Integrating In Silico and In Vitro Approaches to Identify Natural Peptides with Selective Cytotoxicity against Cancer Cells
Abstract
:1. Introduction
2. Results
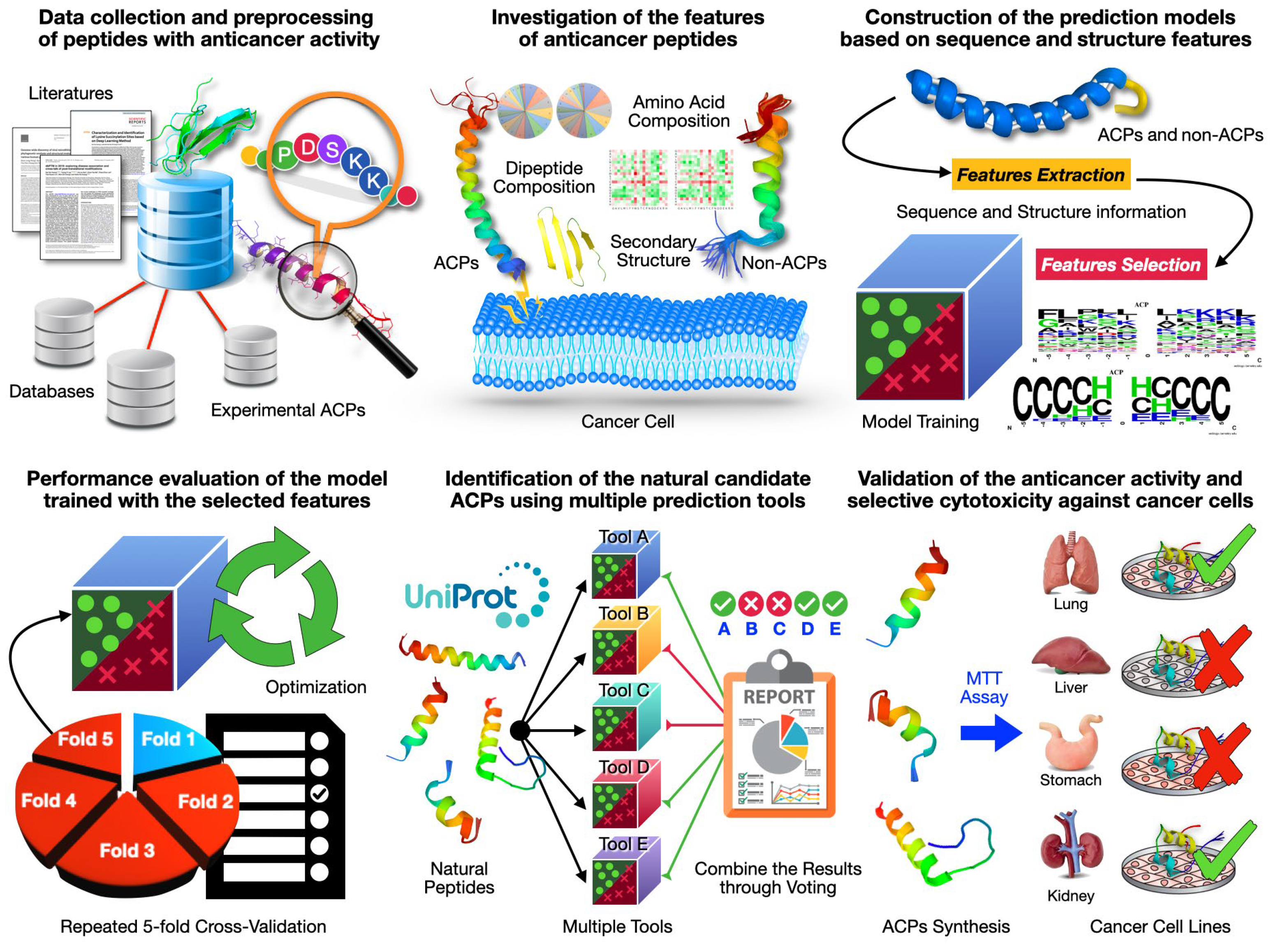
2.1. Data Collection and Preprocessing of Peptides with Anticancer Activity
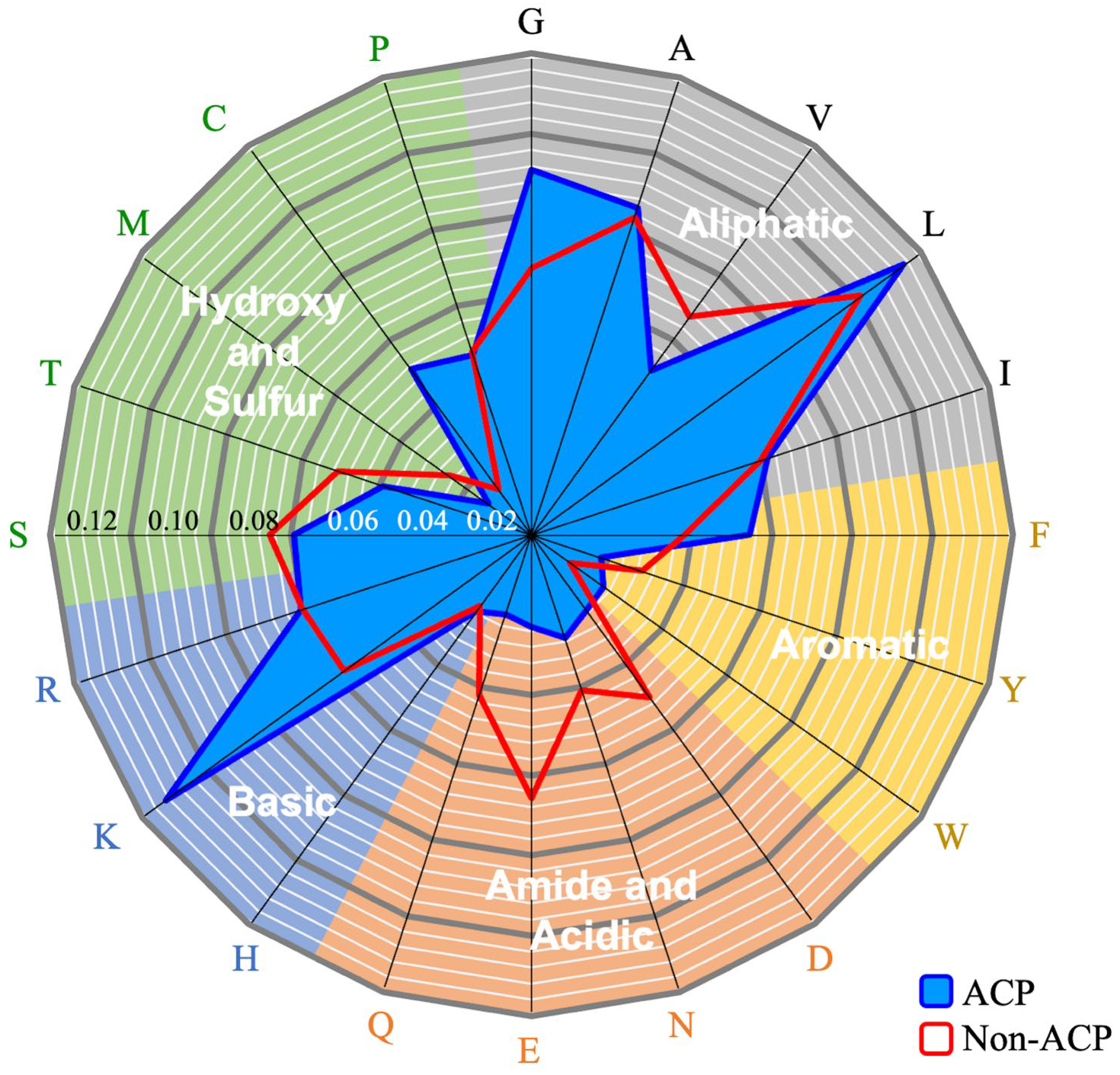
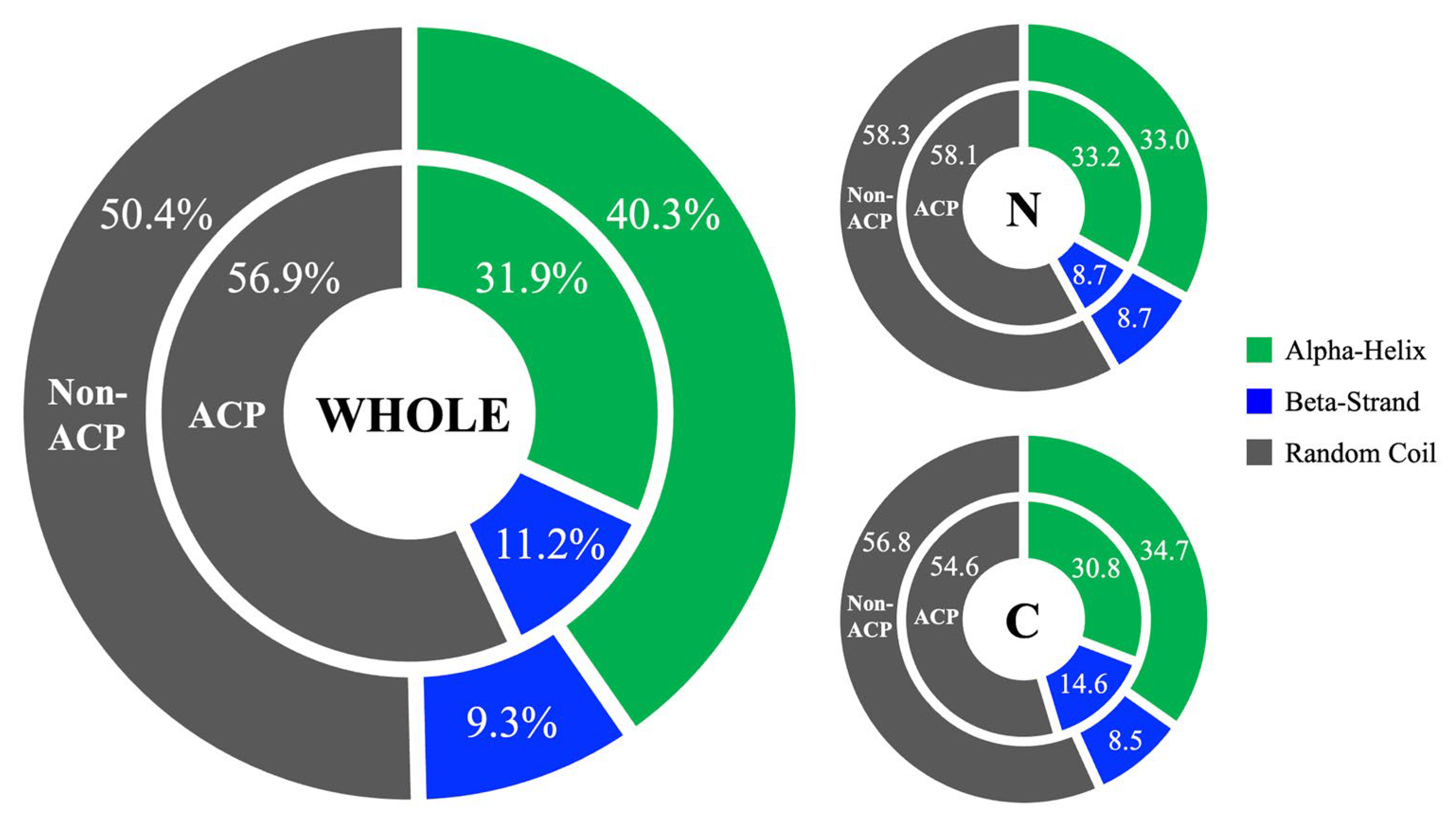
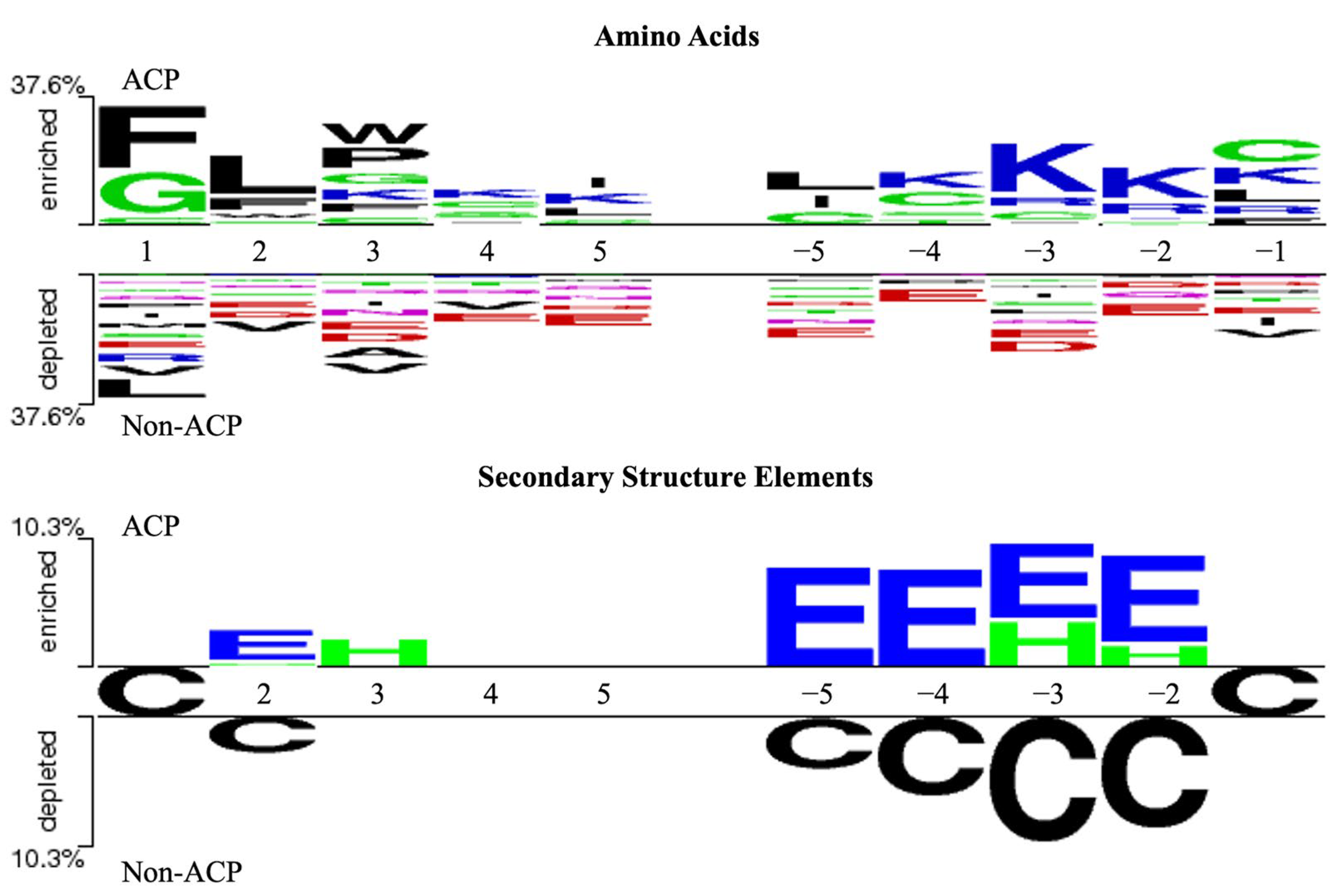
2.2. Investigation of Sequence and Structural Features of Anticancer Peptides
2.3. Construction of Prediction Models Based on Sequence and Structural Features
2.4. Performance Evaluation of Model Trained by Hybrid Feature Sets
2.5. Identification of Natural ACPs by Integrating Multiple Tools
2.6. Validating the Selective Cytotoxicity of ACPs against Specific Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Redundancy Removal
4.2. Feature Investigation
4.3. Construction of Prediction Models
4.4. Performance Evaluation
4.5. Identification of Candidate ACPs Using Multiple Prediction Tools
4.6. Functional Enrichment Analysis
4.7. Evaluation of Anticancer Activity and Selective Cytotoxicity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahase, E. Cancer overtakes CVD to become leading cause of death in high income countries. BMJ 2019, 366, l5368. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Yu, H. Global burden of cancer. Yale J. Biol. Med. 2006, 79, 85–94. [Google Scholar] [PubMed]
- Tohme, S.; Simmons, R.L.; Tsung, A. Surgery for Cancer: A Trigger for Metastases. Cancer Res. 2017, 77, 1548–1552. [Google Scholar] [CrossRef] [PubMed]
- Lamson, D.W.; Brignall, M.S. Antioxidants in cancer therapy; their actions and interactions with oncologic therapies. Altern. Med. Rev. 1999, 4, 304–329. [Google Scholar] [PubMed]
- Potmesil, M. Camptothecins: From bench research to hospital wards. Cancer Res. 1994, 54, 1431–1439. [Google Scholar] [PubMed]
- Coates, A.; Abraham, S.; Kaye, S.B.; Sowerbutts, T.; Frewin, C.; Fox, R.M.; Tattersall, M.H. On the receiving end--patient perception of the side-effects of cancer chemotherapy. Eur. J. Cancer Clin. Oncol. 1983, 19, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [PubMed]
