Investigation on the Potential Functions of ZmEPF/EPFL Family Members in Response to Abiotic Stress in Maize
Abstract
1. Introduction
2. Results
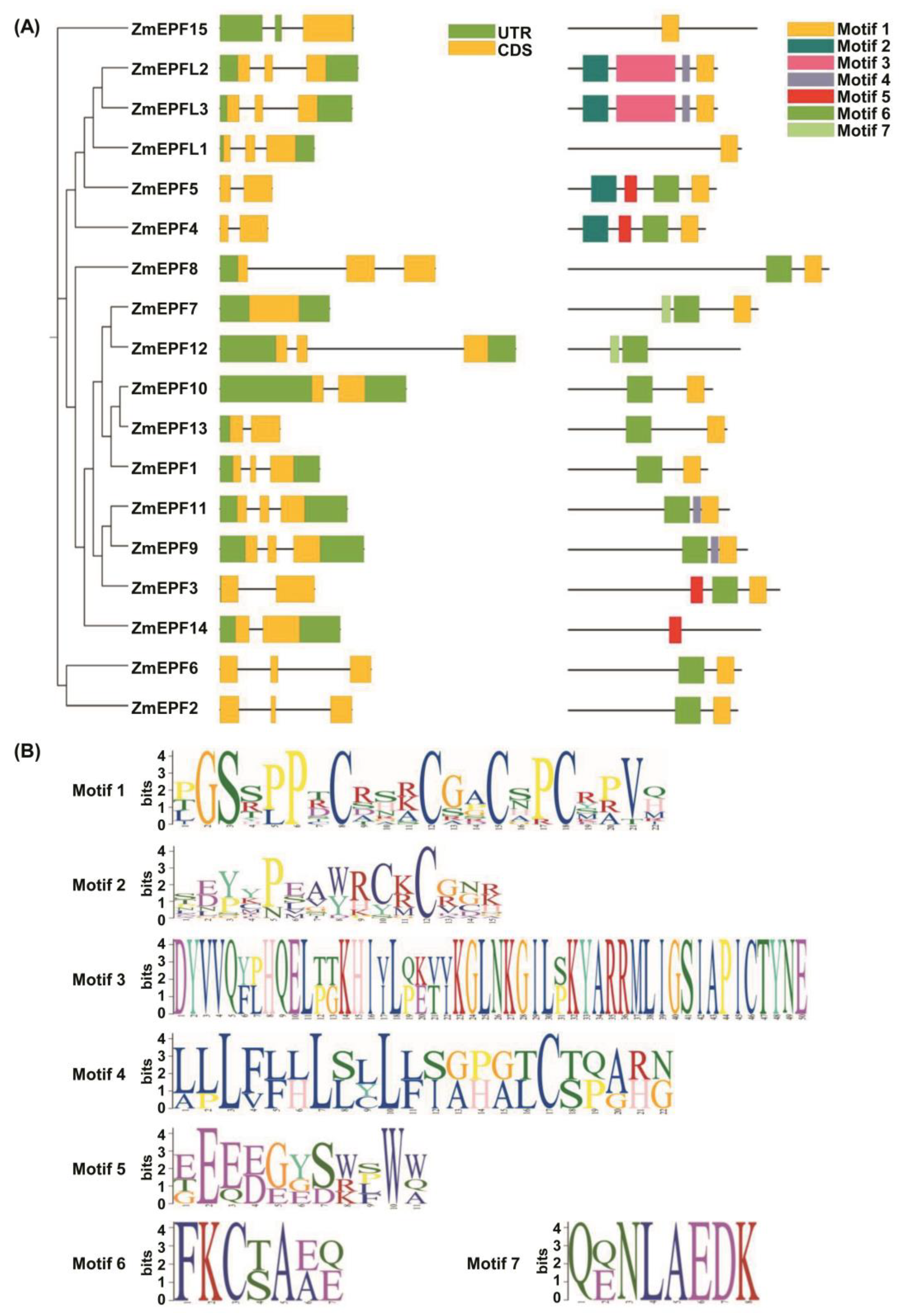
2.1. Gene Structure Analysis and Protein-Conserved Motif and Domain Prediction
2.2. Phylogenetic Relationship Analysis
2.3. Physicochemical Property Prediction
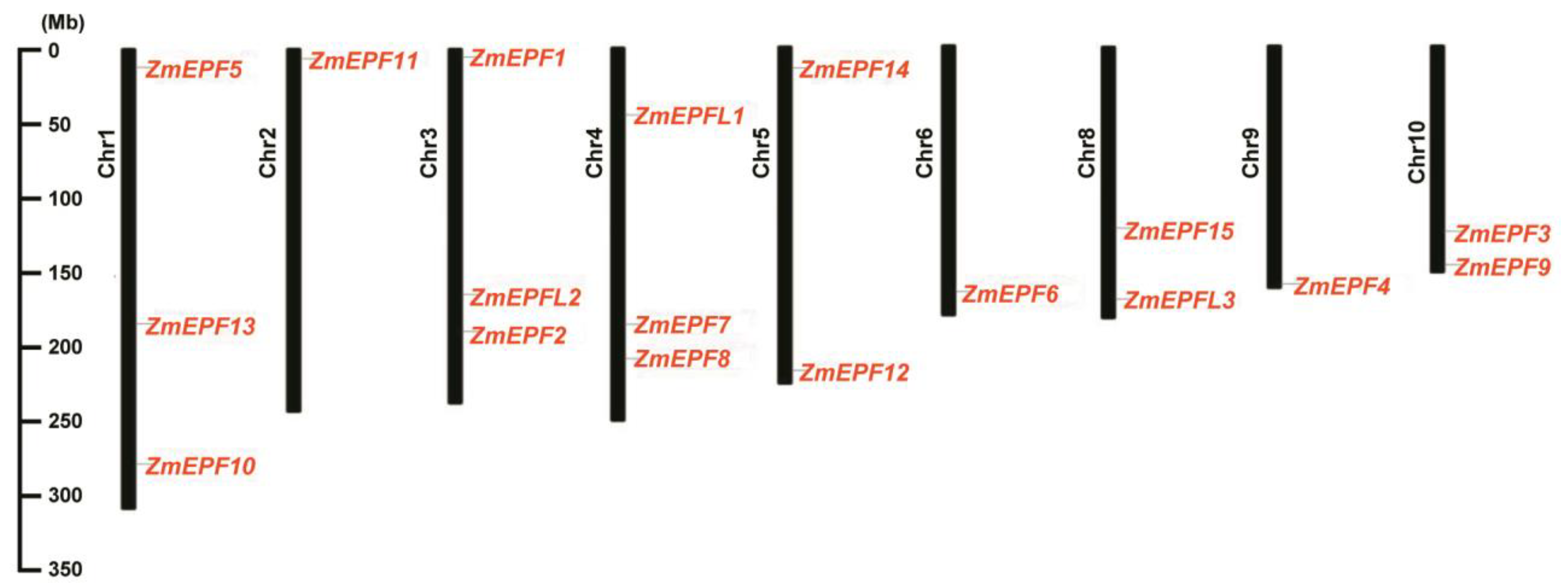
2.4. Identification and Chromosome Distribution
2.5. Analysis of cis-Acting Elements in Promoters
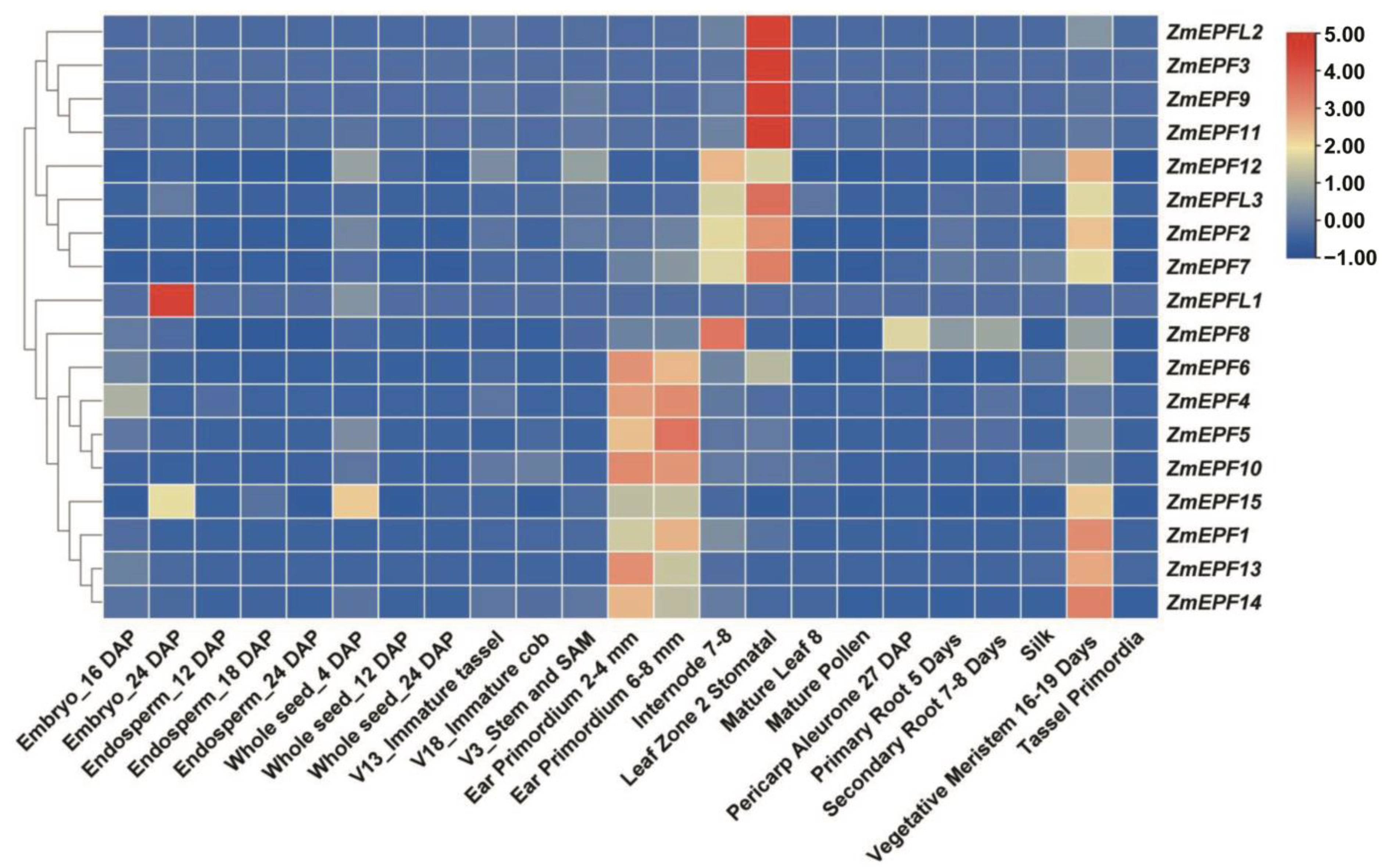
2.6. Expression Pattern Analysis
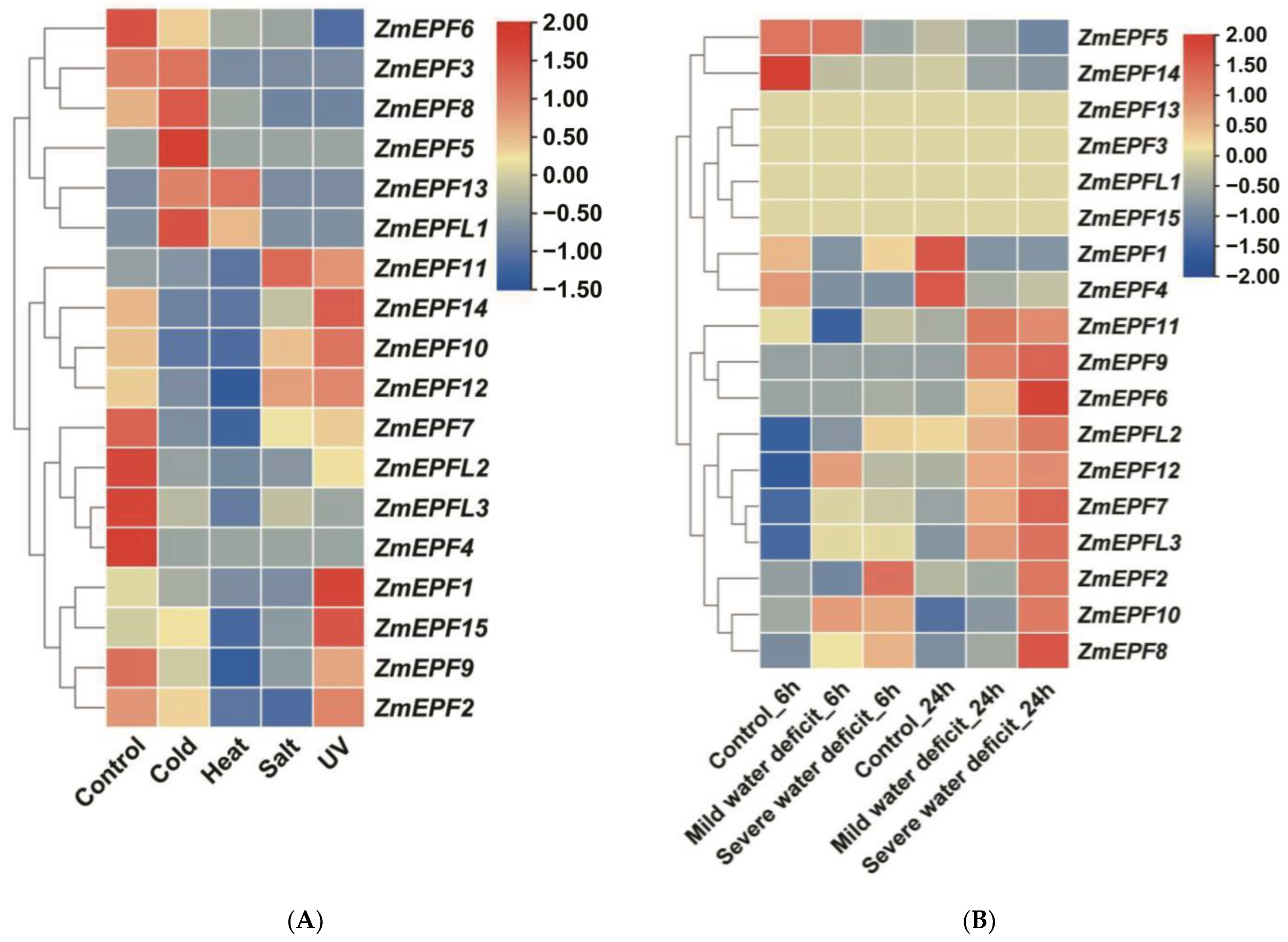
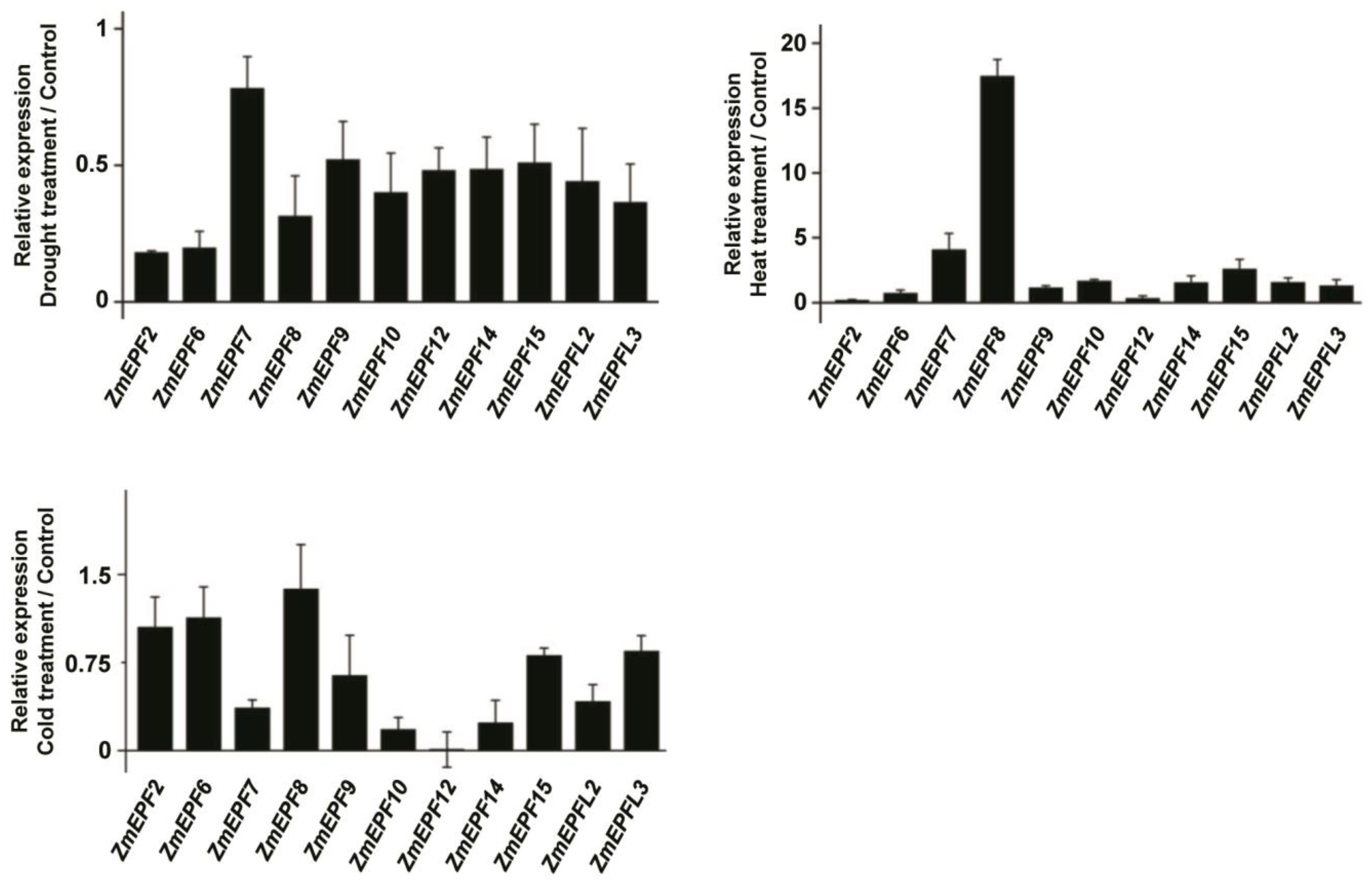
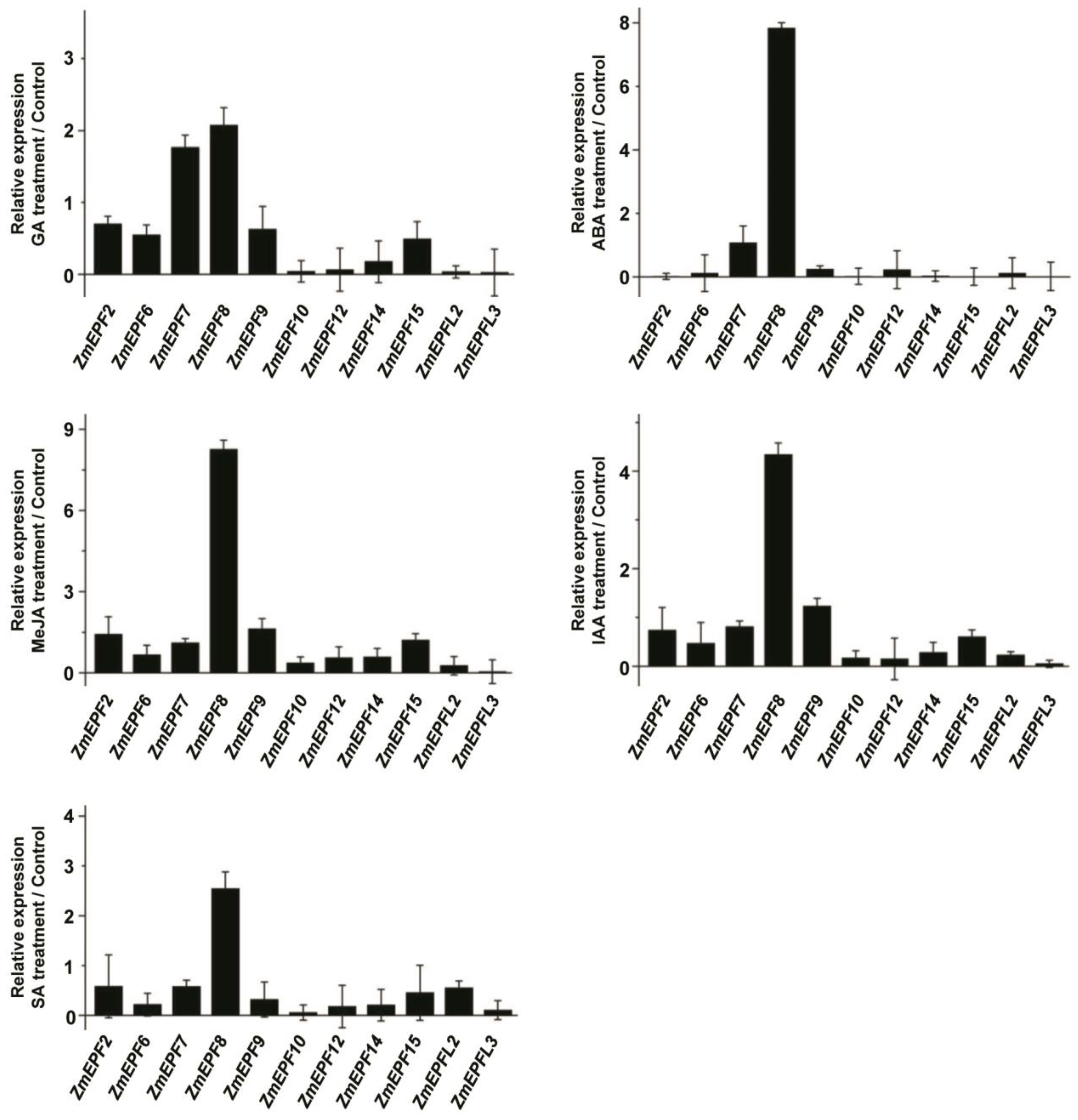
2.7. Expression Levels of ZmEPF/EPFL Genes in Response to Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Identification of ZmEPF/EPFL Family Members and Analysis of Protein Physicochemical Properties
4.3. Distribution of ZmEPF/EPFL Family Members on the Chromosome
4.4. Gene Structure Analysis of ZmEPF/EPFL Family Members
4.5. Conserved Motif, Domain Prediction, and Phylogenetic Tree Construction of ZmEPF/EPFL Proteins
4.6. Analysis of cis-Acting Elements in the Promoters of ZmEPF/EPFL Family Members
4.7. Expression Patterns of ZmEPF/EPFL Family Members
4.8. Collinearity Analysis
4.9. RNA Extraction and Real-Time Fluorescence Quantitative PCR
