Genome-Wide Identification of GRAS Transcription Factors and Their Functional Analysis in Salt Stress Response in Sugar Beet
Abstract
1. Introduction
2. Results
2.1. BvGRAS TF Family Members
2.2. Structure Analysis of BvGRAS TF Sequence
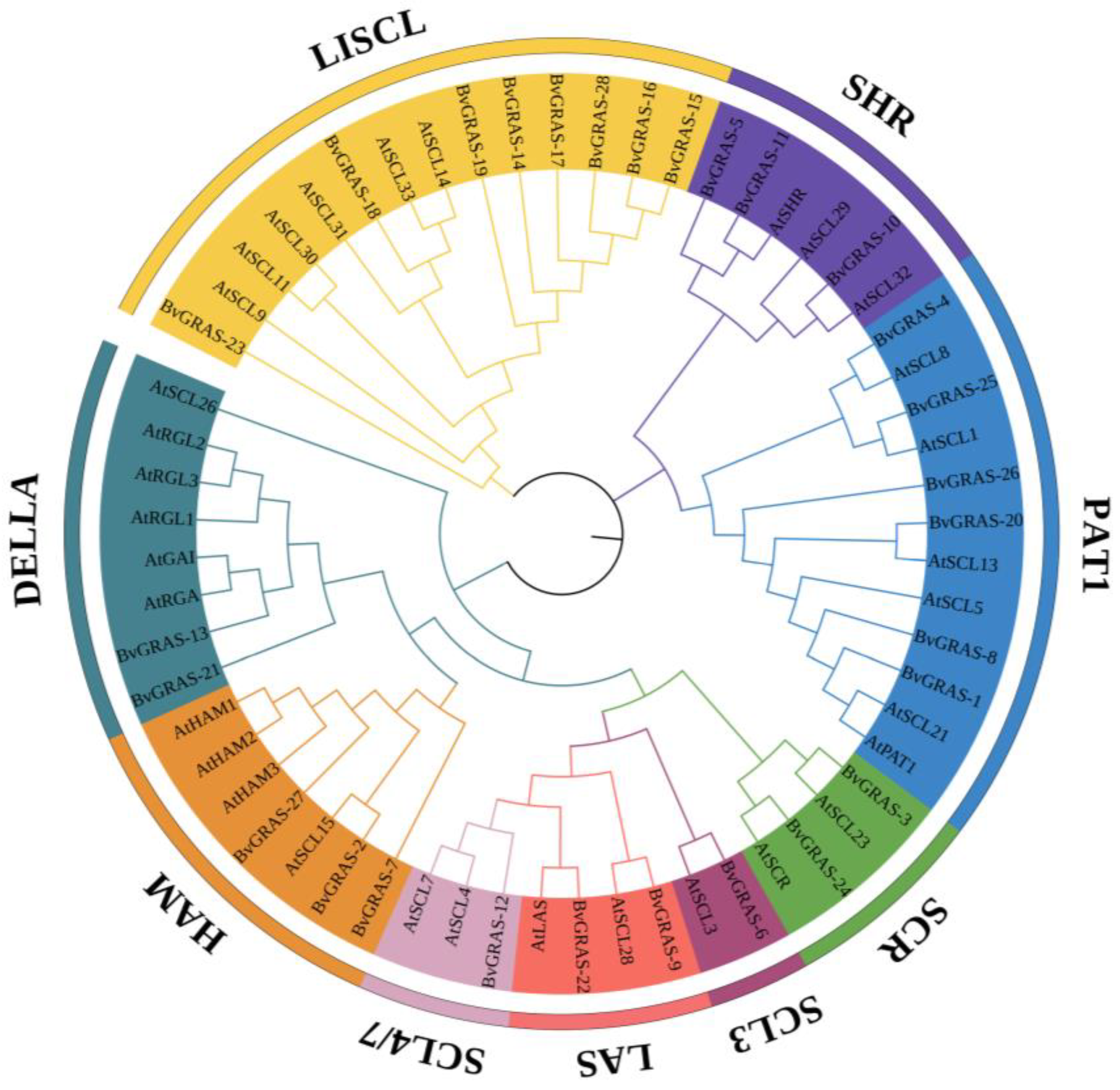
2.3. Phylogenetic Analysis of BvGRAS TF
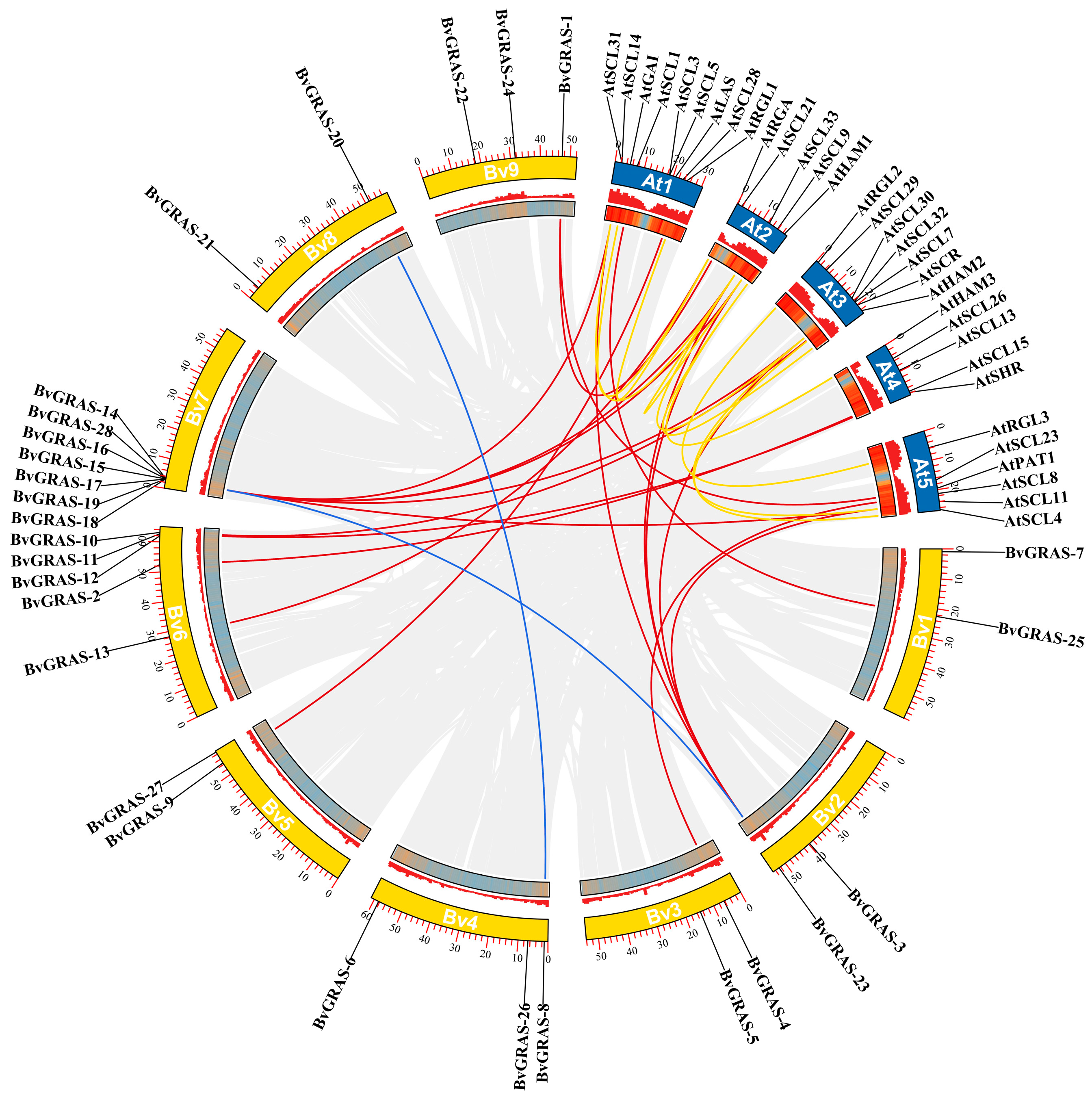
2.4. BvGRAS TF Chromosome Localization and Collinearity Analysis
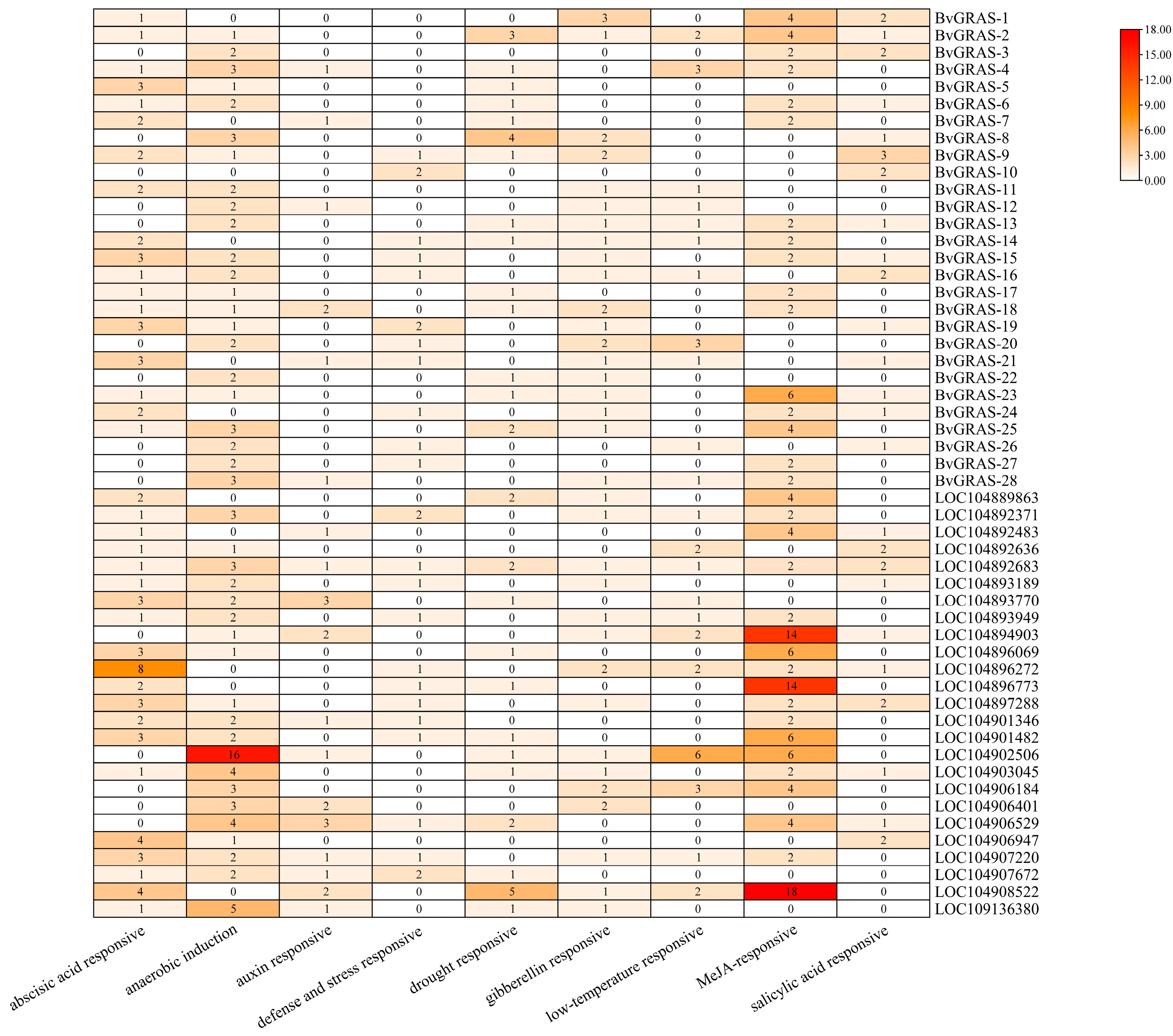
2.5. BvGRAS TF Salt Stress Response Analysis and Co-Expression Analysis
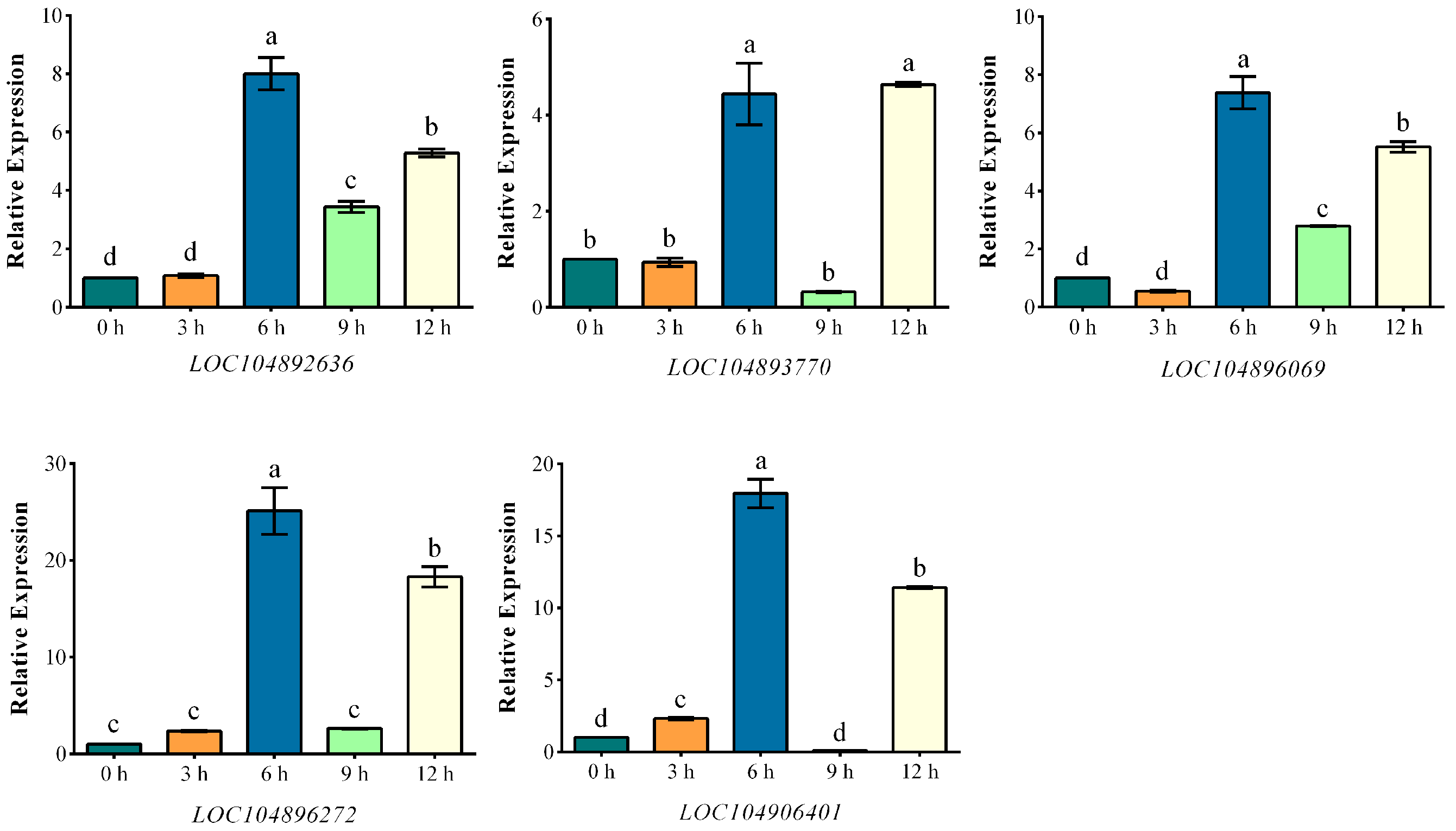
2.6. Promoter Analysis and qRT-PCR Analysis
2.7. Functional Annotation of BvGRAS TF
3. Discussion
4. Materials and Methods
4.1. Plant Material Treatment
4.2. Identification of GRAS TF Family
4.3. Structural Analysis of GRAS TF Sequence
4.4. GRAS TF Phylogenetic Analysis
4.5. GRAS TF Chromosome Localization and Collinearity Analysis
4.6. GRAS TF Salt Stress Response Analysis and Co-Expression Analysis
4.7. GRAS TF Promoter Analysis and qRT-PCR Analysis
4.8. GRAS TF Function Annotation
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 155–169. [Google Scholar] [CrossRef]
- Di Laurenzio, L.; Wysocka-Diller, J.; Malamy, J.E.; Pysh, L.; Helariutta, Y.; Freshour, G.; Hahn, M.G.; Feldmann, K.A.; Benfey, P.N. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 1996, 86, 423–433. [Google Scholar] [CrossRef]
- Li, S.; Zhao, Y.; Zhao, Z.; Wu, X.; Sun, L.; Liu, Q.; Wu, Y. Crystal Structure of the GRAS Domain of SCARECROW-LIKE7 in Oryza sativa. Plant Cell 2016, 28, 1025–1034. [Google Scholar] [CrossRef]
- Sun, X.; Xue, B.; Jones, W.T.; Rikkerink, E.; Dunker, A.K.; Uversky, V.N. A functionally required unfoldome from the plant kingdom: Intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 2011, 77, 205–223. [Google Scholar] [CrossRef]
- Fu, X.; Richards, D.E.; Ait-ali, T.; Hynes, L.W.; Ougham, H.; Peng, J.; Harberd, N.P. Gibberellin-Mediated Proteasome-Dependent Degradation of the Barley DELLA Protein SLN1 Repressor. Plant Cell 2002, 14, 3191–3200. [Google Scholar] [CrossRef]
- Torres-Galea, P.; Hirtreiter, B.; Bolle, C. Two GRAS proteins, SCARECROW-LIKE21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, function cooperatively in phytochrome A signal transduction. Plant Physiol. 2013, 161, 291–304. [Google Scholar] [CrossRef]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef]