- Perez-Tomas, R.; Perez-Guillen, I. Lactate in the Tumor Microenvironment: An Essential Molecule in Cancer Progression and Treatment. Cancers 2020, 12, 3244. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 2009, 625, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, E.G.; Dobroff, A.S.; Taborda, C.P.; Travassos, L.R. Antifungal and antitumor models of bioactive protective peptides. An. Acad. Bras. Ciências 2009, 81, 503–520. [Google Scholar] [CrossRef] [PubMed]
- Droin, N.; Hendra, J.B.; Ducoroy, P.; Solary, E. Human defensins as cancer biomarkers and antitumour molecules. J. Proteom. 2009, 72, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Harris, F.; Dennison, S.R.; Singh, J.; Phoenix, D.A. On the selectivity and efficacy of defense peptides with respect to cancer cells. Med. Res. Rev. 2013, 33, 190–234. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Kapoor, P.; Kumar, R.; Chaudhary, K.; Gautam, A.; Raghava, G.P. In silico models for designing and discovering novel anticancer peptides. Sci. Rep. 2013, 3, 2984. [Google Scholar] [CrossRef] [PubMed]
- Hajisharifi, Z.; Piryaiee, M.; Mohammad Beigi, M.; Behbahani, M.; Mohabatkar, H. Predicting anticancer peptides with Chou’s pseudo amino acid composition and investigating their mutagenicity via Ames test. J. Theor. Biol. 2014, 341, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, S.; Ptv, L. ACPP: A Web Server for Prediction and Design of Anti-cancer Peptides. Int. J. Pept. Res. Ther. 2015, 21, 99–106. [Google Scholar] [CrossRef]
- Chen, W.; Ding, H.; Feng, P.; Lin, H.; Chou, K.C. iACP: A sequence-based tool for identifying anticancer peptides. Oncotarget 2016, 7, 16895–16909. [Google Scholar] [CrossRef] [PubMed]
- Li, F.M.; Wang, X.Q. Identifying anticancer peptides by using improved hybrid compositions. Sci. Rep. 2016, 6, 33910. [Google Scholar] [CrossRef] [PubMed]
- Akbar, S.; Hayat, M.; Iqbal, M.; Jan, M.A. iACP-GAEnsC: Evolutionary genetic algorithm based ensemble classification of anticancer peptides by utilizing hybrid feature space. Artif. Intell. Med. 2017, 79, 62–70. [Google Scholar] [PubMed]
