatt, H.; et al. Olfactory Receptors as Biomarkers in Human Breast Carcinoma Tissues. Front. Oncol. 2018, 8, 33. [Google Scholar] [CrossRef]
- Fehniger, T.A.; Krysiak, K.; White, B.S.; Matlock, M.; Miller, C.; Fulton, R.; Kreisel, F.; Fronick, C.; Cook, L.; Veizer, J.; et al. Recurrent Somatic Genomic Alterations in Follicular NHL (FL) Revealed by Exome and Custom-Capture Next Generation Sequencing. Blood 2015, 126, 574. [Google Scholar] [CrossRef]
- Han, D.Y.; Fu, D.; Xi, H.; Li, Q.Y.; Feng, L.J.; Zhang, W.; Ji, G.; Xiao, J.C.; Wei, Q. Genomic expression profiling and bioinformatics analysis of pancreatic cancer. Mol. Med. Rep. 2015, 12, 4133–4140. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tian, R.; Gao, H.; Yang, Y.; Williams, B.R.G.; Gantier, M.P.; McMillan, N.A.J.; Xu, D.; Hu, Y.; Gao, Y. Identification of a histone family gene signature for predicting the prognosis of cervical cancer patients. Sci. Rep. 2017, 7, 16495. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.K.; Lehrer, J.; Alshalalfa, M.; Erho, N.; Davicioni, E.; Lotan, T.L. Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer 2017, 17, 759. [Google Scholar] [CrossRef]
- Zhong, L.K.; Gan, X.X.; Deng, X.Y.; Shen, F.; Feng, J.H.; Cai, W.S.; Liu, Q.Y.; Miao, J.H.; Zheng, B.X.; Xu, B. Potential five-mRNA signature model for the prediction of prognosis in patients with papillary thyroid carcinoma. Oncol. Lett. 2020, 20, 2302–2310. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Wang, X.; Yin, Y.; Zhang, Y.; Jia, J.; Lu, B.; Xue, W.; Qu, C.; Qi, J. Identification of a potentially functional circRNA-miRNA-mRNA ceRNA regulatory network in bladder cancer by analysis of microarray data. Transl. Androl. Urol. 2020, 10, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Pärssinen, J.; Alarmo, E.L.; Khan, S.; Karhu, R.; Vihinen, M.; Kallioniemi, A. Identification of differentially expressed genes after PPM1D silencing in breast cancer. Cancer Lett. 2008, 259, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Salhia, B.; Kiefer, J.; Ross, J.T.; Metapally, R.; Martinez, R.A.; Johnson, K.N.; DiPerna, D.M.; Paquette, K.M.; Jung, S.; Nasser, S.; et al. Integrated genomic and epigenomic analysis of breast cancer brain metastasis. PLoS ONE 2014, 9, e85448. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Li, W.; Wang, L.; Hu, J.; Song, Y.; Zhang, B.; Ren, X.; Ji, S.; Li, J.; Xu, P.; et al. Histone-Related Genes Are Hypermethylated in Lung Cancer and Hypermethylated HIST1H4F Could Serve as a Pan-Cancer Biomarker. Cancer Res. 2019, 79, 6101–6112. [Google Scholar] [CrossRef] [PubMed]
- Mamoor, S. HIST1H2BB Is a Differentially Expressed Gene in Human Metastatic Breast Cancer, in the Brain and in the Lymph Nodes. 2021. Available online: https://osf.io/cftxw (accessed on 8 May 2024).