- Wysocka-Diller, J.W.; Helariutta, Y.; Fukaki, H.; Malamy, J.E.; Benfey, P.N. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 2000, 127, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Greb, T.; Clarenz, O.; Schäfer, E.; Müller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of theLATERAL SUPPRESSORgene inArabidopsisreveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Hendelman, A.; Kravchik, M.; Stav, R.; Frank, W.; Arazi, T. Tomato HAIRY MERISTEM genes are involved in meristem maintenance and compound leaf morphogenesis. J. Exp. Bot. 2016, 67, 6187–6200. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, H.; Chen, Y.; Zheng, Q.; Li, B.; Li, Z. TaSCL14, a novel wheat (Triticum aestivum L.) GRAS gene, regulates plant growth, photosynthesis, tolerance to photooxidative stress, and senescence. J. Genet. Genom. 2015, 42, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, K.; Minami, M.; Takase, H.; Hotta, Y.; Hiratsuka, K. Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 2003, 278, 20865–20873. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-X.; Guo, Y.; Wu, H.; Li, X.; Li, Q.; Zhao, X.; Duan, X.; An, Y.; Lv, W.; An, H. Identification and expression of GRAS family genes in maize (Zea mays L.). PLoS ONE 2017, 12, e0185418. [Google Scholar] [CrossRef]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef]
- Wang, L.; Ding, X.; Gao, Y.; Yang, S. Genome-wide identification and characterization of GRAS genes in soybean (Glycine max). BMC Plant Biol. 2020, 20, 415. [Google Scholar] [CrossRef]
- Wang, N.; Wang, K.; Li, S.; Jiang, Y.; Li, L.; Zhao, M.; Jiang, Y.; Zhu, L.; Wang, Y.; Su, Y.; et al. Transcriptome-Wide Identification, Evolutionary Analysis, and GA Stress Response of the GRAS Gene Family in Panax ginseng C. A. Meyer. Plants 2020, 9, 190. [Google Scholar] [CrossRef]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef]
- Heo, J.-O.; Chang, K.S.; Kim, I.A.; Lee, M.-H.; Lee, S.A.; Song, S.-K.; Lee, M.M.; Lim, J. Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2011, 108, 2166–2171. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, J.; Weng, S.; Zhang, Y.; Zhang, D.; Shi, C. Identification and characterization of dwarf 62, a loss-of-function mutation in DLT/OsGRAS-32 affecting gibberellin metabolism in rice. Planta 2010, 232, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Feltus, F.A.; Bai, Z.; Xia, P.; Wang, R.; Jiao, J.; Ru, M.; Liu, J.; Liang, Z. Molecular cloning and characterization of five SmGRAS genes associated with tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. PLoS ONE 2017, 12, e0185322. [Google Scholar] [CrossRef]
