Analysis of Circulating miRNA Expression Profiles in Type 2 Diabetes Patients with Diabetic Foot Complications
Abstract
:1. Introduction
2. Results
2.1. Characteristics of the Study Population
2.2. miR Expression in Plasma
2.3. Correlation between miR Expression Levels and Biochemical and Anthropometric Values
2.4. Comorbidities and miR Expression
2.5. Association of Clinical Parameters and miR Expression with Diabetic Foot
3. Discussion
4. Materials and Methods
4.1. Patient Population
4.2. Blood and Serum Samples
4.3. Lipid Profile
4.4. Definition
4.5. miRs Extraction
4.6. Quantification of miRs by Real-Time PCR
4.7. In Silico Analyses
4.8. Statical Analyses
4.9. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Ozdemir, D.; Feinberg, M.W. MicroRNAs in diabetic wound healing: Pathophysiology and therapeutic opportunities. Trends. Cardiovasc. Med. 2019, 29, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Van Netten, J.J.; Baba, M.; Lazzarini, P.A. Epidemiology of diabetic foot disease and diabetes-related lower-extremity amputation in Australia: A systematic review protocol. Syst. Rev. 2017, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Boulton, A.J.M. The Pathway to Foot Ulceration in Diabetes. Med. Clin. N. Am. 2013, 97, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Ascencio-Montiel, I.d.J. 10 years Analysis of Diabetes-related Major Lower Extremity Amputations in Mexico. Arch. Med. Res. 2018, 49, 58–64. [Google Scholar] [CrossRef]
- Martínez-Escalante, J.E.; Romero-Ibargüengoitia, M.E.; Plata-Álvarez, H.; López-Betancourt, G.; Otero-Rodríguez, R.; Garza-Cantú, A.A.; Corral-Guerrero, M.E. Pie diabético en México: Factores de riesgo para mortalidad posterior a una amputación mayor, a 5 años, en un hospital de salud pública de segundo nivel. Cirugía Cir. 2021, 89, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.G.; Wrobel, J.; Robbins, J.M. Guest Editorial: Are diabetes-related wounds and amputations worse than cancer? Int. Wound J. 2007, 4, 286–287. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.Y.; Prins, J.B.; Marwick, T.H. Diabetic Cardiomyopathy: Evidence, Mechanisms, and Therapeutic Implications. Endocr. Rev. 2004, 25, 543–567. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wu, N.N.; Wang, S.; Sowers, J.R.; Zhang, Y. Obesity cardiomyopathy: Evidence, mechanisms, and therapeutic implications. Physiol. Rev. 2021, 101, 1745–1807. [Google Scholar] [CrossRef]
- Tang, Y.-B.; Uwimana, M.M.P.; Zhu, S.-Q.; Zhang, L.-X.; Wu, Q.; Liang, Z.-X. Non-coding RNAs: Role in diabetic foot and wound healing. World J. Diabetes 2022, 13, 1001–1013. [Google Scholar] [CrossRef]
- Yan, C.; Chen, J.; Wang, C.; Yuan, M.; Kang, Y.; Wu, Z.; Li, W.; Zhang, G.; Machens, H.G.; Rinkevich, Y.; et al. Milk exosomes-mediated miR-31-5p delivery accelerates diabetic wound healing through promoting angiogenesis. Drug Deliv. 2022, 29, 214–228. [Google Scholar] [CrossRef]
- Petkovic, M.; Sørensen, A.E.; Leal, E.C.; Carvalho, E.; Dalgaard, L.T. Mechanistic Actions of microRNAs in Diabetic Wound Healing. Cells 2020, 9, 2228. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, M.; Iwamoto, K.; Naito, A.; Imasato, M.; Hyuga, S.; Nakahara, Y.; Mikamori, M.; Furukawa, K.; Moon, J.; Asaoka, T.; et al. Circulating MicroRNAs in Gastrointestinal Cancer. Cancers 2021, 13, 3348. [Google Scholar] [CrossRef] [PubMed]
- Deiuliis, J.A. MicroRNAs as regulators of metabolic disease: Pathophysiologic significance and emerging role as biomarkers and therapeutics. Int. J. Obes. 2016, 40, 88–101. [Google Scholar] [CrossRef]