- Zhang, L.; Zhang, W.; Sun, J.; Liu, K.N.; Gan, Z.X.; Liu, Y.Z.; Chang, J.F.; Yang, X.M.; Sun, F. Nucleotide variation in histone H2BL drives crossalk of histone modification and promotes tumour cell proliferation by upregulating c-Myc. Life Sci. 2021, 271, 119127. [Google Scholar] [CrossRef]
- Thakar, A.; Gupta, P.; Ishibashi, T.; Finn, R.; Silva-Moreno, B.; Uchiyama, S.; Fukui, K.; Tomschik, M.; Ausio, J.; Zlatanova, J. H2A.Z and H3.3 histone variants affect nucleosome structure: Biochemical and biophysical studies. Biochemistry 2009, 48, 10852–10857. [Google Scholar] [CrossRef]
- Yi Leong, H.J. Identification of Potentially Therapeutic Target Genes in Metastatic Breast Cancer via Integrative Network Analysis. Eurasian J. Med. Oncol. 2023, 7, 371–387. [Google Scholar] [CrossRef]
- Delaney, K.; Mailler, J.; Wenda, J.M.; Gabus, C.; Steiner, F.A. Differential Expression of Histone H3.3 Genes and Their Role in Modulating Temperature Stress Response in Caenorhabditis elegans. Genetics 2018, 209, 551–565. [Google Scholar] [CrossRef]
- Yamatani, Y.; Nakai, K. Comprehensive comparison of gene expression diversity among a variety of human stem cells. NAR Genom. Bioinform. 2022, 4, lqac087. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liang, Y.; Sun, C.; Zhai, Y.; Li, X.; Jiang, M.; Yang, R.; Li, X.; Shu, Q.; Kai, G.; et al. Dihydrotanshinone I Inhibits the Lung Metastasis of Breast Cancer by Suppressing Neutrophil Extracellular Traps Formation. Int. J. Mol. Sci. 2022, 23, 15180. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ye, Y.; Peng, K.; Zeng, Z.; Chen, L.; Zeng, Y. Histones: The critical players in innate immunity. Front. Immunol. 2022, 13, 1030610. [Google Scholar] [CrossRef] [PubMed]
- Fraschilla, I.; Amatullah, H.; Jeffrey, K.L. One genome, many cell states: Epigenetic control of innate immunity. Curr. Opin. Immunol. 2022, 75, 102173. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Pang, M.Y.H.; Sun, X.; Ausió, J.; Ishibashi, T. Histone H4 variant, H4G, drives ribosomal RNA transcription and breast cancer cell proliferation by loosening nucleolar chromatin structure. J. Cell Physiol. 2020, 235, 9601–9608. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Nguyen, T.T.; Pang, M.Y.H.; Ishibashi, T. Primate-specific histone variants. Genome 2021, 64, 337–346. [Google Scholar] [CrossRef]
- Nacev, B.A.; Feng, L.; Bagert, J.D.; Lemiesz, A.E.; Gao, J.; Soshnev, A.A.; Kundra, R.; Schultz, N.; Muir, T.W.; Allis, C.D. The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 2019, 567, 473–478. [Google Scholar] [CrossRef]
- Bagert, J.D.; Mitchener, M.M.; Patriotis, A.L.; Dul, B.E.; Wojcik, F.; Nacev, B.A.; Feng, L.; Allis, C.D.; Muir, T.W. Oncohistone mutations enhance chromatin remodeling and alter cell fates. Nat. Chem. Biol. 2021, 17, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Vogler, C.; Huber, C.; Waldmann, T.; Ettig, R.; Braun, L.; Izzo, A.; Daujat, S.; Chassignet, I.; Lopez-Contreras, A.J.; Fernandez-Capetillo, O.; et al. Histone H2A C-terminus regulates chromatin dynamics, remodeling, and histone H1 binding. PLoS Genet. 2010, 6, e1001234. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.L.; Byrum, S.D.; Namjoshi, S.; Graves, H.K.; Dennehey, B.K.; Tackett, A.J.; Tyler, J.K. Mitotic phosphorylation of histone H3 threonine 80. Cell Cycle 2014, 13, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Freitas, M.A.; Sklenar, A.R.; Parthun, M.R. Application of mass spectrometry to the identification and quantification of histone post-translational modifications. J. Cell Biochem. 2004, 92, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Brewis, H.T.; Wang, A.Y.; Gaub, A.; Lau, J.J.; Stirling, P.C.; Kobor, M.S. What makes a histone variant a variant: Changing H2A to become H2A.Z. PLoS Genet. 2021, 17, e1009950. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.L.; Bele, A.; Small, E.C.; Will, C.M.; Nabet, B.; Oyer, J.A.; Huang, X.; Ghosh, R.P.; Grzybowski, A.T.; Yu, T.; et al. A Mutation in Histone H2B Represents a New Class of Oncogenic Driver. Cancer Discov. 2019, 9, 1438–1451. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.R.; Shah, T.; Torabifard, H. Histone H3 Orchestrates the Ubiquitination of Nucleosomal H2A by BRCA1/BARD1-UbcH5c Complex. bioRxiv 2024. [Google Scholar] [CrossRef]
- Eickbush, T.H.; Godfrey, J.E.; Elia, M.C.; Moudrianakis, E.N. H2a-specific proteolysis as a unique probe in the analysis of the histone octamer. J. Biol. Chem. 1988, 263, 18972–18978. [Google Scholar] [CrossRef] [PubMed]
- Jain, K.; Strahl, B.D. Oncohistones: Corruption at the core. Nat. Chem. Biol. 2021, 17, 370–371. [Google Scholar] [CrossRef]
- Espinoza Pereira, K.N.; Shan, J.; Licht, J.D.; Bennett, R.L. Histone mutations in cancer. Biochem. Soc. Trans. 2023, 51, 1749–1763. [Google Scholar] [CrossRef]
- Kang, T.Z.E.; Zhu, L.; Yang, D.; Ding, D.; Zhu, X.; Wan, Y.C.E.; Liu, J.; Ramakrishnan, S.; Chan, L.L.; Chan, S.Y.; et al. The elevated transcription of ADAM19 by the oncohistone H2BE76K contributes to oncogenic properties in breast cancer. J. Biol. Chem. 2021, 296, 100374. [Google Scholar] [CrossRef] [PubMed]
- Messner, S.; Hottiger, M.O. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 2011, 21, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Ogata, N.; Ueda, K.; Hayaishi, O. ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J. Biol. Chem. 1980, 255, 7610–7615. [Google Scholar] [CrossRef] [PubMed]
- Kujirai, T.; Ehara, H.; Fujino, Y.; Shirouzu, M.; Sekine, S.I.; Kurumizaka, H. Structural basis of the nucleosome transition during RNA polymerase II passage. Science 2018, 362, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Tessadori, F.; Giltay, J.C.; Hurst, J.A.; Massink, M.P.; Duran, K.; Vos, H.R.; van Es, R.M.; Scott, R.H.; van Gassen, K.L.I.; Bakkers, J.; et al. Germline mutations affecting the histone H4 core cause a developmental syndrome by altering DNA damage response and cell cycle control. Nat. Genet. 2017, 49, 1642–1646. [Google Scholar] [CrossRef] [PubMed]
- Dao, H.T.; Pham, L.T.D. Acidic patch histone mutations and their effects on nucleosome remodeling. Biochem. Soc. Trans. 2022, 50, 907–919. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Camacho, C.V.; Martire, S.; Nagari, A.; Setlem, R.; Gong, X.; Edwards, A.D.; Chiu, S.P.; Banaszynski, L.A.; Kraus, W.L. Oncohistone Mutations Occur at Functional Sites of Regulatory ADP-Ribosylation. Cancer Res. 2022, 82, 2361–2377. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z.; Li, Y.; Hüttmann, N.; Uppal, G.K.; D’Mello, R.; Berezovski, M.V. Lysine Acetylome of Breast Cancer-Derived Small Extracellular Vesicles Reveals Specific Acetylation Patterns for Metabolic Enzymes. Biomedicines 2023, 11, 1076. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-M.; Chen, H.Y.; Davie, J.R. Effect of Estradiol on Histone Acetylation Dynamics in Human Breast Cancer Cells*. J. Biol. Chem. 2001, 276, 49435–49442. [Google Scholar] [CrossRef]