- Tanabe, S.; Onodera, H.; Hara, N.; Ishii-Minami, N.; Day, B.; Fujisawa, Y.; Hagio, T.; Toki, S.; Shibuya, N.; Nishizawa, Y.; et al. The elicitor-responsive gene for a GRAS family protein, CIGR2, suppresses cell death in rice inoculated with rice blast fungus via activation of a heat shock transcription factor, OsHsf23. Biosci. Biotechnol. Biochem. 2016, 80, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yang, Q.; Fu, T.; Zhou, Y. Overexpression of the Brassica napus BnLAS gene in Arabidopsis affects plant development and increases drought tolerance. Plant Cell Rep. 2011, 30, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.S.; Liang, D.; Shuai, P.; Xia, X.L.; Yin, W.L. The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 2010, 61, 4011–4019. [Google Scholar] [CrossRef] [PubMed]
- Habib, S.; Waseem, M.; Li, N.; Yang, L.; Li, Z. Overexpression of SlGRAS7 Affects Multiple Behaviors Leading to Confer Abiotic Stresses Tolerance and Impacts Gibberellin and Auxin Signaling in Tomato. Int. J. Genom. 2019, 2019, 4051981. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, J.; Yang, Z.E.; Chen, E.Y.; Zhang, C.J.; Zhang, X.Y.; Li, F.G. Genome-wide analysis of GRAS transcription factor gene family in Gossypium hirsutum L. BMC Genom. 2018, 19, 348. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, P.; Wu, S.; Lu, Y.; Sun, J.; Cao, Q.; Li, Z.; Xu, T. Identification and expression analysis of GRAS transcription factors in the wild relative of sweet potato Ipomoea trifida. BMC Genom. 2019, 20, 911. [Google Scholar] [CrossRef]
- Stuurman, J.; Jäggi, F.; Kuhlemeier, C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002, 16, 2213–2218. [Google Scholar] [CrossRef]
- Yuan, Y.; Fang, L.; Karungo, S.K.; Zhang, L.; Gao, Y.; Li, S.; Xin, H. Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis. Plant Cell Rep. 2016, 35, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yu, L.; Zhou, F.; Ma, H.; Yang, K.; Wu, G. Synthesis and characterization of activated carbon from sugar beet residue for the adsorption of hexavalent chromium in aqueous solutions. RSC Adv. 2021, 11, 8025–8032. [Google Scholar] [CrossRef] [PubMed]