- Eliasson, L.; Esguerra, J.L.S. MicroRNA Networks in Pancreatic Islet Cells: Normal Function and Type 2 Diabetes. Diabetes 2020, 69, 804–812. [Google Scholar] [CrossRef]
- Morales-Sánchez, P.; Lambert, C.; Ares-Blanco, J.; Suárez-Gutiérrez, L.; Villa-Fernández, E.; Garcia, A.V.; García-Villarino, M.; Tejedor, J.R.; Fraga, M.F.; Torre, E.M.; et al. Circulating miRNA expression in long-standing type 1 diabetes mellitus. Sci. Rep. 2023, 13, 8611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Q.; Gui, L.; Cai, Y.; Deng, X.; Li, C.; Guo, Q.; He, X.; Huang, J. Let-7e inhibits TNF-α expression by targeting the methyl transferase EZH2 in DENV2-infected THP-1 cells. J. Cell Physiol. 2018, 233, 8605–8616. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, T.; Wang, S.; Wei, J.; Fan, J.; Li, J.; Han, Q.; Liao, L.; Shao, C.; Zhao, R.C.; et al. miR-17-5p and miR-106a are involved in the balance between osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Stem. Cell Res. 2013, 10, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 Contributes to the Regulation of Cholesterol Homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Torres-Paz, Y.E.; Huesca-Gómez, C.; Sánchez-Muñoz, F.; Martínez-Alvarado, R.; Soto, M.A.E.; Torres-Tamayo, M.; Fuentevilla-Álvarez, G.; Gamboa, R. Increased expression of miR-33a in monocytes from Mexican hypertensive patients in elevated carotid intima-media thickness. J. Hum. Hypertens. 2018, 32, 681–690. [Google Scholar] [CrossRef]
- Li, H.; Zhou, J.; Wei, X.; Chen, R.; Geng, J.; Zheng, R.; Chai, J.; Li, F.; Jiang, S. miR-144 and targets, c-fos and cyclooxygenase-2 (COX2), modulate synthesis of PGE2 in the amnion during pregnancy and labor. Sci. Rep. 2016, 6, 27914. [Google Scholar] [CrossRef]
- Su, Z.; Si, W.; Li, L.; Zhou, B.; Li, X.; Xu, Y.; Xu, C.; Jia, H.; Wang, Q.K. MiR-144 regulates hematopoiesis and vascular development by targeting meis1 during zebrafish development. Int. J. Biochem. Cell Biol. 2014, 49, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Falanga, V. Wound healing and its impairment in the diabetic foot. Lancet 2005, 366, 1736–1743. [Google Scholar] [CrossRef]
- Agbu, P.; Carthew, R.W. MicroRNA-mediated regulation of glucose and lipid metabolism. Nat. Rev. Mol. Cell Biol. 2021, 22, 425–438. [Google Scholar] [CrossRef]
- Nishiga, M.; Horie, T.; Kuwabara, Y.; Nagao, K.; Baba, O.; Nakao, T.; Nishino, T.; Hakuno, D.; Nakashima, Y.; Nishi, H.; et al. MicroRNA-33 Controls Adaptive Fibrotic Response in the Remodeling Heart by Preserving Lipid Raft Cholesterol. Circ. Res. 2017, 120, 835–847. [Google Scholar] [CrossRef]
- Ouimet, M.; Ediriweera, H.N.; Gundra, U.M.; Sheedy, F.J.; Ramkhelawon, B.; Hutchison, S.B.; Rinehold, K.; van-Solingen, C.; Fullerton, M.D.; Cecchini, K.; et al. MicroRNA-33–dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Investig. 2015, 125, 4334–4348. [Google Scholar] [CrossRef]
- Ho, P.-C.; Chang, K.-C.; Chuang, Y.-S.; Wei, L.-N. Cholesterol regulation of receptor-interacting protein 140 via microRNA-33 in inflammatory cytokine production. FASEB J. 2011, 25, 1758–1766. [Google Scholar] [CrossRef]
- Price, N.L.; Singh, A.K.; Rotllan, N.; Goedeke, L.; Wing, A.; Canfrán-Duque, A.; Diaz-Ruiz, A.; Araldi, E.; Baldán, Á.; Camporez, J.P.; et al. Genetic Ablation of miR-33 Increases Food Intake, Enhances Adipose Tissue Expansion, and Promotes Obesity and Insulin Resistance. Cell Rep. 2018, 22, 2133–2145. [Google Scholar] [CrossRef]