- Lu, C.; Ramirez, D.; Hwang, S.; Jungbluth, A.; Frosina, D.; Ntiamoah, P.; Healey, J.; Zhu, G.; Chen, W.; Klein, M.; et al. Histone H3K36M mutation and trimethylation patterns in chondroblastoma. Histopathology 2019, 74, 291–299. [Google Scholar] [CrossRef]
- Fang, J.; Huang, Y.; Mao, G.; Yang, S.; Rennert, G.; Gu, L.; Li, H.; Li, G.M. Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3-MutSα interaction. Proc. Natl. Acad. Sci. USA 2018, 115, 9598–9603. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Gong, J.; Yu, T.; Zou, Y.; Zhang, M.; Nie, L.; Chen, X.; Yue, Q.; Liu, Y.; Mao, Q.; et al. Diffuse Midline Gliomas With Histone H3 K27M Mutation in Adults and Children: A Retrospective Series of 164 Cases. Am. J. Surg. Pathol. 2022, 46, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.C.E.; Liu, J.; Chan, K.M. Histone H3 Mutations in Cancer. Curr. Pharmacol. Rep. 2018, 4, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Voon, H.P.J.; Hii, L.; Garvie, A.; Udugama, M.; Krug, B.; Russo, C.; Chüeh, A.C.; Daly, R.J.; Morey, A.; Bell, T.D.M.; et al. Pediatric glioma histone H3.3 K27M/G34R mutations drive abnormalities in PML nuclear bodies. Genome Biol. 2023, 24, 284. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhu, Y.; Feng, X.; Liu, X.; Zhou, K.; Wang, Q.; Zhang, H.; Shi, H. H3F3A K27M Mutation Promotes the Infiltrative Growth of High-Grade Glioma in Adults by Activating β-Catenin/USP1 Signaling. Cancers 2022, 14, 4836. [Google Scholar] [CrossRef] [PubMed]
- DiNapoli, S.E.; Martinez-McFaline, R.; Shen, H.; Doane, A.S.; Perez, A.R.; Verma, A.; Simon, A.; Nelson, I.; Balgobin, C.A.; Bourque, C.T.; et al. Histone 3 Methyltransferases Alter Melanoma Initiation and Progression Through Discrete Mechanisms. Front. Cell Dev. Biol. 2022, 10, 814216. [Google Scholar] [CrossRef] [PubMed]
- Lehnertz, B.; Zhang, Y.W.; Boivin, I.; Mayotte, N.; Tomellini, E.; Chagraoui, J.; Lavallée, V.P.; Hébert, J.; Sauvageau, G. H3(K27M/I) mutations promote context-dependent transformation in acute myeloid leukemia with RUNX1 alterations. Blood 2017, 130, 2204–2214. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.Y.-T.; Piunti, A.; Qi, J.; Morgan, M.; Bartom, E.; Shilatifard, A.; Saratsis, A.M. Effects of H3.3G34V mutation on genomic H3K36 and H3K27 methylation patterns in isogenic pediatric glioma cells. Acta Neuropathol. Commun. 2020, 8, 219. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Farnung, L.; Dienemann, C.; Cramer, P. Structure of H3K36-methylated nucleosome-PWWP complex reveals multivalent cross-gyre binding. Nat. Struct. Mol. Biol. 2020, 27, 8–13. [Google Scholar] [CrossRef]
- Bhattarai, A.M.; Mainali, G.; Jha, P.; Karki, P.; Adhikari, A.; Pandit, A.; Bhattarai, A.M. Diffuse midline glioma H3K27M mutation in adult: A case report. Ann. Med. Surg. 2022, 76, 103567. [Google Scholar] [CrossRef]
- El-Hashash, A.H.K. Histone H3K27M Mutation in Brain Tumors. Adv. Exp. Med. Biol. 2021, 1283, 43–52. [Google Scholar] [PubMed]