- Aljabri, M.; Alharbi, S.; Al-Qthanin, R.N.; Ismaeil, F.M.; Chen, J.; Abou-Elwafa, S.F. Recycling of beet sugar byproducts and wastes enhances sugar beet productivity and salt redistribution in saline soils. Environ. Sci. Pollut. Res. Int. 2021, 28, 45745–45755. [Google Scholar] [CrossRef] [PubMed]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, F.; Bamagoos, A.A.; Ismaeil, F.M.; Zhang, W.; Abou-Elwafa, S.F. Application of beet sugar byproducts improves sugar beet biofortification in saline soils and reduces sugar losses in beet sugar processing. Environ. Sci. Pollut. Res. Int. 2021, 28, 30303–30311. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef]
- Liu, X.Y.; Widmer, A. Genome-wide Comparative Analysis of the GRAS Gene Family in Populus, Arabidopsis and Rice. Plant Mol. Biol. Report. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Fode, B.; Siemsen, T.; Thurow, C.; Weigel, R.; Gatz, C. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 2008, 20, 3122–3135. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Mizokami, Y.; Légeret, B.; Havaux, M. The Apocarotenoid β-Cyclocitric Acid Elicits Drought Tolerance in Plants. iScience 2019, 19, 461–473. [Google Scholar] [CrossRef]
- Cui, H.; Levesque, M.P.; Vernoux, T.; Jung, J.W.; Paquette, A.J.; Gallagher, K.L.; Wang, J.Y.; Blilou, I.; Scheres, B.; Benfey, P.N. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 2007, 316, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, W.; Li, C.; Yu, G.; Li, J.; Wang, Y.; Wang, X. A multilayered cross-species analysis of GRAS transcription factors uncovered their functional networks in plant adaptation to the environment. J. Adv. Res. 2021, 29, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wong, D.C.J.; Wang, Y.; Xu, G.; Ren, C.; Liu, Y.; Kuang, Y.; Fan, P.; Li, S.; Xin, H.; et al. GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 2021, 186, 1660–1678. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, N.; Itoh, J.; Morikami, A.; Nagato, Y.; Matsuoka, M. The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J. 2003, 36, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.E.; Langdale, J.A. SCARECROW is deployed in distinct contexts during rice and maize leaf development. Development 2022, 149, dev200410. [Google Scholar] [CrossRef]
