DNA Repair Protein XRCC1 Stimulates Activity of DNA Polymerase λ under Conditions of Microphase Separation
Abstract
1. Introduction
2. Results
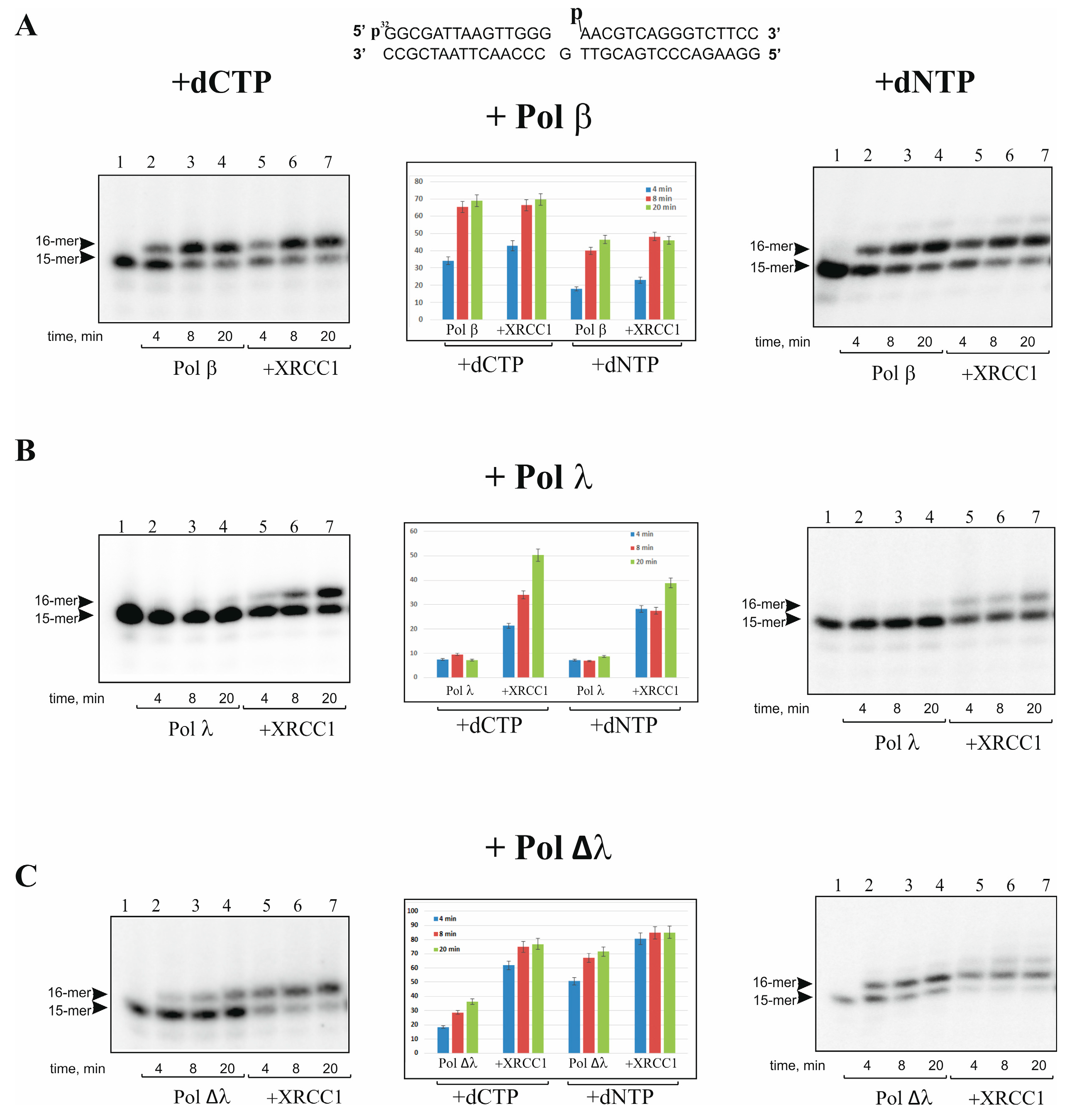
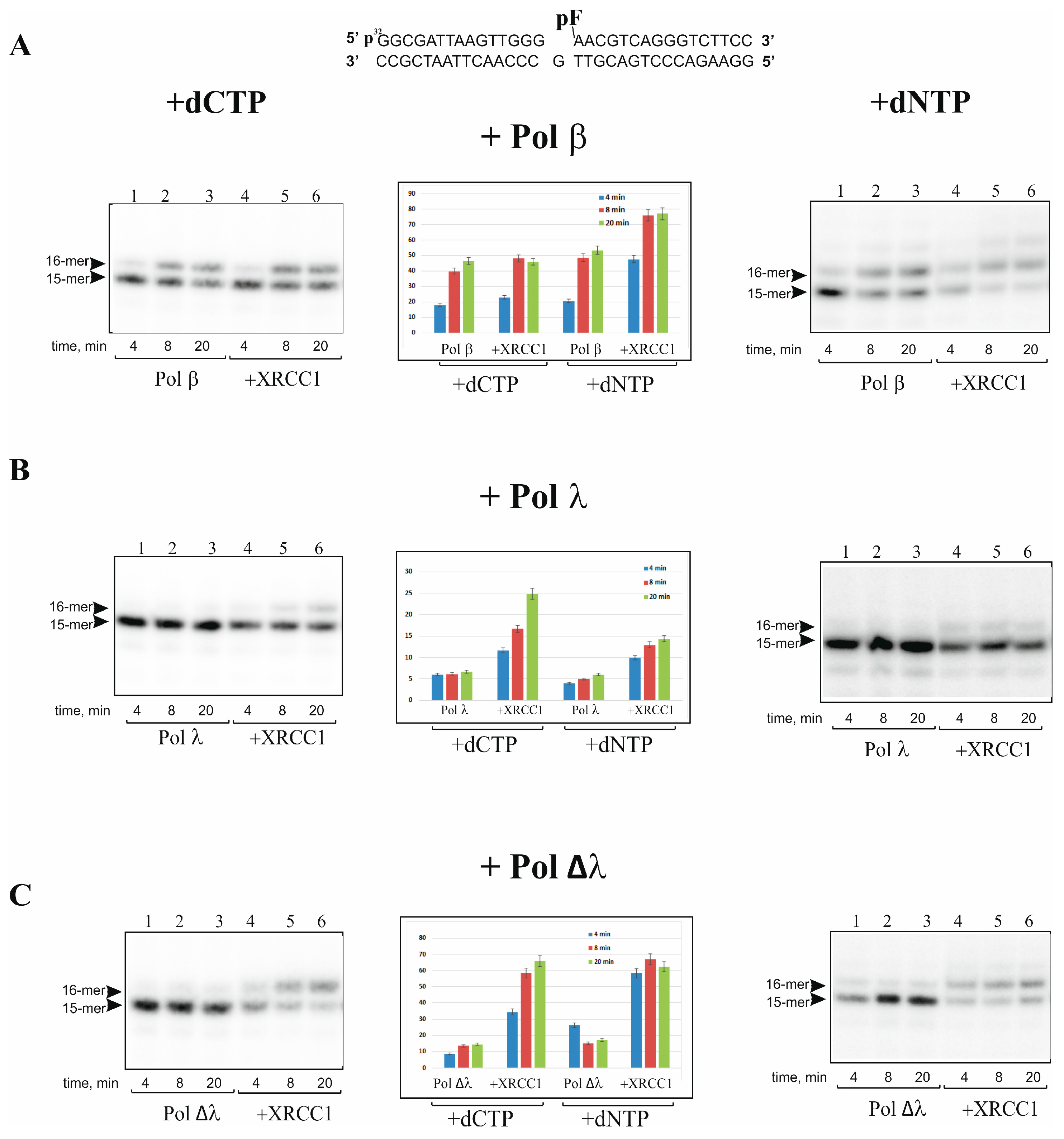
2.1. XRCC1 Stimulates Gap-Filling DNA Synthesis Catalyzed with Pol λ
2.2. Pol λ Binds the C-Terminal Region of XRCC1
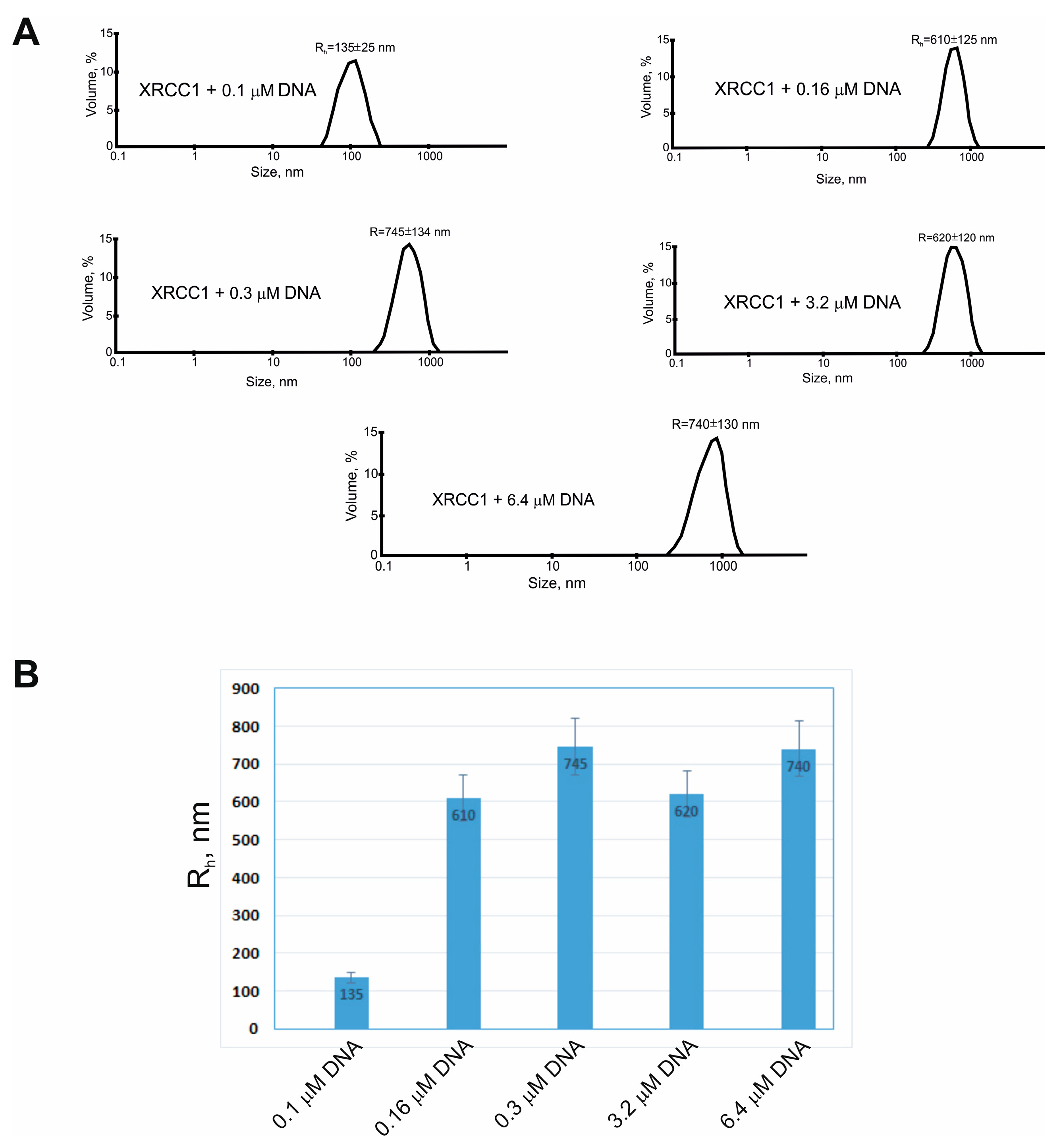
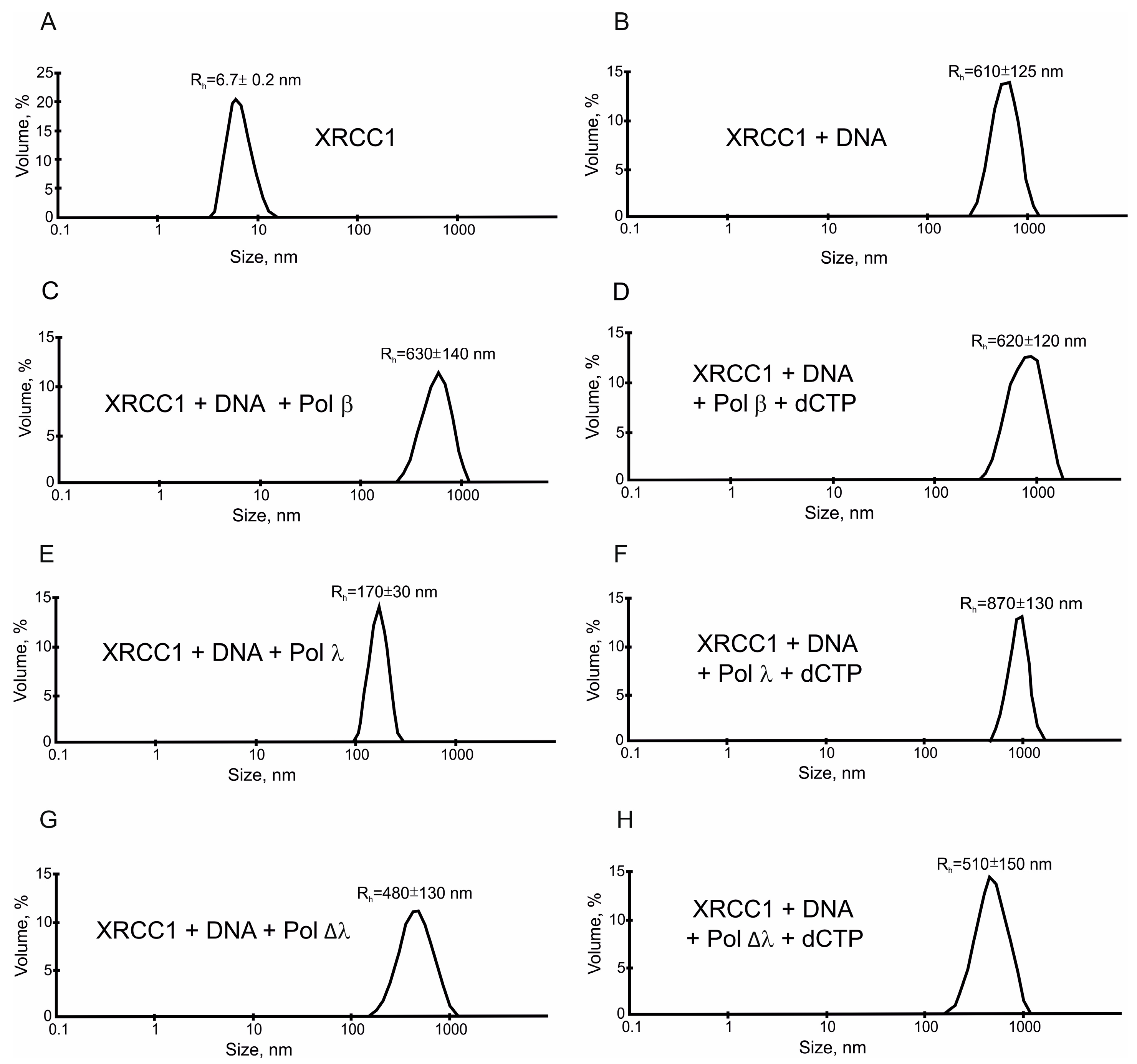
2.3. XRCC1 Can Form Microphases with DNA and DNA Polymerases
3. Discussion
4. Materials and Methods
4.1. Reagents, Plasmids, and Proteins
4.2. Radioactive Labeling of Oligonucleotides and Preparation of DNA Duplexes
4.3. The DNA Synthesis Assay
4.4. Fluorescent Labeling of XRCC1 and Pol β, λ, and Δλ
4.5. FRET Measurements
4.6. The Fluorescence Microscopy Assay
4.7. DLS Measurements
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beard, W.A.; Horton, J.K.; Prasad, R.; Wilson, S.H. Eukaryotic base excision repair: New approaches shine light on mechanism. Annu. Rev. Biochem. 2019, 88, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Singhal, R.K.; Prasad, R.; Wilson, S.H. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J. Biol. Chem. 1995, 270, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Sobol, R.W.; Horton, J.K.; Kuhn, R.; Gu, H.; Singhal, R.K.; Prasad, R.; Rajewsky, K.; Wilson, S.H. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature 1996, 379, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Dianov, G.L.; Prasad, R.; Wilson, S.H.; Bohr, V.A. Role of DNA polymerase beta in the excision step of long patch mammalian base excision repair. J. Biol. Chem. 1999, 274, 13741–13743. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.K.; Baker, A.; Berg, B.J.; Sobol, R.W.; Wilson, S.H. Involvement of DNA polymerase beta in protection against the cytotoxicity of oxidative DNA damage. DNA Repair 2002, 1, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Sukhanova, M.V.; Khodyreva, S.N.; Lebedeva, N.A.; Prasad, R.; Wilson, S.H.; Lavrik, O.I. Human base excision repair enzymes apurinic/apyrimidinic endonuclease1 (APE1), DNA polymerase beta and poly(ADP-ribose) polymerase 1: Interplay between strand-displacement DNA synthesis and proofreading exonuclease activity. Nucleic Acids Res. 2005, 33, 1222–1229. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Gao, Y. Translesion and Repair DNA Polymerases: Diverse Structure and Mechanism. Annu. Rev. Biochem. 2018, 87, 239–261. [Google Scholar] [CrossRef]