4.10. Subcellular Localization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.M.; Zhu, J.H.; Gong, Z.Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Bertolino, L.T.; Caine, R.S.; Gray, J.E. Impact of stomatal density and morphology on water-use efficiency in a changing world. Front. Plant Sci. 2019, 10, 225. [Google Scholar] [CrossRef] [PubMed]
- Bawa, G.; Yu, X.; Liu, Z.; Zhou, Y.; Sun, X. Surviving the enemies: Regulatory mechanisms of stomatal function in response to drought and salt stress. Environ. Exp. Bot. 2023, 209, 105291. [Google Scholar] [CrossRef]
- Naithani, S.; Mohanty, B.; Elser, J.; D’Eustachio, P.; Jaiswal, P. Biocuration of a transcription factors network involved in submergence tolerance during seed germination and coleoptile elongation in rice (Oryza sativa). Plants 2023, 12, 2146. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.S.; Song, C.P.; Wang, B.S.; Zhou, J.M.; Kangasjärvi, J.; Zhu, J.K.; Gong, Z.Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.N.; Jiang, Y.X.; Wang, X.F.; Huang, S.J.; Yuan, M.; Guo, Y. AP3M harbors actin filament binding activity that is crucial for vacuole morphology and stomatal closure in. Proc. Natl. Acad. Sci. USA 2019, 116, 18132–18141. [Google Scholar] [CrossRef] [PubMed]
- Chater, C.C.C.; Caine, R.S.; Fleming, A.J.; Gray, J.E. Origins and evolution of stomatal development. Plant Physiol. 2017, 174, 624–638. [Google Scholar] [CrossRef] [PubMed]