- Shahouzehi, B.; Eghbalian, M.; Fallah, H.; Aminizadeh, S.; Masoumi-Ardakani, Y. Serum microRNA-33 levels in pre-diabetic and diabetic patients. Mol. Biol. Rep. 2021, 48, 4121–4128. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Sheedy, F.J.; Esau, C.C.; Hussain, F.N.; Temel, R.E.; Parathath, S.; van-Gils, J.M.; Rayner, A.J.; Chang, A.N.; Suarez, Y.; et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Investig. 2011, 121, 2921–2931. [Google Scholar] [CrossRef]
- Li, Z.; Rong, Y.; Zhang, Y. MiR-33-5p alleviates spinal cord injury in rats and protects <scp>PC12</scp> cells from lipopolysaccharide-induced apoptosis. Kaohsiung J. Med. Sci. 2023, 39, 52–60. [Google Scholar] [CrossRef]
- Pan, Q.; Xu, X.; He, W.; Wang, Y.; Xiang, Z.; Jin, X.; Tang, Q.; Zhao, T.; Ma, X. Enrichment of miR-17-5p enhances the protective effects of EPC-EXs on vascular and skeletal muscle injury in a diabetic hind limb ischemia model. Biol. Res. 2023, 56, 16. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Chen, K.; Sun, R.; Lin, K.; Qian, X.; Yuan, M.; Wang, Y.; Ma, J.; Qing, Y.; Xu, J.; et al. Glucose impairs angiogenesis and promotes ventricular remodelling following myocardial infarction via upregulation of microRNA-17. Exp. Cell Res. 2019, 381, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Hromadnikova, I.; Kotlabova, K.; Dvorakova, L.; Krofta, L. Postpartum profiling of microRNAs involved in pathogenesis of cardiovascular/cerebrovascular diseases in women exposed to pregnancy-related complications. Int. J. Cardiol. 2019, 291, 158–167. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Kotlabova, K.; Hympanova, L.; Krofta, L. Cardiovascular and Cerebrovascular Disease Associated microRNAs Are Dysregulated in Placental Tissues Affected with Gestational Hypertension, Preeclampsia and Intrauterine Growth Restriction. PLoS ONE 2015, 10, e0138383. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, I.; Sotoda, Y.; Eguchi, R. Contribution of platelet-derived microRNAs to serum microRNAs in healthy men. Platelets 2021, 32, 984–987. [Google Scholar] [CrossRef] [PubMed]
- Dangwal, S.; Stratmann, B.; Bang, C.; Lorenzen, J.M.; Kumarswamy, R.; Fiedler, J.; Falk, C.S.; Scholz, C.J.; Thum, T.; Tschoepe, D. Impairment of Wound Healing in Patients With Type 2 Diabetes Mellitus Influences Circulating MicroRNA Patterns via Inflammatory Cytokines. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1480–1488. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Ampofo, E.; Menger, M.D.; Laschke, M.W. miR-191 suppresses angiogenesis by activation of NF-kB signaling. FASEB J. 2017, 31, 3321–3333. [Google Scholar] [CrossRef] [PubMed]
- Hsu, A.; Chen, S.-J.; Chang, Y.-S.; Chen, H.-C.; Chu, P.-H. Systemic Approach to Identify Serum microRNAs as Potential Biomarkers for Acute Myocardial Infarction. BioMed Res. Int. 2014, 2014, 418628. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, X.; Huang, J.; Sun, Q.; Wang, L. Clinical impact of circulating miR-26a, miR-191, and miR-208b in plasma of patients with acute myocardial infarction. Eur. J. Med. Res. 2015, 20, 58. [Google Scholar] [CrossRef]
- Yang, X.; Du, X.; Ma, K.; Li, G.; Liu, Z.; Rong, W.; Miao, H.; Zhu, F.; Cui, Q.; Wu, S.; et al. Circulating miRNAs Related to Long-term Adverse Cardiovascular Events in STEMI Patients: A Nested Case-Control Study. Can. J. Cardiol. 2021, 37, 77–85. [Google Scholar] [CrossRef]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma MicroRNA Profiling Reveals Loss of Endothelial MiR-126 and Other MicroRNAs in Type 2 Diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014, 51, 823–831. [Google Scholar] [CrossRef]