- Lee, J.W.; Blanco, L.; Zhou, T.; Garcia-Diaz, M.; Bebenek, K.; Kunkel, T.A.; Wang, Z.; Povirk, L.F. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004, 279, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Wu, X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem. Biophys. Res. Commun. 2004, 323, 1328–1333. [Google Scholar] [CrossRef]
- Braithwaite, E.K.; Kedar, P.S.; Lan, L.; Polosina, Y.Y.; Asagoshi, K.; Poltoratsky, V.P.; Horton, J.K.; Miller, H.; Teebor, G.W.; Yasui, A.; et al. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J. Biol. Chem. 2005, 280, 31641–31647. [Google Scholar] [CrossRef]
- Thapar, U.; Demple, B. Deployment of DNA polymerases beta and lambda in single-nucleotide and multinucleotide pathways of mammalian base excision DNA repair. DNA Repair 2019, 76, 11–19. [Google Scholar] [CrossRef]
- Braithwaite, E.K.; Prasad, R.; Shock, D.D.; Hou, E.W.; Beard, W.A.; Wilson, S.H. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J. Biol. Chem. 2005, 280, 18469–18475. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Diaz, M.; Dominguez, O.; Lopez-Fernandez, L.A.; de Lera, L.T.; Saniger, M.L.; Ruiz, J.F.; Parraga, M.; Garcia-Ortiz, M.J.; Kirchhoff, T.; del Mazo, J.; et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 2000, 301, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Aoufouchi, S.; Flatter, E.; Dahan, A.; Faili, A.; Bertocci, B.; Storck, S.; Delbos, F.; Cocea, L.; Gupta, N.; Weill, J.C.; et al. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 2000, 28, 3684–3693. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, K.; Kitamura, K.; Yasui, A.; Nimura, Y.; Ikeda, K.; Hirai, M.; Matsukage, A.; Nakanishi, M. Identification and characterization of human DNA polymerase beta 2, a DNA polymerase beta-related enzyme. J. Biol. Chem. 2000, 275, 31233–31238. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Diaz, M.; Bebenek, K.; Sabariegos, R.; Dominguez, O.; Rodriguez, J.; Kirchhoff, T.; Garcia-Palomero, E.; Picher, A.J.; Juarez, R.; Ruiz, J.F.; et al. DNA polymerase lambda, a novel DNA repair enzyme in human cells. J. Biol. Chem. 2002, 277, 13184–13191. [Google Scholar] [CrossRef] [PubMed]
- Shimazaki, N.; Yoshida, K.; Kobayashi, T.; Toji, S.; Tamai, K.; Koiwai, O. Over-expression of human DNA polymerase lambda in E. coli and characterization of the recombinant enzyme. Genes Cells 2002, 7, 639–651. [Google Scholar] [CrossRef]