- Bowles, A.M.C.; Williamson, C.J.; Williams, T.A.; Lenton, T.M.; Donoghue, P.C.J. The origin and early evolution of plants. Trends Plant Sci. 2023, 28, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Bergmann, D.C. Stomatal patterning and development. Curr. Top. Dev. Biol. 2010, 91, 267–297. [Google Scholar]
- Hara, K.; Kajita, R.; Torii, K.U.; Bergmann, D.C.; Kakimoto, T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007, 2, 1720–1725. [Google Scholar] [CrossRef]
- Zhao, L.; Sack, F.D. Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am. J. Bot. 1999, 86, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Sugano, S.S.; Hara-Nishimura, I. Positive and negative peptide signals control stomatal density. Cell. Mol. Life Sci. 2011, 68, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Caine, R.S.; Chater, C.C.C.; Fleming, A.J.; Gray, J.E. Stomata and sporophytes of the model moss Physcomitrium patens. Front. Plant Sci. 2020, 11, 643. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.; Hepworth, C.; Dutton, C.; Dunn, J.A.; Hunt, L.; Stephens, J.; Waugh, R.; Cameron, D.D.; Gray, J.E. Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 2017, 174, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; He, J.; Zhou, X.; Zhong, J.; Li, J.; Liang, Y.K. Homologous genes of epidermal patterning factor regulate stomatal development in rice. J. Plant Physiol. 2019, 234, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Yokoo, T.; Kajita, R.; Onishi, T.; Yahata, S.; Peterson, K.M.; Torii, K.U.; Kakimoto, T. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009, 50, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Chang, P.; Liu, J.; Yang, Y.; Zhang, X.; Chen, L.; Hu, Y. Genome-wide identification of EPF/EPFL gene family in wheat (Triticum aestivum) and analysis of TaEPF1-2B associated with stomatal traits. J. Trit. Crop. 2021, 41, 1317–1329. [Google Scholar]
- Caine, R.S.; Chater, C.C.; Kamisugi, Y.; Cuming, A.C.; Beerling, D.J.; Gray, J.E.; Fleming, A.J. An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 2016, 143, 3306–3314. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, U.; Caine, R.S.; Atkinson, J.A.; Harrison, E.L.; Wells, D.; Chater, C.C.; Gray, J.E.; Swarup, R.; Murchie, E.H. Rice plants overexpressing OsEPF1 show reduced stomatal density and increased root cortical aerenchyma formation. Sci. Rep. 2019, 9, 5584. [Google Scholar] [CrossRef]
- Hunt, L.; Gray, J.E. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 2009, 19, 864–869. [Google Scholar] [CrossRef]
- Takata, N.; Yokota, K.; Ohki, S.; Mori, M.; Taniguchi, T.; Kurita, M. Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS ONE 2013, 8, e65183. [Google Scholar] [CrossRef]
- Hunt, L.; Bailey, K.J.; Gray, J.E. The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 2010, 186, 609–614. [Google Scholar] [CrossRef]
