The Antimicrobial Activity of Human Defensins at Physiological Non-Permeabilizing Concentrations Is Caused by the Inhibition of the Plasma Membrane H+-ATPases
Abstract
1. Introduction
2. Results
2.1. Determination of Antimicrobial Non-Permeabilizing Concentrations of Human Defensins
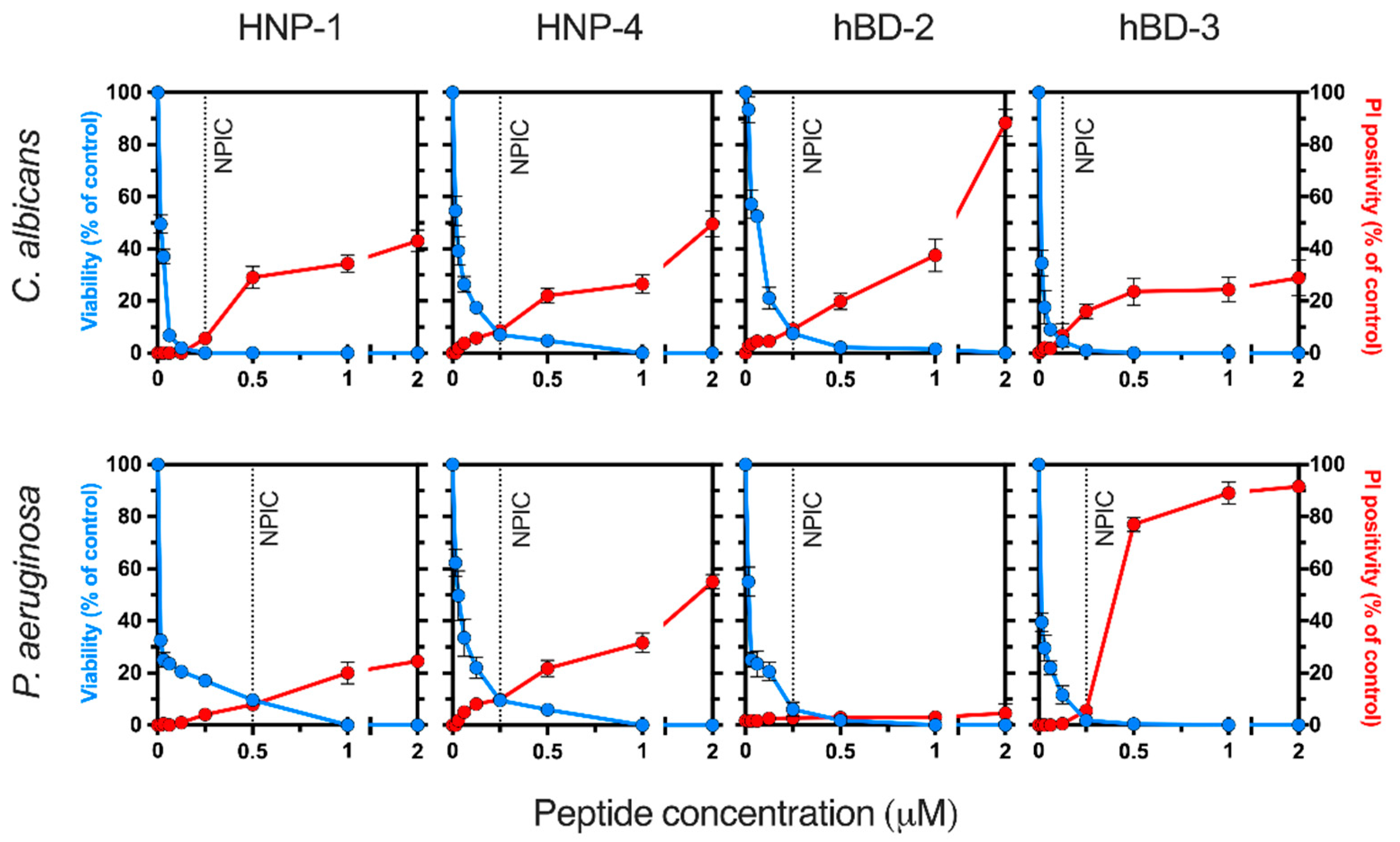
2.2. Effect of Human Defensins on Cellular K+-Homeostasis