- Kabir, M.; Arif, M.; Ahmad, S.; Ali, Z.; Swati, Z.N.K.; Yu, D.-J. Intelligent computational method for discrimination of anticancer peptides by incorporating sequential and evolutionary profiles information. Chemom. Intell. Lab. Syst. 2018, 182, 158–165. [Google Scholar] [CrossRef]
- Wei, L.; Zhou, C.; Chen, H.; Song, J.; Su, R. ACPred-FL: A sequence-based predictor using effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics 2018, 34, 4007–4016. [Google Scholar] [CrossRef] [PubMed]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. ACPred: A Computational Tool for the Prediction and Analysis of Anticancer Peptides. Molecules 2019, 24, 1973. [Google Scholar] [CrossRef]
- Boopathi, V.; Subramaniyam, S.; Malik, A.; Lee, G.; Manavalan, B.; Yang, D.C. mACPpred: A Support Vector Machine-Based Meta-Predictor for Identification of Anticancer Peptides. Int. J. Mol. Sci. 2019, 20, 1964. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Bhagat, D.; Mahalwal, M.; Sharma, N.; Raghava, G.P.S. AntiCP 2.0: An updated model for predicting anticancer peptides. Brief. Bioinform. 2021, 22, bbaa153. [Google Scholar] [CrossRef]
- Ge, R.; Feng, G.; Jing, X.; Zhang, R.; Wang, P.; Wu, Q. EnACP: An Ensemble Learning Model for Identification of Anticancer Peptides. Front. Genet. 2020, 11, 760. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010, 38, D774–D780. [Google Scholar] [CrossRef]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P. CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res. 2015, 43, D837–D843. [Google Scholar] [CrossRef] [PubMed]
- Novkovic, M.; Simunic, J.; Bojovic, V.; Tossi, A.; Juretic, D. DADP: The database of anuran defense peptides. Bioinformatics 2012, 28, 1406–1407. [Google Scholar] [CrossRef]
- Jhong, J.H.; Chi, Y.H.; Li, W.C.; Lin, T.H.; Huang, K.Y.; Lee, T.Y. dbAMP: An integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 2019, 47, D285–D297. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6, 148. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Chaudhary, K.; Dhanda, S.K.; Bhalla, S.; Usmani, S.S.; Gautam, A.; Tuknait, A.; Agrawal, P.; Mathur, D.; Raghava, G.P. SATPdb: A database of structurally annotated therapeutic peptides. Nucleic Acids Res. 2016, 44, D1119–D1126. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar]