- Bessho-Uehara, K.; Wang, D.R.; Furuta, T.; Minami, A.; Nagai, K.; Gamuyao, R.; Asano, K.; Angeles-Shim, R.B.; Shimizu, Y.; Ayano, M.; et al. Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc. Natl. Acad. Sci. USA 2016, 113, 8969–8974. [Google Scholar] [CrossRef]
- Xiong, L.; Huang, Y.; Liu, Z.; Li, C.; Yu, H.; Shahid, M.Q.; Lin, Y.; Qiao, X.; Xiao, J.; Gray, J.E.; et al. Small EPIDERMAL PATTERNING FACTOR-LIKE2 peptides regulate awn development in rice. Plant Physiol. 2022, 190, 516–531. [Google Scholar] [CrossRef]
- Guo, T.; Lu, Z.Q.; Xiong, Y.; Shan, J.X.; Ye, W.W.; Dong, N.Q.; Kan, Y.; Yang, Y.B.; Zhao, H.Y.; Yu, H.X.; et al. Optimization of rice panicle architecture by specifically suppressing ligand-receptor pairs. Nat. Commun. 2023, 14, 1640. [Google Scholar] [CrossRef]
- Liu, S.; Jia, F.L.; Jiao, Z.Y.; Wang, J.J.; Xia, X.L.; Yin, W.L. Ectopic expression of secretory peptide PdEPF3 in confers drought tolerance with reduced stomatal density. Acta Soc. Bot. Pol. 2019, 88, 12. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, K.; Zhang, H.; Bi, M.; He, Y.; Cui, Y.; Tan, C.; Ma, J.; Qi, M. Tomato short internodes and pedicels encode an LRR receptor-like serine/threonine-protein kinase ERECTA regulating stem elongation through modulating gibberellin metabolism. Front. Plant Sci. 2023, 14, 1283489. [Google Scholar] [CrossRef]
- Xia, Y.; Du, K.; Ling, A.; Wu, W.; Li, J.; Kang, X. Overexpression of PagSTOMAGEN, a positive regulator of stomatal density, promotes vegetative growth in poplar. Int. J. Mol. Sci. 2022, 23, 10165. [Google Scholar] [CrossRef]