- Rossmann, S.; Kohlen, W.; Hasson, A.; Theres, K. Lateral suppressor and Goblet act in hierarchical order to regulate ectopic meristem formation at the base of tomato leaflets. Plant J. 2015, 81, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Huang, L.; Ma, Z.; Sun, W.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, expression analysis and functional study of the GRAS gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 2019, 19, 342. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Guo, L.; Han, H.; Liu, X.; Banks, J.A.; Wisecaver, J.H.; Zhou, Y. Conservation and diversification of HAIRY MERISTEM gene family in land plants. Plant J. 2021, 106, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Zhou, Y. HAM Gene Family and Shoot Meristem Development. Front. Plant Sci. 2021, 12, 800332. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef]
- Wild, M.; Davière, J.M.; Cheminant, S.; Regnault, T.; Baumberger, N.; Heintz, D.; Baltz, R.; Genschik, P.; Achard, P. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 2012, 24, 3307–3319. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Okada, K.; Fukazawa, J.; Takahashi, Y. DELLA-dependent and -independent gibberellin signaling. Plant Signal. Behav. 2018, 13, e1445933. [Google Scholar] [CrossRef] [PubMed]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. GROWTH-REGULATING FACTORS Interact with DELLAs and Regulate Growth in Cold Stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Ogawa, M.; Fleet, C.M.; Zentella, R.; Hu, J.; Heo, J.O.; Lim, J.; Kamiya, Y.; Yamaguchi, S.; Sun, T.P. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 2160–2165. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Ksas, B.; Havaux, M. Decoding β-Cyclocitral-Mediated Retrograde Signaling Reveals the Role of a Detoxification Response in Plant Tolerance to Photooxidative Stress. Plant Cell 2018, 30, 2495–2511. [Google Scholar] [CrossRef] [PubMed]
- Arain, S.; Meer, M.; Sajjad, M.; Yasmin, H. Light contributes to salt resistance through GAI protein regulation in Arabidopsis thaliana. Plant Physiol. Biochem. PPB 2021, 159, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, D.K.; Wang, Q.Q.; Ke, S.; Li, Y.; Zhang, D.; Zheng, Q.; Zhang, C.; Liu, Z.J.; Lan, S. Genome-wide identification and expression analysis of the GRAS gene family in Dendrobium chrysotoxum. Front. Plant Sci. 2022, 13, 1058287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Xu, Y.; He, G.; He, K.; Xiao, L.; Hu, R.; Li, S. Genome-Wide Characterization and Expression Profiling of the GRAS Gene Family in Salt and Alkali Stresses in Miscanthus sinensis. Int. J. Mol. Sci. 2022, 23, 14521. [Google Scholar] [CrossRef] [PubMed]