2.2.1. Effect of High Extracellular K+-Concentration on Antimicrobial Activity
2.2.2. Evaluation of K+-Efflux Induced by Defensins
2.2.3. Effect of the Inhibition of the K+-Channel Tok1p on Candidacidal Activity
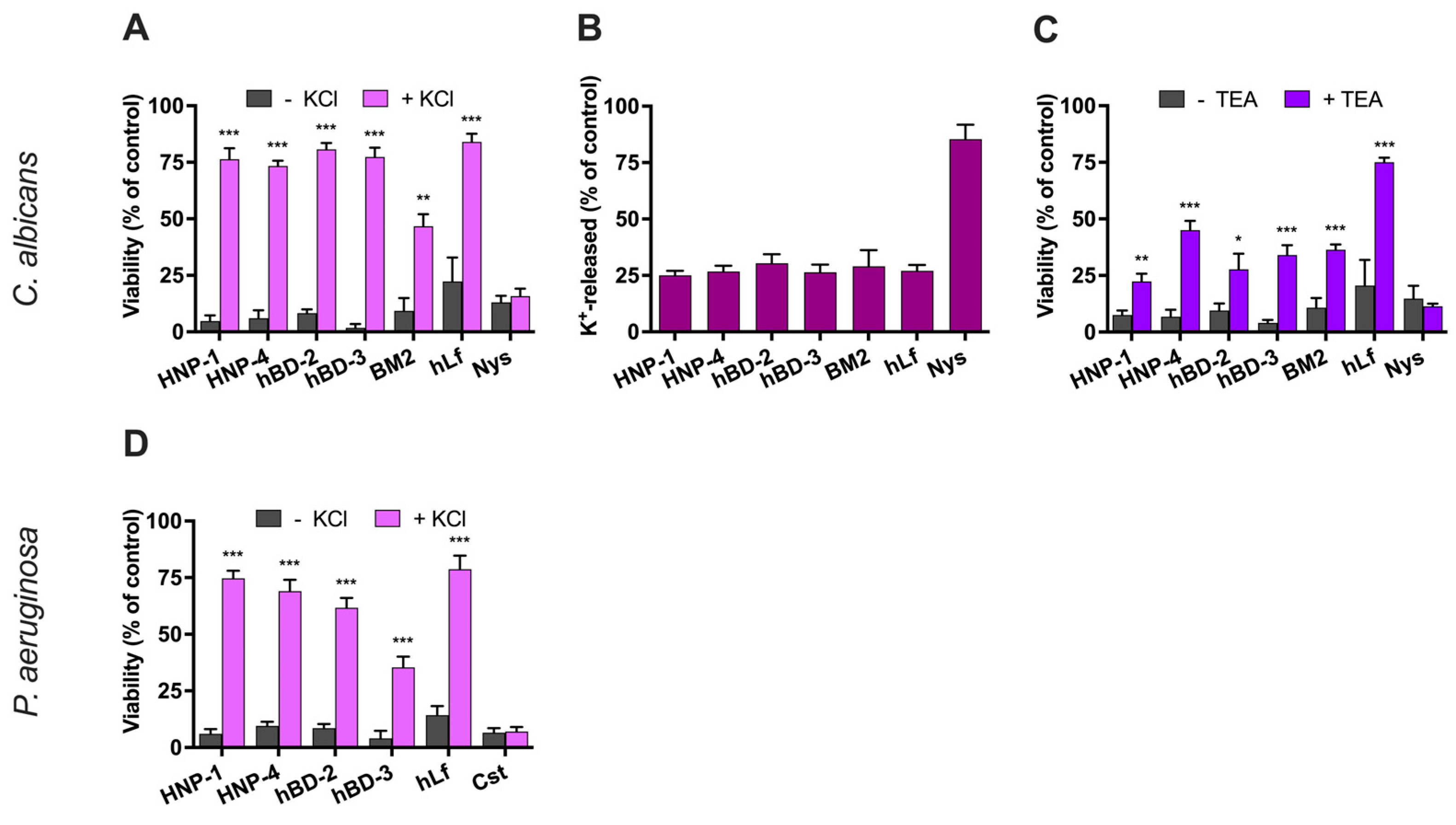
2.3. Influence of Bioenergetics on the Microbicidal Activity of Defensins
2.3.1. Influence of Cellular Respiration on the Microbicidal Activity of Defensins
2.3.2. Effect of CCCP on the Defensin Bactericidal Activity