- Franco, J. BZR1, you have an invite: EPFL-ERECTA wants to join your female germline specification network. Plant Cell 2023, 35, 1298–1299. [Google Scholar] [CrossRef]
- Vaghela, P.; Gandhi, G.; Trivedi, K.; Anand, K.G.V.; Chavda, D.; Manna, M.; Seth, T.; Seth, A.; Shanmugam, M.; Ghosh, A. Underpinning beneficial maize response to application of minimally processed homogenates of red and brown seaweeds. Front. Plant Sci. 2023, 14, 1273355. [Google Scholar] [CrossRef]
- Tameshige, T.; Ikematsu, S.; Torii, K.U.; Uchida, N. Stem development through vascular tissues: EPFL-ERECTA family signaling that bounces in and out of phloem. J. Exp. Bot. 2017, 68, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhang, H.; Shi, K. Plant peptides in plant defense responses. Plant Signal. Behav. 2018, 13, e1475175. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Luan, Q.L.; Zhu, Y.X.; Ma, S.; Sun, Y.; Liu, X.Y.; Liu, M.; Balint-Kurti, P.J.; Wang, G.F. Maize metacaspases modulate the defense response mediated by the NLR protein Rp1-D21 likely by affecting its subcellular localization. Plant J. 2021, 105, 151–166. [Google Scholar] [CrossRef]



| Gene ID | Protein Name | Subcellular Localization | Protein Length (aa) | Molecular Mass (Da) | Theoretical pI | Instability Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|---|
| Zm00001eb121050 | ZmEPF1 | Nucleus | 115 | 12,115.97 | 9.30 | 54.00 | −0.203 |