- Barutta, F.; Bruno, G.; Matullo, G.; Chaturvedi, N.; Grimaldi, S.; Schalkwijk, C.; Stehouwer, C.D.; Fuller, J.H.; Gruden, G. MicroRNA-126 and micro-/macrovascular complications of type 1 diabetes in the EURODIAB Prospective Complications Study. Acta Diabetol. 2017, 54, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Lucena, R.; Camargo, A.; Alcalá-Diaz, J.F.; Romero-Baldonado, C.; Luque, R.M.; van Ommen, B.; Delgado-Lista, J.; Ordovás, J.M.; Pérez-Martínez, P.; Rangel-Zúñiga, O.A.; et al. A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: From the CORDIOPREV study. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef]
- De Rosa, S.; Arcidiacono, B.; Chiefari, E.; Brunetti, A.; Indolfi, C.; Foti, D.P. Type 2 Diabetes Mellitus and Cardiovascular Disease: Genetic and Epigenetic Links. Front. Endocrinol. 2018, 9, 334630. [Google Scholar] [CrossRef]
- Ueta, M.; Nishigaki, H.; Komai, S.; Mizushima, K.; Tamagawa-Mineoka, R.; Naito, Y.; Katoh, N.; Sotozono, C.; Kinoshita, S. Positive regulation of innate immune response by miRNA-let-7a-5p. Front. Genet. 2022, 13, 1025539. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yoon, S.R.; Kim, O.Y. miR-Let7A Controls the Cell Death and Tight Junction Density of Brain Endothelial Cells under High Glucose Condition. Oxid. Med. Cell Longev. 2017, 2017, 6051874. [Google Scholar] [CrossRef] [PubMed]
- Mononen, N.; Lyytikäinen, L.-P.; Seppälä, I.; Mishra, P.P.; Juonala, M.; Waldenberger, M.; Klopp, N.; Illig, T.; Leiviskä, J.; Loo, B.M.; et al. Whole blood microRNA levels associate with glycemic status and correlate with target mRNAs in pathways important to type 2 diabetes. Sci. Rep. 2019, 9, 8887. [Google Scholar] [CrossRef]
- Karolina, D.S.; Armugam, A.; Tavintharan, S.; Wong, M.T.K.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS ONE 2011, 6, e22839. [Google Scholar] [CrossRef]
- Okamura, T.; Okada, H.; Hashimoto, Y.; Majima, S.; Senmaru, T.; Nakanishi, N.; Asano, M.; Yamazaki, M.; Hamaguchi, M.; Fukui, M. Let-7e-5p Regulates IGF2BP2, and Induces Muscle Atrophy. Front. Endocrinol. 2021, 12, 791363. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, B.; Shang, J.; Wang, Y.; Jia, L.; She, X.; Xu, X.; Zhang, D.; Guo, J.; Zhang, F. Diabetic and nondiabetic BMSC–derived exosomes affect bone regeneration via regulating miR-17-5p/SMAD7 axis. Int. Immunopharmacol. 2023, 125, 111190. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Gupta, V.; Mahapatra, N.R. Key regulatory miRNAs in lipid homeostasis: Implications for cardiometabolic diseases and development of novel therapeutics. Drug Discov. Today 2022, 27, 2170–2180. [Google Scholar] [CrossRef] [PubMed]
- Fachim, H.A.; Loureiro, C.M.; Siddals, K.; Dalton, C.F.; Reynolds, G.P.; Gibson, J.M.; Chen, Z.B.; Heald, A.H. Circulating microRNA changes in patients with impaired glucose regulation. Adipocyte 2020, 9, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Ortega, R.; Liu, B.; Persaud, S.J. Effects of miR-33 Deficiency on Metabolic and Cardiovascular Diseases: Implications for Therapeutic Intervention. Int. J. Mol. Sci. 2023, 24, 10777. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association. World Medical Association Declaration of Helsinki. JAMA 2013, 310, 2191. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Salinas, C.A.; Olaiz, G.; Valles, V.; Torres, J.M.R.; Pérez, F.J.G.; Rull, J.A.; Rojas, R.; Franco, A.; Sepulveda, J. High prevalence of low HDL cholesterol concentrations and mixed hyperlipidemia in a Mexican nationwide survey. J. Lipid Res. 2001, 42, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Mexican Official Norm NOM-037-SSA2-2012. For the prevention, treatment and control of dyslipidemias. Rev. Mex. Cardiol. 2012, 23, 91–124.