2.3.3. Role of Mitochondrial ATP Synthase in the Candidacidal Activity of Defensins

2.4. Human Defensins Inhibit Fungal Pma1p H+-ATPase
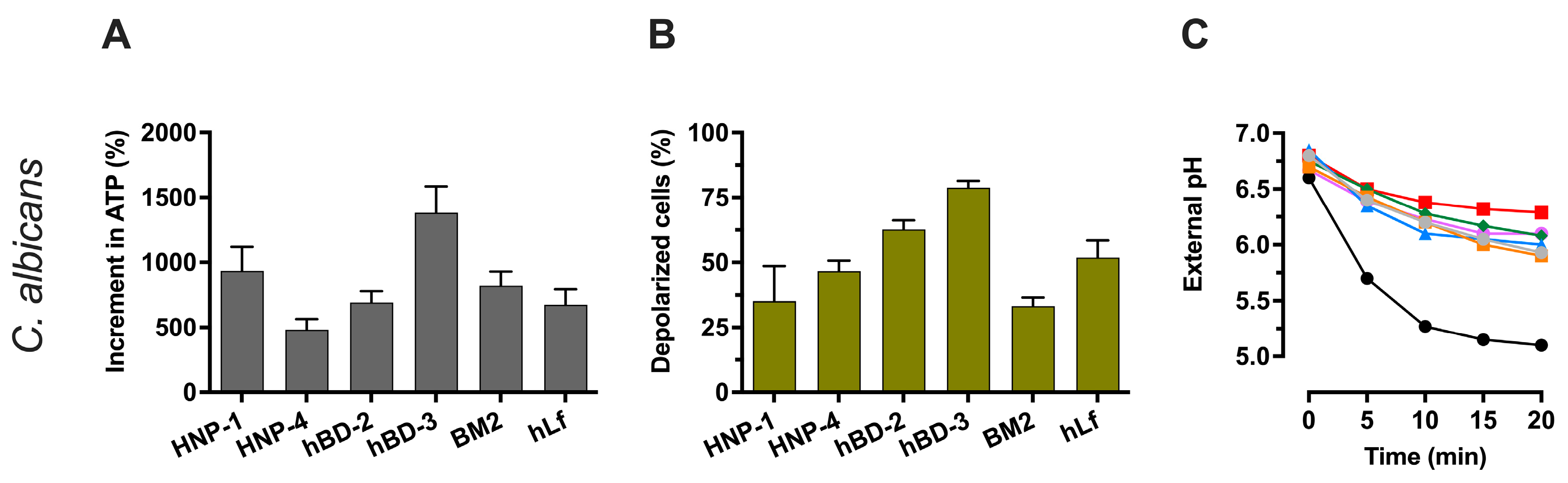
2.4.1. Evaluation of Cellular ATP Levels in Relation to Candidacidal Activity of Defensins
2.4.2. Effect of Defensins on The Electrical Potential of the Plasma Membrane
2.4.3. Effect of Defensins on H+-Extrusion by Pma1p H+-ATPase