- Navrot, N.; Finnie, C.; Svensson, B.; Hägglund, P. Plant redox proteomics. J. Proteom. 2011, 74, 1450–1462. [Google Scholar] [CrossRef]
- Licht, A.; Schneider, E. ATP binding cassette systems: Structures, mechanisms, and functions. Open Life Sci. 2011, 6, 785–801. [Google Scholar] [CrossRef]
- Nguyen, V.N.; Moon, S.; Jung, K.H. Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J. Plant Physiol. 2014, 171, 1276–1288. [Google Scholar] [CrossRef]
- Saha, J.; Sengupta, A.; Gupta, K.; Gupta, B. Molecular phylogenetic study and expression analysis of ATP-binding cassette transporter gene family in Oryza sativa in response to salt stress. Comput. Biol. Chem. 2015, 54, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Champion, E.A.; Lane, B.H.; Jackrel, M.E.; Regan, L.; Baserga, S.J. A direct interaction between the Utp6 half-a-tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient pre-rRNA processing. Mol. Cell. Biol. 2008, 28, 6547–6556. [Google Scholar] [CrossRef]
- Hammani, K.; Cook, W.B.; Barkan, A. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl. Acad. Sci. USA 2012, 109, 5651–5656. [Google Scholar] [CrossRef]
- Moghadam, A.A.; Ebrahimie, E.; Taghavi, S.M.; Niazi, A.; Babgohari, M.Z.; Deihimi, T.; Djavaheri, M.; Ramezani, A. How the nucleus and mitochondria communicate in energy production during stress: Nuclear MtATP6, an early-stress responsive gene, regulates the mitochondrial F1F0-ATP synthase complex. Mol. Biotechnol. 2013, 54, 756–769. [Google Scholar] [CrossRef]
- Yamada, N.; Takahashi, H.; Kitou, K.; Sahashi, K.; Tamagake, H.; Tanaka, Y.; Takabe, T. Suppressed expression of choline monooxygenase in sugar beet on the accumulation of glycine betaine. Plant Physiol. Biochem. PPB 2015, 96, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Kanehara, K.; Nakamura, Y. Arabidopsis CHOLINE/ETHANOLAMINE KINASE 1 (CEK1) is a primary choline kinase localized at the endoplasmic reticulum (ER) and involved in ER stress tolerance. New Phytol. 2019, 223, 1904–1917. [Google Scholar] [CrossRef]
- Jain, N.; Farhat, S.; Kumar, R.; Singh, N.; Singh, S.; Sreevathsa, R.; Kalia, S.; Singh, N.K.; Teruhiro, T.; Rai, V. Alteration of proteome in germinating seedlings of piegonpea (Cajanus cajan) after salt stress. Physiol. Mol. Biol. Plants 2021, 27, 2833–2848. [Google Scholar] [CrossRef]