- Nagaraja, S.; Quezada, M.A.; Gillespie, S.M.; Arzt, M.; Lennon, J.J.; Woo, P.J.; Hovestadt, V.; Kambhampati, M.; Filbin, M.G.; Suva, M.L.; et al. Histone Variant and Cell Context Determine H3K27M Reprogramming of the Enhancer Landscape and Oncogenic State. Mol. Cell 2019, 76, 965–980.e12. [Google Scholar] [CrossRef]
- Chen, H.; Lorton, B.; Gupta, V.; Shechter, D. A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 2017, 36, 373–386. [Google Scholar] [CrossRef]
- Pal, S.; Baiocchi, R.A.; Byrd, J.C.; Grever, M.R.; Jacob, S.T.; Sif, S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007, 26, 3558–3569. [Google Scholar] [CrossRef]
- Tanikawa, C.; Espinosa, M.; Suzuki, A.; Masuda, K.; Yamamoto, K.; Tsuchiya, E.; Ueda, K.; Daigo, Y.; Nakamura, Y.; Matsuda, K. Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nat. Commun. 2012, 3, 676. [Google Scholar] [CrossRef]
- Nozawa, K.; Takizawa, Y.; Pierrakeas, L.; Sogawa-Fujiwara, C.; Saikusa, K.; Akashi, S.; Luk, E.; Kurumizaka, H. Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B. Proc. Natl. Acad. Sci. USA 2022, 119, e2206542119. [Google Scholar] [CrossRef]
- Sahu, V.; Lu, C. Oncohistones: Hijacking the histone code. Annu. Rev. Cancer Biol. 2022, 6, 293–312. [Google Scholar] [CrossRef] [PubMed]
- Dan, H.; Zhang, S.; Zhou, Y.; Guan, Q. DNA Methyltransferase Inhibitors: Catalysts For Antitumour Immune Responses. Onco Targets Ther. 2019, 12, 10903–10916. [Google Scholar] [CrossRef] [PubMed]
- Feehley, T.; O’Donnell, C.W.; Mendlein, J.; Karande, M.; McCauley, T. Drugging the epigenome in the age of precision medicine. Clin. Epigenet. 2023, 15, 6. [Google Scholar] [CrossRef]
- Christman, J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 2002, 21, 5483–5495. [Google Scholar] [CrossRef]
- Daher-Reyes, G.S.; Merchan, B.M.; Yee, K.W.L. Guadecitabine (SGI-110): An investigational drug for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Expert. Opin. Investig. Drugs 2019, 28, 835–849. [Google Scholar] [CrossRef] [PubMed]
- Papadatos-Pastos, D.; Yuan, W.; Pal, A.; Crespo, M.; Ferreira, A.; Gurel, B.; Prout, T.; Ameratunga, M.; Chénard-Poirier, M.; Curcean, A.; et al. Phase 1, dose-escalation study of guadecitabine (SGI-110) in combination with pembrolizumab in patients with solid tumors. J. Immunother. Cancer 2022, 10, e004495. [Google Scholar] [CrossRef]
- Kaufman, D.W.; Kelly, J.P.; Rosenberg, L.; Stolley, P.D.; Schottenfeld, D.; Shapiro, S. Hydralazine and breast cancer. J. Natl. Cancer Inst. 1987, 78, 243–246. [Google Scholar]
- Plummer, R.; Vidal, L.; Griffin, M.; Lesley, M.; de Bono, J.; Coulthard, S.; Sludden, J.; Siu, L.L.; Chen, E.X.; Oza, A.M.; et al. Phase I Study of MG98, an Oligonucleotide Antisense Inhibitor of Human DNA Methyltransferase 1, Given as a 7-Day Infusion in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2009, 15, 3177–3183. [Google Scholar] [CrossRef]
- Dahn, M.L.; Cruickshank, B.M.; Jackson, A.J.; Dean, C.; Holloway, R.W.; Hall, S.R.; Coyle, K.M.; Maillet, H.; Waisman, D.M.; Goralski, K.B.; et al. Decitabine Response in Breast Cancer Requires Efficient Drug Processing and Is Not Limited by Multidrug Resistance. Mol. Cancer Ther. 2020, 19, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Billam, M.; Sobolewski, M.D.; Davidson, N.E. Effects of a novel DNA methyltransferase inhibitor zebularine on human breast cancer cells. Breast Cancer Res. Treat. 2010, 120, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.C.S.; Chan, A.H.Y.; Ganesan, A. Thirty Years of HDAC Inhibitors: 2020 Insight and Hindsight. J. Med. Chem. 2020, 63, 12460–12484. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.S.; Parmigiani, R.B.; Marks, P.A. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 2007, 26, 5541–5552. [Google Scholar] [CrossRef] [PubMed]