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]

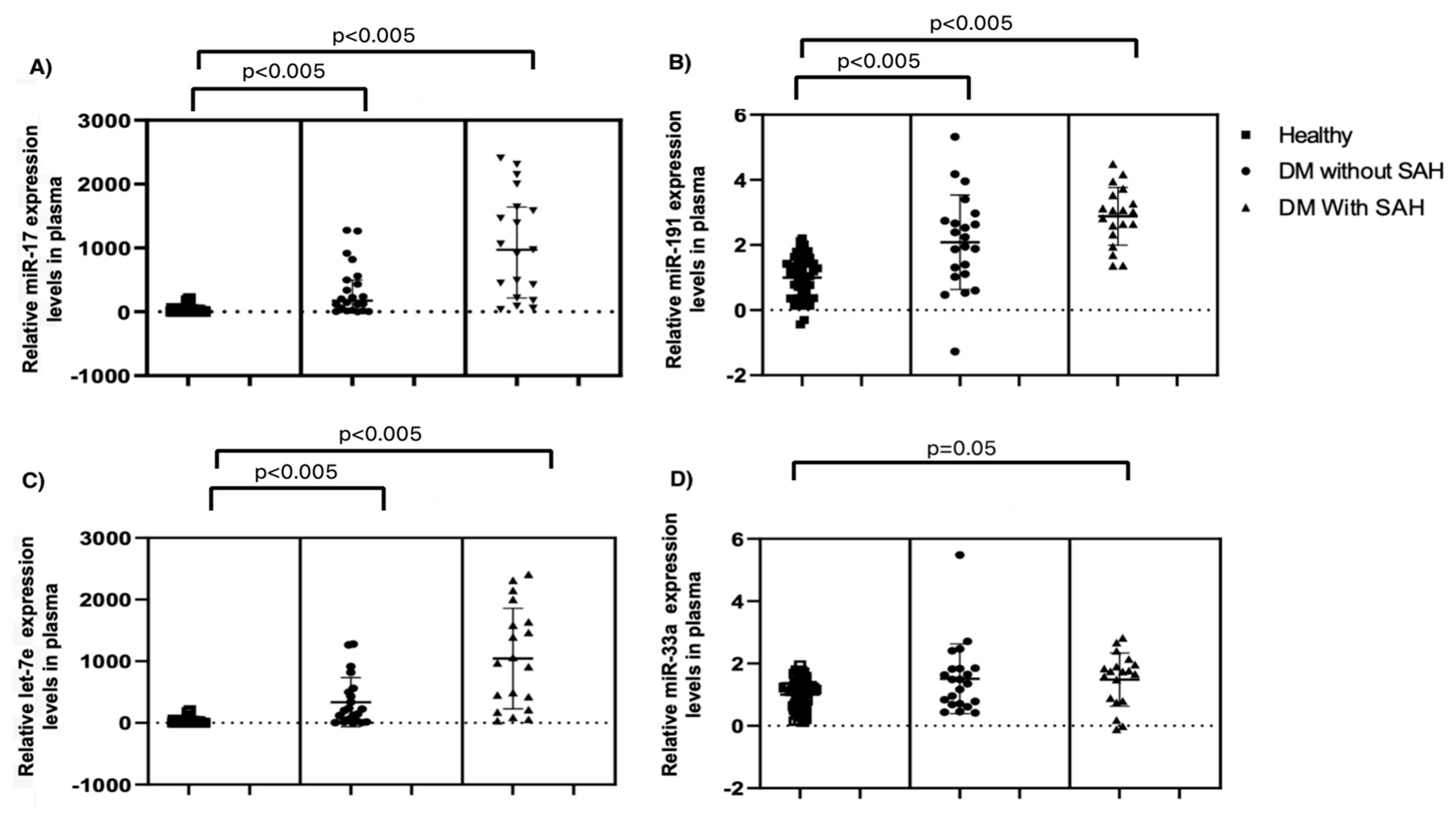

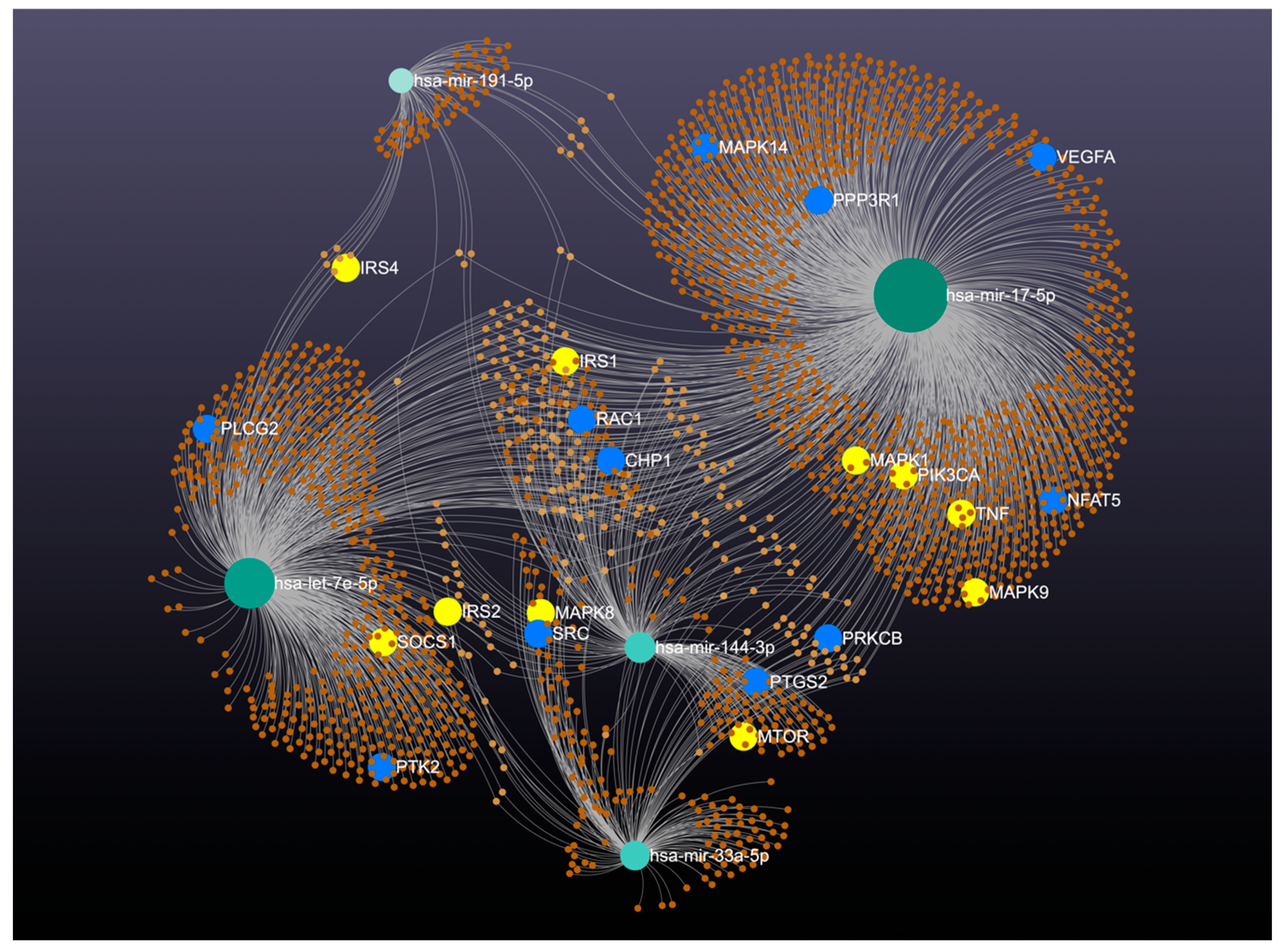
| Variables | Diabetic Foot (n = 41) | Control (n = 50) | p |
|---|---|---|---|
| Age | 55.71 ± 13.50 | 47.8 ± 5.11 | 0.0001 |
| Weight (kg) | 76.48 ± 16.12 | 69.90 ± 11.59 | 0.028 |
| BMI | 28.92 ± 5.56 | 25.10 ± 2.91 | 0.001 |
| Total Cholesterol (mg/dL) | 228.54 ± 45.37 | 186.22± 37.57 | 0.001 |
| HDL-C (mg/dL) | 34.61 ± 2.95 | 46.48 ± 12.61 | 0.001 |
| LDL-C (mg/dL) | 132. 71 ± 41.16 | 113.47 ± 32.25 | 0.01 |
| Triglycerides (mg/dL) | 136.05 ± 18.48 | 137.28 ± 77.97 | 0.92 |
| Glucose (mg/dL) | 134.78 ± 58.00 | 92.60 ± 8.86 | 0.001 |
| Diabetes % (n) | 100% | 0% | - |
| DMH | 65% | 0% | |
| HTA % (n) | 57.5% | 0% | - |
| SBP (mmHg) | 124.46 ± 15.32 | 127.30 ± 20.22 | 0.476 |
| DBP (mmHg) | 73.78 ± 9.53 | 86.40 ± 20.40 | 0.001 |
| HR (beats/min) | 105.02 ± 30.21 | 77.02 ± 12.11 | 0.001 |
| Smoking % (n) | 24.8% | 8.2% | - |
| Alcoholism % (n) | 35% | 69% | - |
| Controls without Comorbidities | Patients with Diabetes Median (min.–max.) Q1, Q2, Q3 | |||||
|---|---|---|---|---|---|---|
| Without Obesity n = 12 | With Obesity n = 25 | p1 | p2 | p3 | ||
| miR-17 | 0.82 (0.36–2.32) 0.54-0.82-1.50 | 2.34 (1.2–3.3) 2.1-2.3-3.1 | 2.69 (0.72–3.33) 1.7-2.6-3.1 | 0.000 | 0.000 | 0.000 |
| miR-191 | 1.10 (−0.44–2.20) 0.37-1.10-1.45 | 2.6 (1.2–5.3) 1.6-2.6-3.8 | 2.6 (0.47–4.4) 1.6-2.6-3.0 | 0.0001 | 0.0001 | 0.000 |
| let-7e | 4.0 (−0.52–2.03) 0.71-1.0-1.48 | 2.3 (1.5–4.5) 1.3-2.3-3.2 | 1.9 (0.16–3.59) 0.99-1.9-2.6 | 0.0001 | 0.0001 | 0.001 |