| Zm00001eb149400 | ZmEPF2 | Plasma membrane, Nucleus | 140 | 15,319.77 | 9.44 | 71.03 | −0.309 |
| Zm00001eb423950 | ZmEPF3 | Nucleus | 175 | 18,452.86 | 7.57 | 61.90 | −0.069 |
| Zm00001eb403370 | ZmEPF4 | Nucleus | 113 | 12,325.37 | 9.91 | 69.28 | −0.174 |
| Zm00001eb004650 | ZmEPF5 | Plasma membrane, Nucleus | 122 | 13,264.23 | 9.39 | 75.99 | −0.313 |
| Zm00001eb290530 | ZmEPF6 | Chloroplast, Nucleus, Cytoplasm | 143 | 14,827.99 | 9.49 | 54.09 | −0.031 |
| Zm00001eb194040 | ZmEPF7 | Chloroplast, Nucleus | 157 | 16,999.45 | 9.13 | 55.22 | −0.265 |
| Zm00001eb200540 | ZmEPF8 | Chloroplast, Nucleus | 216 | 23,070.52 | 10.7 | 76.84 | −0.278 |
| Zm00001eb431780 | ZmEPF9 | Plasma membrane | 148 | 15,291.78 | 8.07 | 46.20 | 0.194 |
| Zm00001eb055900 | ZmEPF10 | Nucleus | 119 | 13,069.02 | 9.78 | 63.50 | −0.680 |
| Zm00001eb069150 | ZmEPF11 | Plasma membrane | 133 | 13,454.54 | 8.43 | 52.04 | 0.180 |
| Zm00001eb255380 | ZmEPF12 | Nucleus | 143 | 15,175.36 | 9.04 | 56.26 | −0.267 |
| Zm00001eb032920 | ZmEPF13 | Chloroplast | 131 | 13,665.28 | 9.98 | 59.87 | −0.582 |
| Zm00001eb217540 | ZmEPF14 | Nucleus | 159 | 17,542.06 | 8.36 | 55.92 | −0.233 |
| Zm00001eb352020 | ZmEPF15 | Nucleus, Cytoplasm | 156 | 16,344.11 | 10.00 | 52.98 | −0.724 |
| Zm00001eb174960 | ZmEPFL1 | Plasma membrane | 143 | 15,131.95 | 6.81 | 61.21 | −0.376 |
| Zm00001eb143570 | ZmEPFL2 | Chloroplast, Nucleus, Cytoplasm | 123 | 13,623.8 | 8.83 | 47.80 | −0.062 |