- Sahu, S.S.; Panda, G. A novel feature representation method based on Chou’s pseudo amino acid composition for protein structural class prediction. Comput. Biol. Chem. 2010, 34, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Park, K.J.; Kanehisa, M. Prediction of protein subcellular locations by support vector machines using compositions of amino acids and amino acid pairs. Bioinformatics 2003, 19, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Tang, Y.R.; Sheng, Z.Y.; Zhang, Z. Prediction of mucin-type O-glycosylation sites in mammalian proteins using the composition of k-spaced amino acid pairs. BMC Bioinform. 2008, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.D.; Won, H.S.; Kim, J.H.; Mishig-Ochir, T.; Lee, B.J. Antimicrobial peptides for therapeutic applications: A review. Molecules 2012, 17, 12276–12286. [Google Scholar] [CrossRef] [PubMed]
- Baindara, P.; Gautam, A.; Raghava, G.P.S.; Korpole, S. Anticancer properties of a defensin like class IId bacteriocin Laterosporulin10. Sci. Rep. 2017, 7, 46541. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.K.; McCormick, T.S.; Weinberg, A. Human Beta Defensins and Cancer: Contradictions and Common Ground. Front. Oncol. 2019, 9, 341. [Google Scholar] [CrossRef] [PubMed]
- Oelkrug, C.; Hartke, M.; Schubert, A. Mode of action of anticancer peptides (ACPs) from amphibian origin. Anticancer Res. 2015, 35, 635–643. [Google Scholar]
- Baindara, P.; Kapoor, A.; Korpole, S.; Grover, V. Cysteine-rich low molecular weight antimicrobial peptides from Brevibacillus and related genera for biotechnological applications. World J. Microbiol. Biotechnol. 2017, 33, 124. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Gao, Y.; Qi, Y.; Chen, L.; Ma, Y.; Li, Y. Peptide-based cancer therapy: Opportunity and challenge. Cancer Lett. 2014, 351, 13–22. [Google Scholar] [CrossRef]
- Martin, A.C.; Orengo, C.A.; Hutchinson, E.G.; Jones, S.; Karmirantzou, M.; Laskowski, R.A.; Mitchell, J.B.; Taroni, C.; Thornton, J.M. Protein folds and functions. Structure 1998, 6, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Singh, S.; Singh Raghava, G.P. Peptide Secondary Structure Prediction using Evolutionary Information. bioRxiv 2019, 558791. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1999, 1462, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Papo, N.; Shahar, M.; Eisenbach, L.; Shai, Y. A novel lytic peptide composed of DL-amino acids selectively kills cancer cells in culture and in mice. J. Biol. Chem. 2003, 278, 21018–21023. [Google Scholar] [CrossRef] [PubMed]
- Papo, N.; Shai, Y. New lytic peptides based on the D,L-amphipathic helix motif preferentially kill tumor cells compared to normal cells. Biochemistry 2003, 42, 9346–9354. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.C.; Mi, Z.; Kim, S.H.; Ng, B.; Robbins, P.D. A proapoptotic peptide for the treatment of solid tumors. Cancer Res. 2001, 61, 7709–7712. [Google Scholar] [PubMed]
- Papo, N.; Oren, Z.; Pag, U.; Sahl, H.G.; Shai, Y. The consequence of sequence alteration of an amphipathic alpha-helical antimicrobial peptide and its diastereomers. J. Biol. Chem. 2002, 277, 33913–33921. [Google Scholar] [CrossRef] [PubMed]
- Dennison, S.R.; Whittaker, M.; Harris, F.; Phoenix, D.A. Anticancer alpha-helical peptides and structure/function relationships underpinning their interactions with tumour cell membranes. Curr. Protein Pept. Sci. 2006, 7, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Feng, Q.; Yan, Q.; Hao, X.; Chen, Y. Alpha-helical cationic anticancer peptides: A promising candidate for novel anticancer drugs. Mini Rev. Med. Chem. 2015, 15, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Chiangjong, W.; Chutipongtanate, S.; Hongeng, S. Anticancer peptide: Physicochemical property, functional aspect and trend in clinical application (Review). Int. J. Oncol. 2020, 57, 678–696. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, B.; Basith, S.; Shin, T.H.; Choi, S.; Kim, M.O.; Lee, G. MLACP: Machine-learning-based prediction of anticancer peptides. Oncotarget 2017, 8, 77121–77136. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27:1–27:27. [Google Scholar] [CrossRef]
- Huang, K.Y.; Tseng, Y.J.; Kao, H.J.; Chen, C.H.; Yang, H.H.; Weng, S.L. Identification of subtypes of anticancer peptides based on sequential features and physicochemical properties. Sci. Rep. 2021, 11, 13594. [Google Scholar] [PubMed]