- Lee, Y.K.; Rhee, J.Y.; Lee, S.H.; Chung, G.C.; Park, S.J.; Segami, S.; Maeshima, M.; Choi, G. Functionally redundant LNG3 and LNG4 genes regulate turgor-driven polar cell elongation through activation of XTH17 and XTH24. Plant Mol. Biol. 2018, 97, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Li, J.; Cui, J.; Dai, C.; Liu, T.; Cheng, D.; Luo, C. Whole-Transcriptome RNA Sequencing Reveals the Global Molecular Responses and CeRNA Regulatory Network of mRNAs, lncRNAs, miRNAs and circRNAs in Response to Salt Stress in Sugar Beet (Beta vulgaris). Int. J. Mol. Sci. 2020, 22, 289. [Google Scholar] [CrossRef]
- Magali, L.; Patrice, D.; Gert, T.; Kathleen, M.; Yves, M.; Yves, V.P.; Pierre, R.; Stephane, R. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Rio, D.; Ares, M.; Hannon, G.; Nilsen, T. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Sehrish, S.; Sumbal, W.; Xie, M.; Zhao, C.; Zuo, R.; Gao, F.; Liu, S. Genome-Wide Identification and Characterization of SET Domain Family Genes in Brassica napus L. Int. J. Mol. Sci. 2022, 23, 1936. [Google Scholar] [CrossRef]
- Consortium, G.O. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]



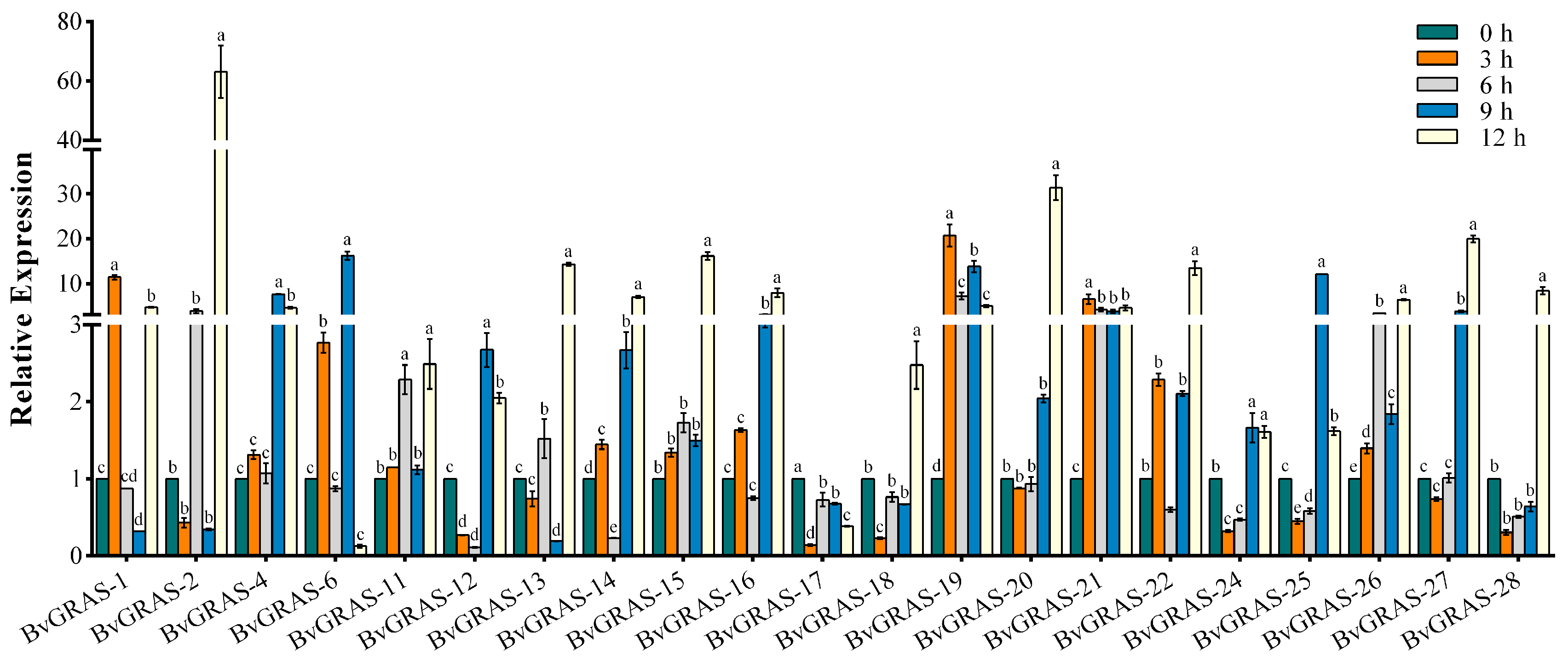

| Gene ID | Gene Name | Chromosome | Location | Length (aa) | pI | MW (kDa) | Instability Index |
|---|---|---|---|---|---|---|---|
| LOC104883303 | BvGRAS-1 | Chr9 | 47272109–47276339 | 521 | 5.84 | 58.36 | 46.25 |
| LOC104884709 | BvGRAS-2 | Chr6 | 52483906–52486547 | 589 | 6.22 | 64.61 | 44 |
| LOC104886598 | BvGRAS-3 | Chr2 | 39617378–39619237 | 427 | 6.11 | 46.73 | 46.8 |
| LOC104888390 | BvGRAS-4 | Chr3 | 6251492–6254730 | 783 | 5.58 | 86.54 | 52.97 |
| LOC104888856 | BvGRAS-5 | Chr3 | 14855492–14857205 | 452 | 6.81 | 51.96 | 44.14 |
| LOC104890059 | BvGRAS-6 | Chr4 | 58393359–58395438 | 456 | 6.04 | 50.93 | 54.7 |
| LOC104891471 | BvGRAS-7 | Chr1 | 943670–945249 | 460 | 6.21 | 52.16 | 40.82 |
| LOC104891820 | BvGRAS-8 | Chr4 | 1276279–1279109 | 466 | 5.29 | 52.07 | 41.11 |
| LOC104892338 | BvGRAS-9 | Chr5 | 54516753–54520401 | 678 | 5.67 | 76.68 | 62.07 |
| LOC104894934 | BvGRAS-10 | Chr6 | 63135438–63137801 | 488 | 5.59 | 54.88 | 42.41 |
| LOC104894957 | BvGRAS-11 | Chr6 | 62691123–62693201 | 555 | 6.12 | 63.65 | 48.38 |
| LOC104895016 | BvGRAS-12 | Chr6 | 61969338–61971599 | 605 | 5.08 | 66.73 | 48.26 |
| LOC104896788 | BvGRAS-13 | Chr6 | 28255757–28258546 | 611 | 5.37 | 67.05 | 44.34 |