| Zm00001eb364920 | ZmEPFL3 | Plasma membrane | 123 | 13,615.88 | 8.92 | 52.74 | −0.032 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Xu, K.; Li, Y.; Zhao, W.; Ji, H.; Lei, X.; Ma, T.; Ye, J.; Zhang, J.; Du, H.; et al. Investigation on the Potential Functions of ZmEPF/EPFL Family Members in Response to Abiotic Stress in Maize. Int. J. Mol. Sci. 2024, 25, 7196. https://doi.org/10.3390/ijms25137196
Liu R, Xu K, Li Y, Zhao W, Ji H, Lei X, Ma T, Ye J, Zhang J, Du H, et al. Investigation on the Potential Functions of ZmEPF/EPFL Family Members in Response to Abiotic Stress in Maize. International Journal of Molecular Sciences. 2024; 25(13):7196. https://doi.org/10.3390/ijms25137196
Chicago/Turabian StyleLiu, Rui, Keli Xu, Yu Li, Wanqing Zhao, Hongjing Ji, Xiongbiao Lei, Tian Ma, Juan Ye, Jianhua Zhang, Hewei Du, and et al. 2024. "Investigation on the Potential Functions of ZmEPF/EPFL Family Members in Response to Abiotic Stress in Maize" International Journal of Molecular Sciences 25, no. 13: 7196. https://doi.org/10.3390/ijms25137196
APA StyleLiu, R., Xu, K., Li, Y., Zhao, W., Ji, H., Lei, X., Ma, T., Ye, J., Zhang, J., Du, H., & Cao, S.-K. (2024). Investigation on the Potential Functions of ZmEPF/EPFL Family Members in Response to Abiotic Stress in Maize. International Journal of Molecular Sciences, 25(13), 7196. https://doi.org/10.3390/ijms25137196