- Lian, B.; Chen, X.; Shen, K. Inhibition of histone deacetylases attenuates tumor progression and improves immunotherapy in breast cancer. Front. Immunol. 2023, 14, 1164514. [Google Scholar] [CrossRef]
- Connolly, R.M.; Rudek, M.A.; Piekarz, R. Entinostat: A promising treatment option for patients with advanced breast cancer. Future Oncol. 2017, 13, 1137–1148. [Google Scholar] [CrossRef]
- Salvador, M.A.; Wicinski, J.; Cabaud, O.; Toiron, Y.; Finetti, P.; Josselin, E.; Lelièvre, H.; Kraus-Berthier, L.; Depil, S.; Bertucci, F.; et al. The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin. Cancer Res. 2013, 19, 6520–6531. [Google Scholar] [CrossRef] [PubMed]
- Linden, H.M.; Kurland, B.F.; Link, J.; Novakova, A.; Chai, X.; Specht, J.M.; Gadi, V.K.; Gralow, J.; Schubert, E.K.; Peterson, L.; et al. A phase II clinical trial of HDACi (vorinostat) and AI therapy in breast cancer with molecular imaging correlates. J. Clin. Oncol. 2014, 32 (Suppl. S15), 556. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Padi, S.K.R.; Tindall, D.J.; Guo, B. Polycomb protein EZH2 suppresses apoptosis by silencing the proapoptotic miR-31. Cell Death Dis. 2014, 5, e1486. [Google Scholar] [CrossRef] [PubMed]
- Stein, E.M.; Garcia-Manero, G.; Rizzieri, D.A.; Tibes, R.; Berdeja, J.G.; Savona, M.R.; Jongen-Lavrenic, M.; Altman, J.K.; Thomson, B.; Blakemore, S.J.; et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 2018, 131, 2661–2669. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qu, J.; Qi, Y.; Duan, Y.; Huang, Y.-W.; Zhou, Z.; Li, P.; Yao, J.; Huang, B.; Zhang, S.; et al. EZH2 engages TGFβ signaling to promote breast cancer bone metastasis via integrin β1-FAK activation. Nat. Commun. 2022, 13, 2543. [Google Scholar] [CrossRef] [PubMed]
- Nassa, G.; Salvati, A.; Tarallo, R.; Gigantino, V.; Alexandrova, E.; Memoli, D.; Sellitto, A.; Rizzo, F.; Malanga, D.; Mirante, T.; et al. Inhibition of histone methyltransferase DOT1L silences ERα gene and blocks proliferation of antiestrogen-resistant breast cancer cells. Sci. Adv. 2019, 5, eaav5590. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chan, Y.-T.; Tan, H.-Y.; Li, S.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol. Cancer 2020, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Cochran, A.G.; Conery, A.R.; Sims, R.J., 3rd. Bromodomains: A new target class for drug development. Nat. Rev. Drug Discov. 2019, 18, 609–628. [Google Scholar] [CrossRef]
- Andrikopoulou, A.; Liontos, M.; Koutsoukos, K.; Dimopoulos, M.A.; Zagouri, F. The emerging role of BET inhibitors in breast cancer. Breast 2020, 53, 152–163. [Google Scholar] [CrossRef]