- The Gene Ontology, C. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar]
- Ghadiri, N.; Javidan, M.; Sheikhi, S.; Tastan, O.; Parodi, A.; Liao, Z.; Tayybi Azar, M.; Ganjalikhani-Hakemi, M. Bioactive peptides: An alternative therapeutic approach for cancer management. Front. Immunol. 2024, 15, 1310443. [Google Scholar] [CrossRef] [PubMed]
- Jafari, A.; Babajani, A.; Sarrami Forooshani, R.; Yazdani, M.; Rezaei-Tavirani, M. Clinical Applications and Anticancer Effects of Antimicrobial Peptides: From Bench to Bedside. Front. Oncol. 2022, 12, 819563. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, G.; Tallima, H.; Dabbish, E.; Badr ElDin, N.; Abd El-Rahman, M.K.; Ibrahim, M.A.A.; Shoeib, T. Anti-Cancer Peptides: Status and Future Prospects. Molecules 2023, 28, 1148. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]

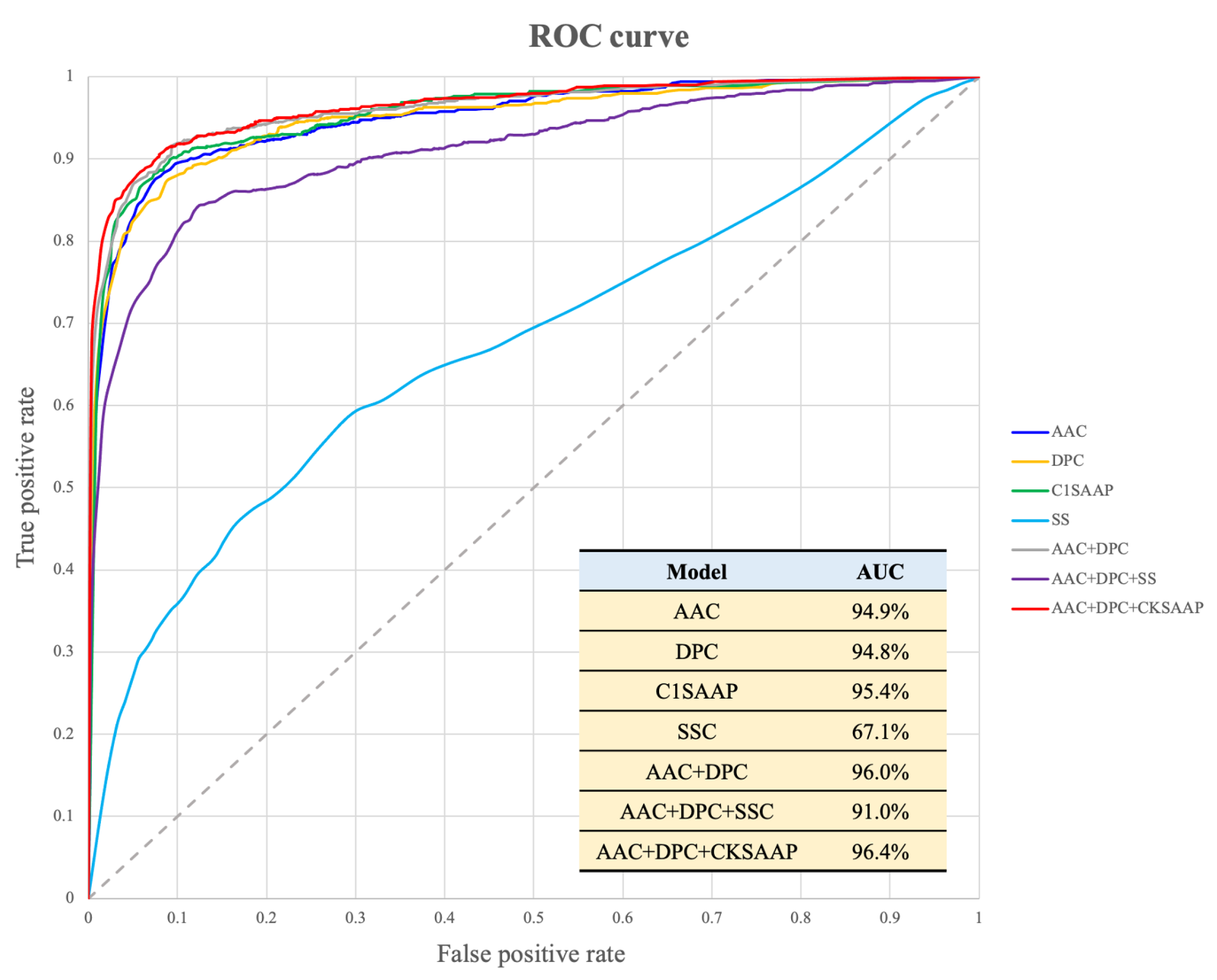
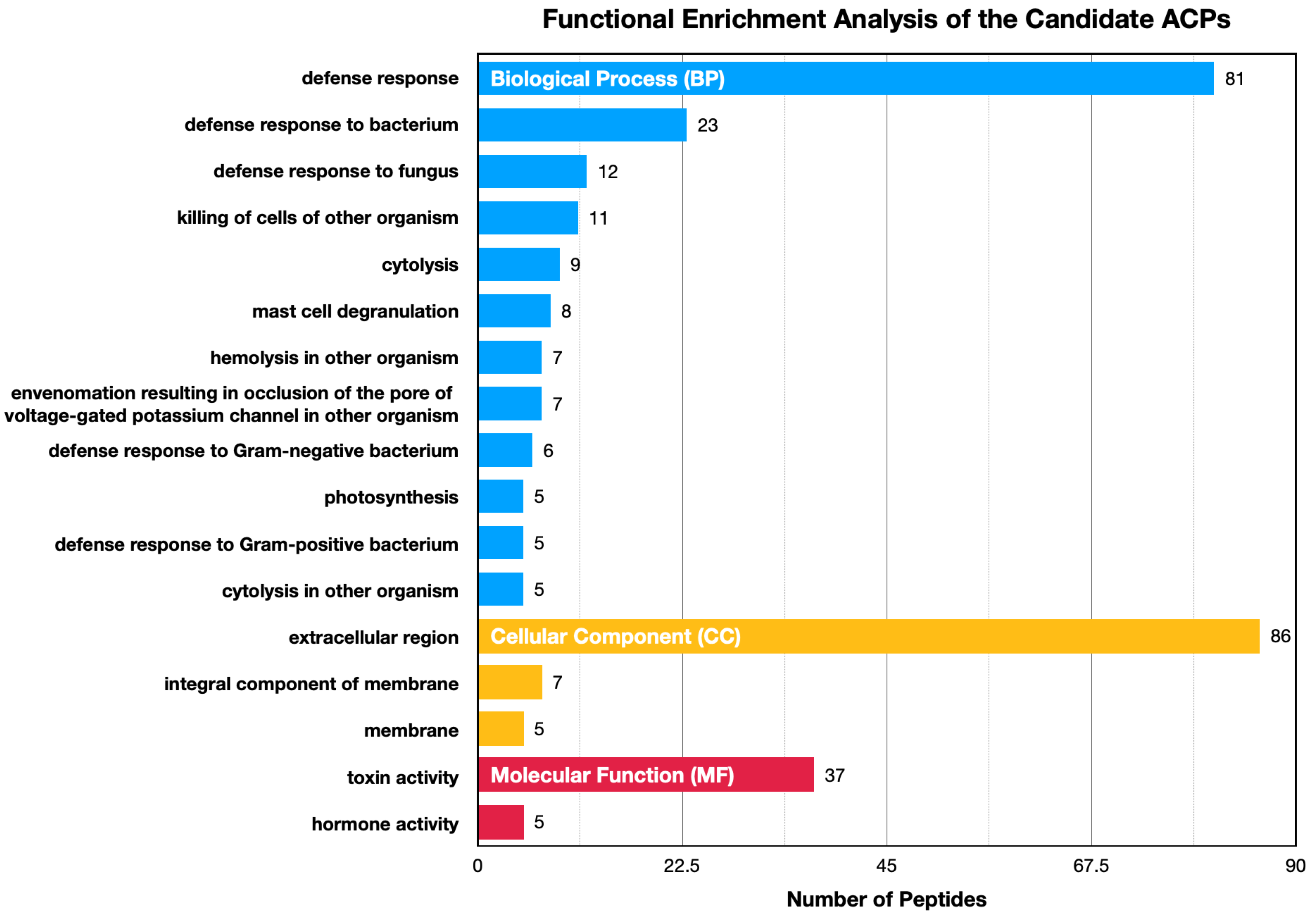
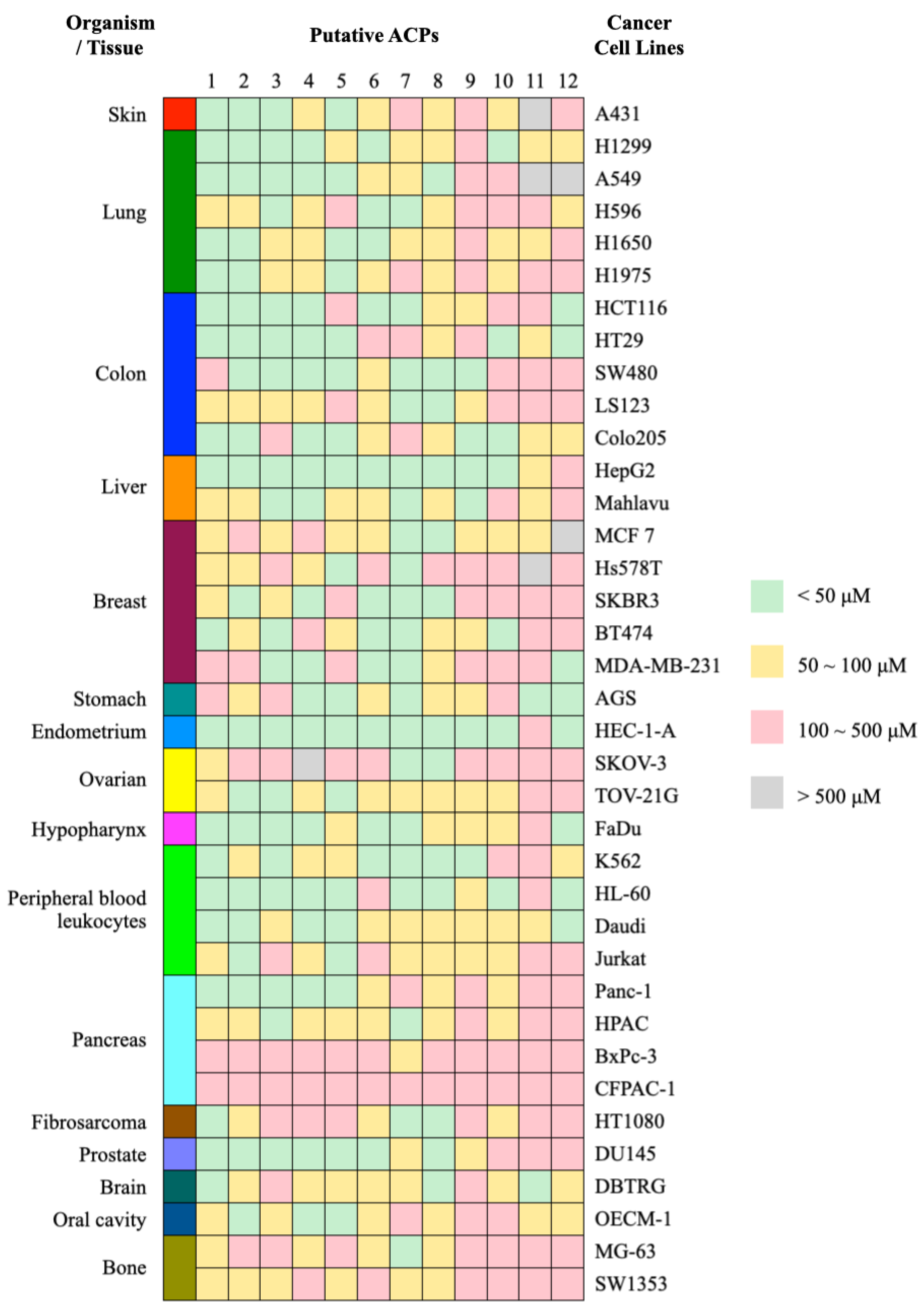
| Dataset | Number of ACPs | Number of Non-ACPs |
|---|---|---|
| Raw data | 1462 | 2875 |
| Length 10–50 aa | 1344 | 2361 |
| Training dataset | 804 | 1494 |
| Testing dataset | 41,489 natural peptides | |
| Model | Sensitivity (%) | Specificity (%) | Accuracy (%) | MCC * |
|---|---|---|---|---|
| AAC | 89.00 ± 0.0038 | 89.48 ± 0.0030 | 89.31 ± 0.0022 | 0.77 ± 0.0046 |
| DPC | 88.71 ± 0.0030 | 88.96 ± 0.0025 | 88.87 ± 0.0008 | 0.76 ± 0.0014 |
| C1SAAP | 90.00 ± 0.0017 | 90.09 ± 0.0035 | 90.06 ± 0.0022 | 0.79 ± 0.0042 |
| C2SAAP | 89.70 ± 0.0046 | 89.53 ± 0.0044 | 89.59 ± 0.0033 | 0.78 ± 0.0068 |
| C3SAAP | 89.10 ± 0.0014 | 90.58 ± 0.0025 | 90.06 ± 0.0016 | 0.79 ± 0.0033 |
| SSEC | 62.29 ± 0.0140 | 64.08 ± 0.0167 | 63.46 ± 0.0070 | 0.25 ± 0.0083 |
| N-AAC | 73.26 ± 0.0052 | 74.67 ± 0.0055 | 74.18 ± 0.0036 | 0.46 ± 0.0065 |
| N-SSEC | 50.92 ± 0.1771 | 50.67 ± 0.1782 | 50.76 ± 0.0539 | 0.02 ± 0.0027 |
| C-AAC | 69.18 ± 0.0077 | 70.08 ± 0.0048 | 69.77 ± 0.0032 | 0.38 ± 0.0069 |
| C-SSEC | 55.50 ± 0.0997 | 54.42 ± 0.1230 | 54.80 ± 0.0468 | 0.10 ± 0.0344 |
| Model | Sensitivity (%) | Specificity (%) | Accuracy (%) | MCC * |
|---|---|---|---|---|
| AAC + DPC | 91.02 ± 0.0025 | 90.12 ± 0.0047 | 90.44 ± 0.0037 | 0.80 ± 0.0074 |