| miR33a | 1.11 (0.12–1.94) 0.61-1.11-1.31 | 1.7 (0.44–2.8) 0.79-1.7-2.6 | 1.4 (0.02–5.4) 0.73-1.4-1.8 | 0.003 | 0.003 | 0.03 |
| miR1-44 | 1.03 (0.23–2.10) 0.67-1.03-1.30 | 1.3 (2.0–2.6) 0.89-1.31-1.9 | 0.96 (0.13–2.5) 0.67-0.96-1.7 | - | - | - |
| Without SAH n = 22 | With SAH n = 19 | |||||
| miR-17 | 0.82 (0.36–2.32) 0.54-0.82-1.50 | 2.2 (1.2–3.1) 1.3-2.2-2.7 | 2.9 (1.5–3.3) 2.3-2.9-3.2 | 0.003 | 0.003 | 0.003 |
| miR-191 | 1.10 (−0.44–2.20) 0.37-1.10-1.45 | 2.09 (1.2–5.3) 1.0-2.0-2.7 | 3.0 (1.3–4.4) 2.3-3.0-3.5 | 0.02 | 0.02 | 0.02 |
| let-7e | 4.0 (−0.52–2.03) 0.71-1.0-1.48 | 1.7 (1.5–4.5) 0.79-1.71-2.3 | 2.5 (0.27–3.59) 1.5-2.5-2.9 | 0.02 | 0.02 | 0.02 |
| miR33a | 1.11 (0.12–1.94) 0.61-1.11-1.31 | 1.4 (0.42–5.4) 0.70-1.4-1.8 | 1.7 (0.11–2.8) 0.79-1.74-1.96 | - | - | - |
| miR1-44 | 1.03 (0.23–2.10) 0.67-1.03-1.30 | 0.91 (2.0–2.02) 0.67-0.91-1.2 | 1.4 (0.3–2.6) 0.78-1.4-2.0 | 0.03 | - | 0.0001 |
| Without dyslipidemia n = 26 | With dyslipidemia n = 15 | |||||
| miR-17 | 0.82 (0.36–2.32) 0.54-0.82-1.50 | 2.5 (1.2–3.3) 1.7-2.5-3.1 | 2.6 (0.72–3.3) 2.0-2.6-3.0 | - | - | - |
| miR-191 | 1.10 (−0.44–2.20) 0.37-1.10-1.45 | 2.5 (1.2–4.1) 1.3-2.5-2.8 | 3.0 (0.47–5.32) 1.9-3.0-3.7 | - | - | - |
| let-7e | 4.0 (−0.52–2.03) 0.71-1.0-1.48 | 1.9 (1.5–3.3) 0.92-1.9-2.6 | 2.4 (0.16–4.5) 1.0-2.4-2.9 | - | - | - |
| miR33a | 1.11 (0.12–1.94) 0.61-1.11-1.31 | 1.2 (0.2–5.4) 0.73-1.2-1.8 | 1.7 (0.11–2.6) 1.4-1.7-1.8 | - | - | - |
| miR1-44 | 1.03 (0.23–2.10) 0.67-1.03-1.30 | 2.1 (2.0–2.2) 0.77-1.1-1.5 | 0.98 (0.16–2.6) 0.53-0.98-2.0 | - | - | - |