- Qi, J. Bromodomain and extraterminal domain inhibitors (BETi) for cancer therapy: Chemical modulation of chromatin structure. Cold Spring Harb. Perspect. Biol. 2014, 6, a018663. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-E.; Park, Y.-K.; Park, S.; Jang, Y.; Waring, N.; Dey, A.; Ozato, K.; Lai, B.; Peng, W.; Ge, K. Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat. Commun. 2017, 8, 2217. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Zhu, M.; Tan, X.; Zhang, Y.; Liu, X.; Yang, L. Super-enhancers and the super-enhancer reader BRD4: Tumorigenic factors and therapeutic targets. Cell Death Discov. 2023, 9, 470. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Deng, W.; Liu, Y.; Wang, C. General mechanism of JQ1 in inhibiting various types of cancer. Mol. Med. Rep. 2020, 21, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, C.-Y.; Wang, S. The Making of I-BET762, a BET Bromodomain Inhibitor Now in Clinical Development. J. Med. Chem. 2013, 56, 7498–7500. [Google Scholar] [CrossRef] [PubMed]
- Roboz, G.J.; Desai, P.; Lee, S.; Ritchie, E.K.; Winer, E.S.; DeMario, M.; Brennan, B.; Nüesch, E.; Chesne, E.; Brennan, L.; et al. A dose escalation study of RO6870810/TEN-10 in patients with acute myeloid leukemia and myelodysplastic syndrome. Leuk. Lymphoma 2021, 62, 1740–1748. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, R.; Riveiro, M.E.; Astorgues-Xerri, L.; Odore, E.; Rezai, K.; Erba, E.; Panini, N.; Rinaldi, A.; Kwee, I.; Beltrame, L.; et al. The bromodomain inhibitor OTX015 (MK-8628) exerts anti-tumor activity in triple-negative breast cancer models as single agent and in combination with everolimus. Oncotarget 2017, 8, 7598–7613. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, N.; Sharma, A.R.; Baylin, S.B. Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu. Rev. Med. 2016, 67, 73–89. [Google Scholar] [CrossRef]
- Fardi, M.; Solali, S.; Farshdousti Hagh, M. Epigenetic mechanisms as a new approach in cancer treatment: An updated review. Genes Dis 2018, 5, 304–311. [Google Scholar] [CrossRef]
- Yoo, C.B.; Jones, P.A. Epigenetic therapy of cancer: Past, present and future. Nat. Rev. Drug Discov. 2006, 5, 37–50. [Google Scholar] [CrossRef]
- Chomiak, A.A.; Tiedemann, R.L.; Liu, Y.; Kong, X.; Cui, Y.; Wiseman, A.K.; Thurlow, K.E.; Cornett, E.M.; Topper, M.J.; Baylin, S.B.; et al. Select EZH2 inhibitors enhance viral mimicry effects of DNMT inhibition through a mechanism involving NFAT:AP-1 signaling. Sci. Adv. 2024, 10, eadk4423. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhahri, H.; Saintilnord, W.N.; Chandler, D.; Fondufe-Mittendorf, Y.N. Beyond the Usual Suspects: Examining the Role of Understudied Histone Variants in Breast Cancer. Int. J. Mol. Sci. 2024, 25, 6788. https://doi.org/10.3390/ijms25126788
Dhahri H, Saintilnord WN, Chandler D, Fondufe-Mittendorf YN. Beyond the Usual Suspects: Examining the Role of Understudied Histone Variants in Breast Cancer. International Journal of Molecular Sciences. 2024; 25(12):6788. https://doi.org/10.3390/ijms25126788
Chicago/Turabian StyleDhahri, Hejer, Wesley N. Saintilnord, Darrell Chandler, and Yvonne N. Fondufe-Mittendorf. 2024. "Beyond the Usual Suspects: Examining the Role of Understudied Histone Variants in Breast Cancer" International Journal of Molecular Sciences 25, no. 12: 6788. https://doi.org/10.3390/ijms25126788
APA StyleDhahri, H., Saintilnord, W. N., Chandler, D., & Fondufe-Mittendorf, Y. N. (2024). Beyond the Usual Suspects: Examining the Role of Understudied Histone Variants in Breast Cancer. International Journal of Molecular Sciences, 25(12), 6788. https://doi.org/10.3390/ijms25126788