| AAC + DPC + SSEC | 84.30 ± 0.0069 | 85.07 ± 0.0028 | 84.80 ± 0.0036 | 0.68 ± 0.0080 |
| AAC + DPC + CKSAAP | 91.17 ± 0.0037 | 90.83 ± 0.0032 | 90.95 ± 0.0021 | 0.81 ± 0.0043 |
| AAC + DPC + CKSAAP + SSEC | 86.52 ± 0.0047 | 87.07 ± 0.0033 | 86.88 ± 0.0015 | 0.72 ± 0.0030 |
| Entry Name | Sequence | ACPred | ACPred-FL | AntiCP | AntiCP2 | iACP | mACPpred | Our Model |
|---|---|---|---|---|---|---|---|---|
| KAB4_OLDAF | GLPVCGETCVGGTCNTPGCTCSWPVCTRD | 98.60% | 98.11% | 72.58% | 96.00% | 99.73% | 98.17% | 99.60% |
| CYO22_VIOOD | GLPICGETCVGGTCNTPGCTCSWPVCTRN | 99.50% | 95.12% | 71.77% | 95.00% | 99.90% | 98.42% | 99.70% |
| THN2_VISAL | KSCCPNTTGRNIYNTCRFGGGSREVCASLSGCKIISASTCPSYPDK | 99.50% | 99.22% | 70.56% | 94.00% | 99.52% | 96.51% | 96.60% |
| CYH3_VIOHE | GLPVCGETCFGGTCNTPGCICDPWPVCTRN | 98.70% | 92.89% | 71.77% | 95.00% | 99.80% | 98.80% | 99.50% |
| MYX_CRODR | YKQCHKKGGHCFPKEKICIPPSSDFGKMDCRWRWKCCKKGSG | 99.60% | 99.22% | 70.56% | 91.00% | 99.83% | 94.71% | 95.10% |
| KAB10_OLDAF | GLPTCGETCFGGTCNTPGCSCSSWPICTRD | 99.40% | 98.11% | 70.56% | 90.00% | 99.93% | 98.48% | 99.60% |
| PROTO_POLPI | ILGTILGLLKSL | 97.80% | 99.22% | 95.16% | 54.00% | 88.85% | 98.35% | 99.50% |
| CYO23_VIOOD | GLPTCGETCFGGTCNTPGCTCDSSWPICTHN | 99.70% | 98.11% | 70.56% | 87.00% | 99.93% | 98.47% | 99.60% |
| CYPLE_PSYLE | SVTPIVCGETCFGGTCNTPGCSCSWPICTK | 99.90% | 99.22% | 68.95% | 87.00% | 99.97% | 96.86% | 99.70% |
| CYPLD_PSYBR | GLPVCGESCFGGTCNTPGCSCTWPVCTRD | 98.10% | 95.12% | 72.18% | 87.00% | 98.28% | 98.01% | 99.50% |
| ATOX_PHYTB | LTWKIPTRFCGVT | 91.90% | 99.22% | 83.47% | 50.00% | 96.44% | 96.64% | 91.10% |
| CR12_RANCA | GLLGVLGSVAKHVLPHVVPVIAEHL | 99.30% | 98.11% | 70.16% | 84.00% | 99.79% | 98.62% | 86.10% |
| KAB14_OLDAF | GLPVCGESCFGGTCNTPGCACDPWPVCTRD | 88.70% | 83.42% | 71.77% | 86.00% | 99.76% | 97.94% | 99.00% |
| CYPLC_PSYLE | GDLPVCGETCFGGTCNTPGCVCAWPVCTR | 95.70% | 98.11% | 68.15% | 83.00% | 99.25% | 98.21% | 99.40% |
| CYPLB_PSYLE | GDLPICGETCFGGTCNTPGCVCAWPVCNR | 95.10% | 98.11% | 67.74% | 83.00% | 99.43% | 97.87% | 99.50% |
| GRAB_GRASX | IGGIISFFKRLF | 100.00% | 99.22% | 85.08% | 69.00% | 82.43% | 96.20% | 98.90% |