- Blanca, G.; Shelev, I.; Ramadan, K.; Villani, G.; Spadari, S.; Hübscher, U.; Maga, G. Human DNA polymerase λ diverged in evolution from DNA polymerase β toward specific Mn++ dependence: A kinetic and thermodynamic study. Biochemistry 2003, 42, 7467–7476. [Google Scholar] [CrossRef]
- Fiala, K.A.; Duym, W.W.; Zhang, J.; Suo, Z. Up-regulation of the fidelity of human DNA polymerase lambda by its non-enzymatic proline-rich domain. J. Biol. Chem. 2006, 281, 19038–19044. [Google Scholar] [CrossRef]
- Mueller, G.A.; Moon, A.F.; DeRose, E.F.; Havener, J.M.; Ramsden, D.A.; Pedersen, L.C.; London, R.E. A comparison of BRCT domains involved in nonhomologous end-joining: Introducing the solution structure of the BRCT domain of polymerase lambda. DNA Repair 2008, 7, 1340–1351. [Google Scholar] [CrossRef]
- Bebenek, K.; Pedersen, L.C.; Kunkel, T.A. Structure-function studies of DNA polymerase λ. Biochemistry 2014, 53, 2781–2792. [Google Scholar] [CrossRef] [PubMed]
- Fiala, K.A.; Abdel-Gawad, W.; Suo, Z. Pre-steady-state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase lambda. Biochemistry 2004, 43, 6751–6762. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, K.; Shevelev, I.V.; Maga, G.; Hubscher, U. DNA polymerase lambda from calf thymus preferentially replicates damaged DNA. J. Biol. Chem. 2002, 277, 18454–18458. [Google Scholar] [CrossRef] [PubMed]
- Bebenek, K.; Garcia-Diaz, M.; Blanco, L.; Kunkel, T.A. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J. Biol. Chem. 2003, 278, 34685–34690. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Diaz, M.; Bebenek, K.; Gao, G.; Pedersen, L.C.; London, R.E.; Kunkel, T.A. Structure-function studies of DNA polymerase lambda. DNA Repair 2005, 4, 1358–1367. [Google Scholar] [CrossRef] [PubMed]
- Vasil’eva, I.A.; Moor, N.A.; Lavrik, O.I. Effect of Human XRCC1 Protein Oxidation on the Functional Activity of Its Complexes with the Key Enzymes of DNA Base Excision Repair. Biochemistry 2020, 85, 288–299. [Google Scholar] [CrossRef]
- Vasil’eva, I.A.; Anarbaev, R.O.; Moor, N.A.; Lavrik, O.I. Dynamic light scattering study of base excision DNA repair proteins and their complexes. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.-F.; Riccio, A.A.; Pascal, J.M. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014, 42, 7762–7775. [Google Scholar] [CrossRef]