| LOC104900088 | BvGRAS-14 | Chr7 | 3155937–3158021 | 694 | 5.69 | 78.84 | 52.41 |
| LOC104900090 | BvGRAS-15 | Chr7 | 3130525–3133015 | 687 | 5.32 | 77.41 | 47.5 |
| LOC104900091 | BvGRAS-16 | Chr7 | 3135597–3138281 | 687 | 5.22 | 77.37 | 47.8 |
| LOC104900093 | BvGRAS-17 | Chr7 | 3118322–3121040 | 696 | 5.46 | 78.81 | 43.7 |
| LOC104900094 | BvGRAS-18 | Chr7 | 3090537–3093719 | 752 | 5.49 | 84.21 | 49.33 |
| LOC104900447 | BvGRAS-19 | Chr7 | 3104795–3107038 | 747 | 5.29 | 84.39 | 52.57 |
| LOC104900825 | BvGRAS-20 | Chr8 | 51348137–51352794 | 548 | 5.84 | 61.54 | 52.03 |
| LOC104902119 | BvGRAS-21 | Chr8 | 4394976–4396924 | 482 | 5.22 | 53.32 | 46.77 |
| LOC104903785 | BvGRAS-22 | Chr9 | 18250458–18253370 | 488 | 6.23 | 55.33 | 61.09 |
| LOC104904278 | BvGRAS-23 | Chr2 | 52598520–52602465 | 795 | 6.08 | 89.47 | 49.86 |
| LOC104904740 | BvGRAS-24 | Chr9 | 31592859–31598730 | 840 | 5.82 | 91.89 | 56.06 |
| LOC104905710 | BvGRAS-25 | Chr1 | 22511536–22513426 | 556 | 5.1 | 62.85 | 46.48 |
| LOC104907103 | BvGRAS-26 | Chr4 | 6676833–6681498 | 579 | 6.17 | 64.80 | 59.03 |
| LOC104907825 | BvGRAS-27 | Chr5 | 58827250–58830908 | 835 | 5.85 | 91.33 | 51.8 |
| LOC109133554 | BvGRAS-28 | Chr7 | 3146288–3149122 | 681 | 5.02 | 76.76 | 46.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, X.; Gong, Y.; Chen, S.; Ma, C.; Duanmu, H. Genome-Wide Identification of GRAS Transcription Factors and Their Functional Analysis in Salt Stress Response in Sugar Beet. Int. J. Mol. Sci. 2024, 25, 7132. https://doi.org/10.3390/ijms25137132
Hao X, Gong Y, Chen S, Ma C, Duanmu H. Genome-Wide Identification of GRAS Transcription Factors and Their Functional Analysis in Salt Stress Response in Sugar Beet. International Journal of Molecular Sciences. 2024; 25(13):7132. https://doi.org/10.3390/ijms25137132
Chicago/Turabian StyleHao, Xiaolin, Yongyong Gong, Sixue Chen, Chunquan Ma, and Huizi Duanmu. 2024. "Genome-Wide Identification of GRAS Transcription Factors and Their Functional Analysis in Salt Stress Response in Sugar Beet" International Journal of Molecular Sciences 25, no. 13: 7132. https://doi.org/10.3390/ijms25137132
APA StyleHao, X., Gong, Y., Chen, S., Ma, C., & Duanmu, H. (2024). Genome-Wide Identification of GRAS Transcription Factors and Their Functional Analysis in Salt Stress Response in Sugar Beet. International Journal of Molecular Sciences, 25(13), 7132. https://doi.org/10.3390/ijms25137132