| Years of evolution <1–10 | Evolution greater than 10 years | |||||
| miR-17 | 0.82 (0.36–2.32) 0.54-0.82-1.50 | 2.3 (1.2–3.3) 1.8-2.3-2.9 | 2.6 (0.6–3.3) 2.1-2.6-3.1 | - | - | - |
| miR-191 | 1.10 (−0.44–2.20) 0.37-1.10-1.45 | 2.5 (1.2–5.3) 1.3-2.5-3.0 | 2.8 (0.6–4.4) 1.8-2.8-3.6 | - | - | - |
| let-7e | 4.0 (−0.52–2.03) 0.71-1.0-1.48 | 1.9 (1.5–4.5) 0.82-1.9-2.6 | 2.3 (0.5–3.5) 1.1-2.3-2.9 | - | - | - |
| miR33a | 1.11 (0.12–1.94) 0.61-1.11-1.31 | 1.3 (0.11–2.8) 0.64-1.4-1.8 | 1.7 (0.18–5.4) 1.0-1.7-2.3 | - | - | - |
| miR1-44 | 1.03 (0.23–2.10) 0.67-1.03-1.30 | 0.96 (2.0–2.6) 0.78-0.96-1.6 | 1.2 (0.13–2.5) 0.50-1.2-1.9 | - | - | - |
| Control Median (min.–max.) Q1, Q2, Q3 | Patients with Diabetes Median (min.–max.) Q1, Q2, Q3 | |||||
|---|---|---|---|---|---|---|
| Without Obesity N = 24 | With Obesity N = 26 | p1 | Without Obesity N = 12 | With Obesity N = 25 | p2 | |
| miR-17 | 0.87 (0.36–2.32) 0.53-0.87-1.5 | 0.79 (0.36–2.259) 0.56-0.79-1.1 | NS | 2.34 (1.2–3.3) 2.1-2.3-3.1 | 2.69 (0.72–3.33) 1.7-2.6-3.1 | 0.000 |
| miR-191 | 1.1 (0.13–2.2) 0.32-1.1-1.5 | 1.0 (0.44–2.1) 0.65-1.0-1.4 | NS | 2.6 (1.2–5.3) 1.6-2.6-3.8 | 2.6 (0.47–4.4) 1.6-2.6-3.0 | 0.0001 |
| let-7e | 1.1 (0.10–2.0) 0.78-1.1-1.4 | 0.92 (0.52–1.9) 0.63-0.92-1.2 | NS | 2.3 (1.5–4.5) 1.3-2.3-3.2 | 1.9 (0.16–3.59) 0.99-1.9-2.6 | 0.0001 |
| miR33a | 0.89 (0.15–1.78) 0.59-0.89-1.2 | 1.1 (0.12–1.9) 0.66-1.1-1.3 | NS | 1.7 (0.44–2.8) 0.79-1.7-2.6 | 1.4 (0.02–5.4) 0.73-1.4-1.8 | 0.003 |