- Kutuzov, M.M.; Khodyreva, S.N.; Amé, J.C.; Ilina, E.S.; Sukhanova, M.V.; Schreiber, V.; Lavrik, O.I. Interaction of PARP-2 with DNA structures mimicking DNA repair intermediates and consequences on activity of base excision repair proteins. Biochimie 2013, 95, 1208–1215. [Google Scholar] [CrossRef]
- Caldecott, K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008, 9, 619–631. [Google Scholar] [CrossRef]
- Moor, N.A.; Vasil’eva, I.A.; Anarbaev, R.O.; Antson, A.A.; Lavrik, O.I. Quantitative characterization of protein-protein complexes involved in base excision DNA repair. Nucleic Acids Res. 2015, 43, 6009–6022. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. XRCC1 protein; Form and function. DNA Repair 2019, 81, 102664. [Google Scholar] [CrossRef] [PubMed]
- Moor, N.A.; Lavrik, O.I. Protein-Protein Interactions in DNA Base Excision Repair. Biochemistry 2018, 83, 411–422. [Google Scholar] [CrossRef]
- Beernink, P.T.; Hwang, M.; Ramirez, M.; Murphy, M.B.; Doyle, S.A.; Thelen, M.P. Specificity of protein interactions mediated by BRCT domains of the XRCC1 DNA repair protein. J. Biol. Chem. 2005, 280, 30206–30213. [Google Scholar] [CrossRef] [PubMed]
- Gerloff, D.L.; Woods, N.T.; Farago, A.A.; Monteiro, A.N. BRCT domains: A little more than kin, and less than kind. FEBS Lett. 2012, 586, 2711–2716. [Google Scholar] [CrossRef] [PubMed]
- Rechkunova, N.I.; Zhdanova, P.V.; Lebedeva, N.A.; Maltseva, E.A.; Koval, V.V.; Lavrik, O.I. Structural features of DNA polymerases β and λ in complex with benzo[a]pyrene-adducted DNA cause a difference in lesion tolerance. DNA Repair 2022, 116, 103353. [Google Scholar] [CrossRef] [PubMed]
- Maltseva, E.A.; Rechkunova, N.I.; Lavrik, O.I. Non-Catalytic Domains of DNA Polymerase λ: Influence on Enzyme Activity and Its Regulation. Dokl. Biochem. Biophys. 2023, 512, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, N.A.; Rechkunova, N.I.; Dezhurov, S.V.; Khodyreva, S.N.; Favre, A.; Blanco, L.; Lavrik, O.I. Comparison of functional properties of mammalian DNA polymerase lambda and DNA polymerase beta in reactions of DNA synthesis related to DNA repair. Biochim. Biophys. Acta 2005, 1751, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Skosareva, L.V.; Lebedeva, N.A.; Rechkunova, N.I.; Kolbanovskiy, A.; Geacintov, N.E.; Lavrik, O.I. Human DNA polymerase λ catalyzes lesion bypass across benzo[a]pyrene-derived DNA adduct during base excision repair. DNA Repair 2012, 11, 367–373. [Google Scholar] [CrossRef]