| CIRF_CHAPA | AIPCGESCVWIPCISAAIGCSCKNKVCYR | 99.60% | 82.67% | 75.81% | 89.00% | 99.80% | 98.54% | 99.50% |
| CYVNA_VIOIN | GIPVCGETCTLGTCYTAGCSCSWPVCTRN | 99.60% | 98.11% | 71.37% | 82.00% | 99.80% | 98.12% | 99.50% |
| PNG1_PANCL | LNWGAILKHIIK | 99.90% | 99.22% | 81.85% | 58.00% | 99.20% | 98.22% | 99.00% |
| PSMA3_STAAN | MEFVAKLFKFFKDLLGKFLGNN | 98.60% | 97.98% | 75.81% | 82.00% | 96.15% | 87.97% | 85.70% |
| Peptide | UniProt ID | Length | Sequence |
|---|---|---|---|
| ACP1 | KAB4_OLDAF | 29 | GLPVCGETCVGGTCNTPGCTCSWPVCTRD |
| ACP2 | CIRF_CHAPA | 29 | AIPCGESCVWIPCISAAIGCSCKNKVCYR |
| ACP3 | PSMA3_STAAN | 22 | MEFVAKLFKFFKDLLGKFLGNN |
| ACP4 | CYMEK_MELDN | 31 | GSIPCGESCVWIPCISSVVGCACKNKVCYKN |
| ACP5 | CYVNA_VIOIN | 29 | GIPVCGETCTLGTCYTAGCSCSWPVCTRN |
| ACP6 | CIRB_CHAPA | 31 | GVIPCGESCVFIPCISTLLGCSCKNKVCYRN |
| ACP7 | THN2_VISAL | 46 | KSCCPNTTGRNIYNTCRFGGGSREVCASLSGCKIISASTCPSYPDK |
| ACP8 | MYX_CRODR | 42 | YKQCHKKGGHCFPKEKICIPPSSDFGKMDCRWRWKCCKKGSG |
| ACP9 | CR12_RANCA | 25 | GLLGVLGSVAKHVLPHVVPVIAEHL |
| ACP10 | CYPLE_PSYLE | 30 | SVTPIVCGETCFGGTCNTPGCSCSWPICTK |
| ACP11 | UT114_PEA | 15 | EQQQQQQPQNRRFRE |
| ACP12 | TL11_SPIOL | 22 | FKGGGPYGQGVTRGQDLSGKDF |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kao, H.-J.; Weng, T.-H.; Chen, C.-H.; Chen, Y.-C.; Chi, Y.-H.; Huang, K.-Y.; Weng, S.-L. Integrating In Silico and In Vitro Approaches to Identify Natural Peptides with Selective Cytotoxicity against Cancer Cells. Int. J. Mol. Sci. 2024, 25, 6848. https://doi.org/10.3390/ijms25136848
Kao H-J, Weng T-H, Chen C-H, Chen Y-C, Chi Y-H, Huang K-Y, Weng S-L. Integrating In Silico and In Vitro Approaches to Identify Natural Peptides with Selective Cytotoxicity against Cancer Cells. International Journal of Molecular Sciences. 2024; 25(13):6848. https://doi.org/10.3390/ijms25136848
Chicago/Turabian StyleKao, Hui-Ju, Tzu-Han Weng, Chia-Hung Chen, Yu-Chi Chen, Yu-Hsiang Chi, Kai-Yao Huang, and Shun-Long Weng. 2024. "Integrating In Silico and In Vitro Approaches to Identify Natural Peptides with Selective Cytotoxicity against Cancer Cells" International Journal of Molecular Sciences 25, no. 13: 6848. https://doi.org/10.3390/ijms25136848