| miR1-44 | 1.0 (0.23–2.1) 0.86-1.0-1.2) | 0.88 (0.28–1.6) 0.66-0.88-1.3 | NS | 1.3 (2.0–2.6) 0.89-1.31-1.9 | 0.96 (0.13–2.5) 0.67-0.96-1.7 | NS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuentevilla-Alvarez, G.; Soto, M.E.; Robles-Herrera, G.J.; Vargas-Alarcón, G.; Sámano, R.; Meza-Toledo, S.E.; Huesca-Gómez, C.; Gamboa, R. Analysis of Circulating miRNA Expression Profiles in Type 2 Diabetes Patients with Diabetic Foot Complications. Int. J. Mol. Sci. 2024, 25, 7078. https://doi.org/10.3390/ijms25137078
Fuentevilla-Alvarez G, Soto ME, Robles-Herrera GJ, Vargas-Alarcón G, Sámano R, Meza-Toledo SE, Huesca-Gómez C, Gamboa R. Analysis of Circulating miRNA Expression Profiles in Type 2 Diabetes Patients with Diabetic Foot Complications. International Journal of Molecular Sciences. 2024; 25(13):7078. https://doi.org/10.3390/ijms25137078
Chicago/Turabian StyleFuentevilla-Alvarez, Giovanny, María Elena Soto, Gustavo Jaziel Robles-Herrera, Gilberto Vargas-Alarcón, Reyna Sámano, Sergio Enrique Meza-Toledo, Claudia Huesca-Gómez, and Ricardo Gamboa. 2024. "Analysis of Circulating miRNA Expression Profiles in Type 2 Diabetes Patients with Diabetic Foot Complications" International Journal of Molecular Sciences 25, no. 13: 7078. https://doi.org/10.3390/ijms25137078
APA StyleFuentevilla-Alvarez, G., Soto, M. E., Robles-Herrera, G. J., Vargas-Alarcón, G., Sámano, R., Meza-Toledo, S. E., Huesca-Gómez, C., & Gamboa, R. (2024). Analysis of Circulating miRNA Expression Profiles in Type 2 Diabetes Patients with Diabetic Foot Complications. International Journal of Molecular Sciences, 25(13), 7078. https://doi.org/10.3390/ijms25137078