- Liu, Y.; Beard, W.A.; Shock, D.D.; Prasad, R.; Hou, E.W.; Wilson, S.H. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J. Biol. Chem. 2005, 280, 3665–3674. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Kim, K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science 1995, 269, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Dianov, G.; Price, A.; Lindahl, T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell. Biol. 1992, 12, 1605–1612. [Google Scholar]
- Matsumoto, Y.; Kim, K.; Bogenhagen, D.F. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: An alternative pathway of base excision DNA repair. Mol. Cell. Biol. 1994, 14, 6187–6197. [Google Scholar] [PubMed]
- Tang, Q.; Çağlayan, M. The scaffold protein XRCC1 stabilizes the formation of polbeta/gap DNA and ligase III alpha/nick DNA complexes in base excision repair. J. Biol. Chem. 2021, 297, 101025. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Mo, W.; Park, J.; Lee, J.W.; Lee, S. Single-Molecule FRET Detection of Sub-Nanometer Distance Changes in the Range below a 3-Nanometer Scale. Biosensors 2020, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Sastre-Moreno, G.; Pryor, J.M.; Moreno-Oñate, M.; Herrero-Ruiz, A.M.; Cortés-Ledesma, F.; Blanco, L.; Ramsden, D.A.; Ruiz, J.F. Regulation of human pollambda by ATM-mediated phosphorylation during non-homologous end joining. DNA Repair 2017, 51, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Sawaya, M.R.; Pelletier, H.; Kumar, A.; Wilson, S.H.; Kraut, J. Crystal structure of rat DNA polymerase beta: Evidence for a common polymerase mechanism. Science 1994, 264, 1930–1935. [Google Scholar] [CrossRef] [PubMed]
- London, R.E. The structural basis of XRCC1-mediated DNA repair. DNA Repair 2015, 30, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Mani, R.S.; Karimi-Busheri, F.; Fanta, M.; Caldecott, K.W.; Cass, C.E.; Weinfeld, M. Biophysical characterization of human XRCC1 and its binding to damaged and undamaged DNA. Biochemistry 2004, 43, 16505–16514. [Google Scholar] [CrossRef]
- Cuneo, M.J.; London, R.E. Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity. Proc. Natl. Acad. Sci. USA 2010, 107, 6805–6810. [Google Scholar] [CrossRef]
- Dianova, I.I.; Sleeth, K.M.; Allinson, S.L.; Parsons, J.L.; Breslin, C.; Caldecott, K.W.; Dianov, G.L. XRCC1-DNA polymerase beta interaction is required for efficient base excision repair. Nucleic Acids Res. 2004, 32, 2550–2555. [Google Scholar] [CrossRef] [PubMed]
- Sukhanova, M.V.; Anarbaev, R.O.; Maltseva, E.A.; Pastré, D.; Lavrik, O.I. FUS Microphase Separation: Regulation by Nucleic Acid Polymers and DNA Repair Proteins. Int. J. Mol. Sci. 2022, 23, 13200. [Google Scholar] [CrossRef] [PubMed]
- Mok, M.C.Y.; Campalans, A.; Pillon, M.C.; Guarné, A.; Radicella, J.P.; Junop, M.S. Identification of an XRCC1 DNA binding activity essential for retention at sites of DNA damage. Sci. Rep. 2019, 9, 3095. [Google Scholar] [CrossRef]
- Wilson, S.H.; Kunkel, T.A. Passing the baton in base excision repair. Nat. Struct. Biol. 2000, 7, 176–178. [Google Scholar] [CrossRef]
- Prasad, R.; Beard, W.A.; Batra, V.K.; Liu, Y.; Shock, D.D.; Wilson, S.H. A review of recent experiments on step-to-step “hand-off” of the DNA intermediates in mammalian base excision repair pathways. Mol. Biol. 2011, 45, 586–600. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Wilson III, D.M. Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 2012, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Lavrik, O.I. PARPs’ impact on base excision DNA repair. DNA Repair 2020, 93, 102911. [Google Scholar] [CrossRef] [PubMed]
- London, R.E. XRCC1—Strategies for coordinating and assembling a versatile DNA damage response. DNA Repair 2020, 93, 102917. [Google Scholar] [CrossRef]
- Chaudhuri, A.R.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodeling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Horton, J.K.; Watson, M.; Stefanick, D.F.; Shaughnessy, D.T.; Taylor, J.A.; Wilson, S.H. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008, 18, 48–63. [Google Scholar] [CrossRef]
- Caldecott, K.W. Mammalian DNA base excision repair: Dancing in the moonlight. DNA Repair 2020, 93, 102921. [Google Scholar] [CrossRef] [PubMed]
- Abbotts, R.; Wilson, D.M. 3rd. Coordination of DNA single strand break repair. Free Radic. Biol. Med. 2017, 107, 228–244. [Google Scholar] [CrossRef]
- Altmeyer, M.; Neelsen, K.J.; Teloni, F.; Pozdnyakova, I.; Pellegrino, S.; Grøfte, M.; Rask, M.D.; Streicher, W.; Jungmichel, S.; Nielsen, M.L.; et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 2015, 19, 8088. [Google Scholar] [CrossRef]
- Singatulina, A.S.; Hamon, L.; Sukhanova, M.V.; Desforges, B.; Joshi, V.; Bouhss, A.; Lavrik, O.I.; Pastré, D. PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep. 2019, 27, 1809–1821.e5. [Google Scholar] [CrossRef] [PubMed]
- Miné-Hattab, J.; Liu, S.; Taddei, A. Repair Foci as Liquid Phase Separation: Evidence and Limitations. Genes 2022, 13, 1846. [Google Scholar] [CrossRef] [PubMed]
- Dall’Agnese, G.; Dall’Agnese, A.; Banani, S.F.; Codrich, M.; Malfatti, M.C.; Antoniali, G.; Tell, G. Role of condensates in modulating DNA repair pathways and its implication for chemoresistance. J. Biol. Chem. 2023, 299, 104800. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.Y.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.M.; et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, D.M.; Kriwacki, R.W. Cell Phase separation in biology; functional organization of a higher order. Commun. Signal. 2016, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Hammel, M.; Rashid, I.; Sverzhinsky, A.; Pourfarjam, Y.; Tsai, M.S.; Ellenberger, T.; Pascal, J.M.; Kim, I.K.; Tainer, J.A.; Tomkinson, A.E. An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex. Nucleic Acids Res. 2021, 49, 306–321. [Google Scholar] [CrossRef]
- Marintchev, A.; Mullen, M.A.; Maciejewski, M.W.; Pan, B.; Gryk, M.R.; Mullen, G.P. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat. Struct. Biol. 1999, 6, 884–893. [Google Scholar]
- El-Khamisy, S.F.; Masutani, M.; Suzuki, H.; Caldecott, K.W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003, 31, 5526–5533. [Google Scholar] [CrossRef] [PubMed]
- Hanzlikova, H.; Gittens, W.; Krejcikova, K.; Zeng, Z.; Caldecott, K.W. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017, 45, 2546–2557. [Google Scholar] [CrossRef] [PubMed]
- Campalans, A.; Kortulewski, T.; Amouroux, R.; Menoni, H.; Vermeulen, W.; Radicella, J.P. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res. 2013, 41, 3115–3129. [Google Scholar] [CrossRef] [PubMed]
- Veith, S.; Schink, A.; Engbrecht, M.; Mack, M.; Rank, L.; Rossatti, P.; Hakobyan, M.; Goly, D.; Hefele, T.; Frensch, M.; et al. PARP1 regulates DNA damage-induced nucleolar-nucleoplasmic shuttling of WRN and XRCC1 in a toxicant and protein-specific manner. Sci. Rep. 2019, 9, 10075. [Google Scholar] [CrossRef] [PubMed]
- Hanssen-Bauer, A.; Solvang-Garten, K.; Sundheim, O.; Peña-Diaz, J.; Andersen, S.; Slupphaug, G.; Krokan, H.E.; Wilson, D.M., 3rd; Akbari, M.; Otterlei, M. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ. Mol. Mutagen. 2011, 52, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Solarczyk, K.J.; Kordon, M.; Berniak, K.; Dobrucki, J.W. Two stages of XRCC1 recruitment and two classes of XRCC1 foci formed in response to low level DNA damage induced by visible light, or stress triggered by heat shock. DNA Repair 2016, 37, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Drachkova, I.A.; Petruseva, I.O.; Safronov, I.V.; Zakharenko, A.I.; Shishkin, G.V.; Lavrik, O.I.; Khodyreva, S.N. Reagents for modification of protein–nucleic acids complexes with primers elongated by the dCTP exo-N-substituted arylazido derivatives. Russ. J. Bioorg. Chem. 2001, 27, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Belousova, E.A.; Vasil’eva, I.A.; Moor, N.A.; Zatsepin, T.S.; Oretskaya, T.S.; Lavrik, O.I. Clustered DNA lesions containing 5-formyluracil and AP site: Repair via the BER system. PLoS ONE 2013, 8, e68576. [Google Scholar] [CrossRef][Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Lalonde, S.; Ehrhardt, D.W.; Loqu’e, D.; Chen, J.; Rhee, S.Y.; Frommer, W.B. Molecular and cellular approaches for the detection of protein-protein interactions: Latest techniques and current limitations. Plant J. 2008, 53, 610–635. [Google Scholar] [CrossRef]
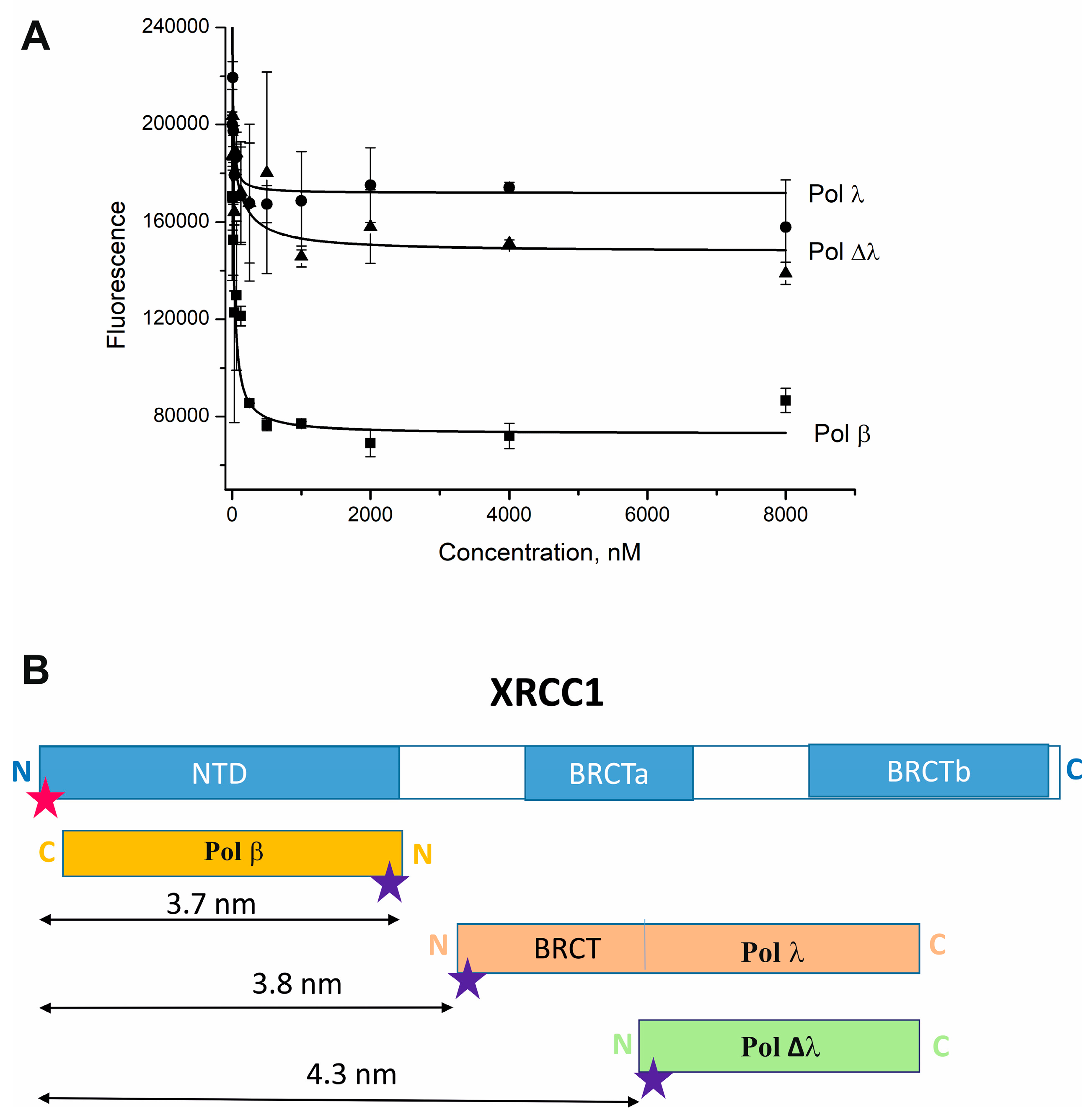


| Labeled Protein | Kd, nM | E | R, nm |
|---|---|---|---|
| Cy5-Pol β | 29 ± 12 | 0.91 ± 0.06 | 3.7 ± 0.2 |
| Cy5-Pol λ | 9.6 ± 4.0 | 0.90 ± 0.05 | 3.8 ± 0.2 |
| Cy5-Pol Δλ | 139 ± 14 | 0.80 ± 0.05 | 4.3 ± 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebedeva, N.A.; Anarbaev, R.O.; Maltseva, E.A.; Sukhanova, M.V.; Rechkunova, N.I.; Lavrik, O.I. DNA Repair Protein XRCC1 Stimulates Activity of DNA Polymerase λ under Conditions of Microphase Separation. Int. J. Mol. Sci. 2024, 25, 6927. https://doi.org/10.3390/ijms25136927
Lebedeva NA, Anarbaev RO, Maltseva EA, Sukhanova MV, Rechkunova NI, Lavrik OI. DNA Repair Protein XRCC1 Stimulates Activity of DNA Polymerase λ under Conditions of Microphase Separation. International Journal of Molecular Sciences. 2024; 25(13):6927. https://doi.org/10.3390/ijms25136927
Chicago/Turabian StyleLebedeva, Natalia A., Rashid O. Anarbaev, Ekaterina A. Maltseva, Maria V. Sukhanova, Nadejda I. Rechkunova, and Olga I. Lavrik. 2024. "DNA Repair Protein XRCC1 Stimulates Activity of DNA Polymerase λ under Conditions of Microphase Separation" International Journal of Molecular Sciences 25, no. 13: 6927. https://doi.org/10.3390/ijms25136927
APA StyleLebedeva, N. A., Anarbaev, R. O., Maltseva, E. A., Sukhanova, M. V., Rechkunova, N. I., & Lavrik, O. I. (2024). DNA Repair Protein XRCC1 Stimulates Activity of DNA Polymerase λ under Conditions of Microphase Separation. International Journal of Molecular Sciences, 25(13), 6927. https://doi.org/10.3390/ijms25136927