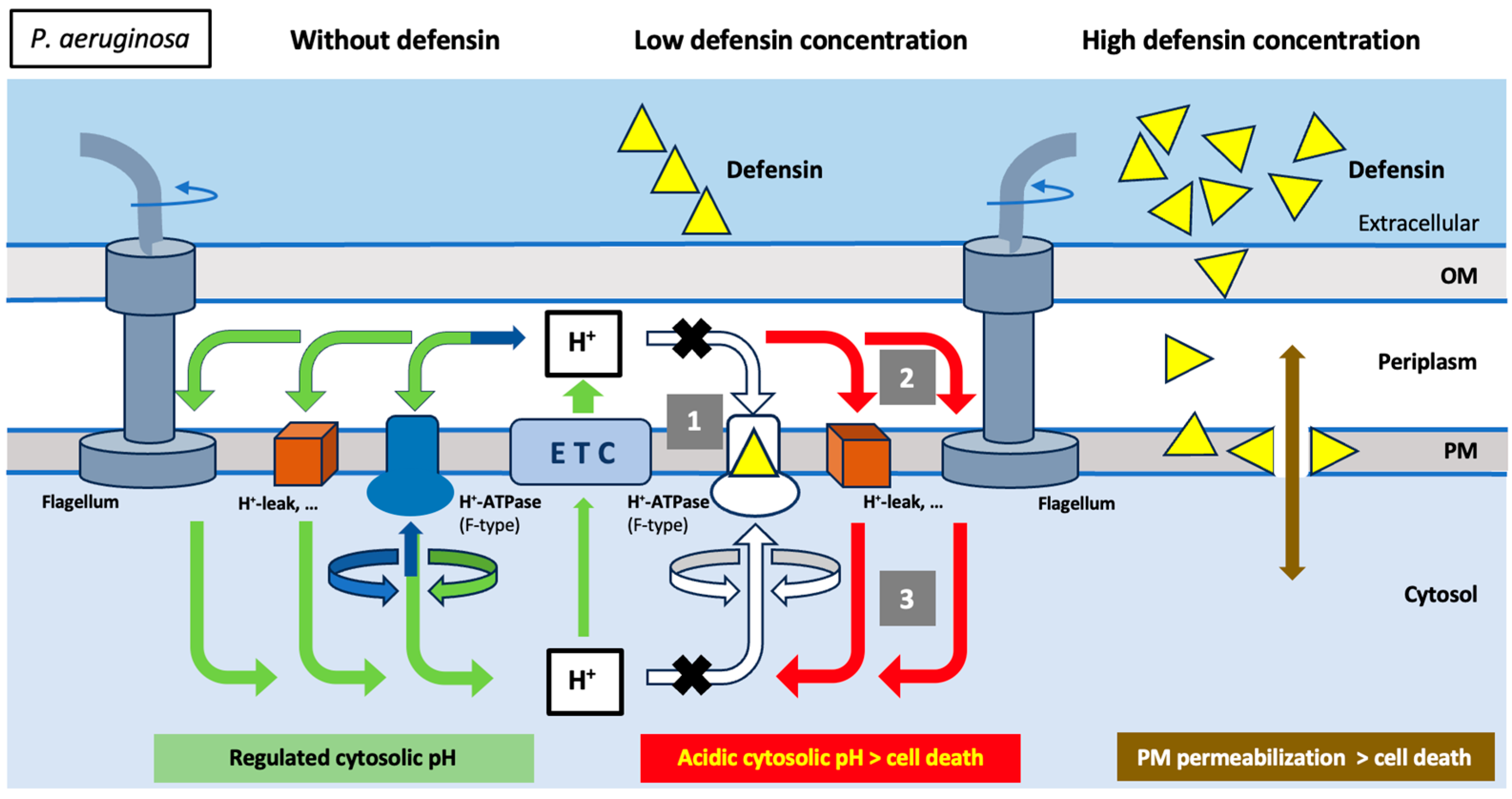
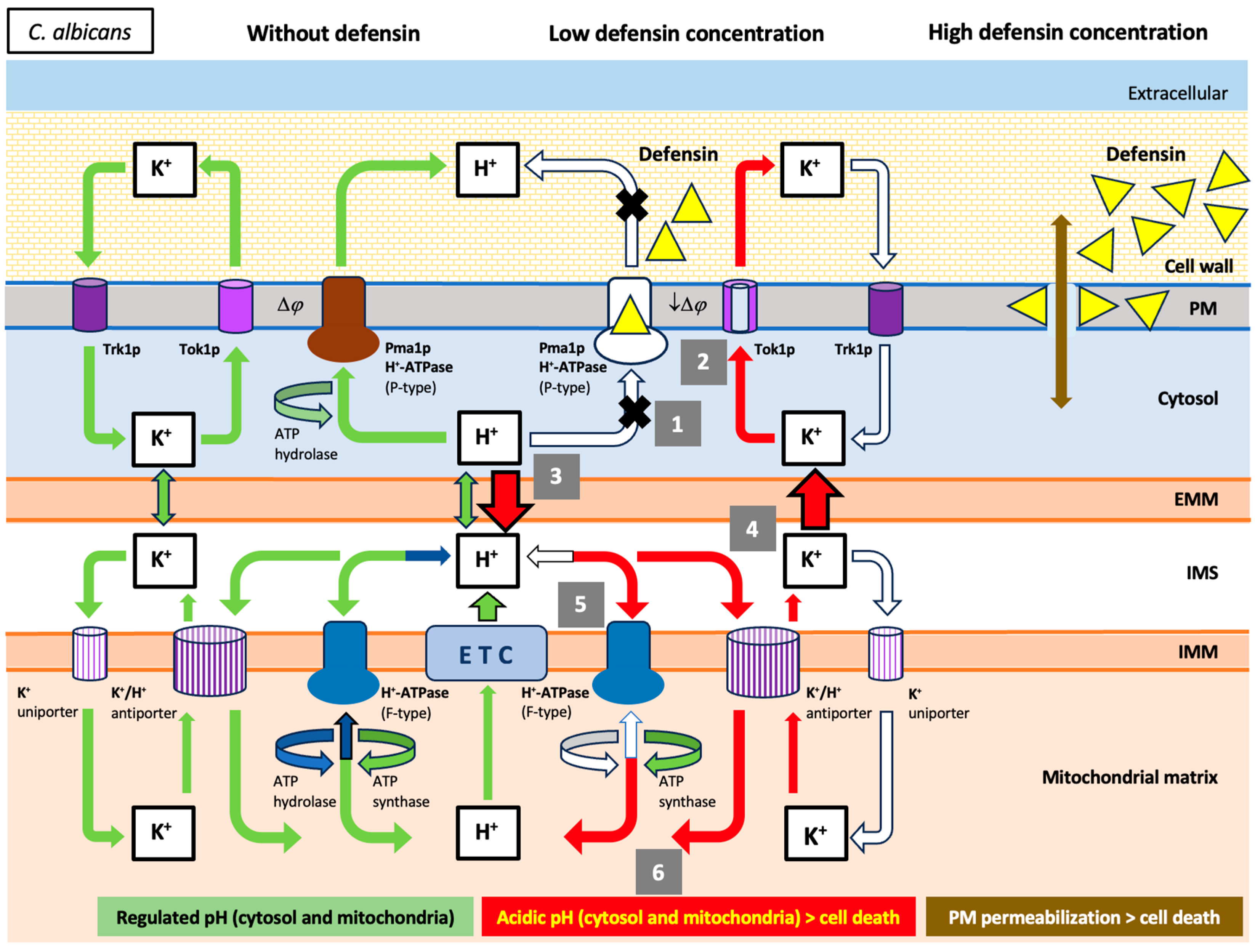
3. Discussion
| Human α-Defensins | Human β-Defensins | Human Transferrins | |||
|---|---|---|---|---|---|
| HNP-1 | HNP-4 | hBD-2 | hBD-3 | Human Lactoferrin | |
| Number of amino acids | 30 | 33 | 41 | 45 | 691 (protein) |
| Archetypal antimicrobial structures γ-core motif (number) Isomeric form (type) | Yes (one) Levomeric (2) | Yes (one) Levomeric (2) | Yes (one) Levomeric (2) | Yes (one) Levomeric (2) | Yes (two) Levomeric (1) |
| Antimicrobial activity NPIC (μM) Cidal/static Membrane depolarization Membrane permeabilization Partial K+-efflux ATP increment/ATP-efflux Inhibition of H+-extrusion (yeast) Antimicrobial range | 0.25 a, 0.5 b Cidal [24,62] Yes [26] Yes [14] Yes Yes [47]/Yes [47] Yes Gm +,Gm −,F,V,P | 0.25 a,b Cidal [62,63] Yes [12] Yes [23] Yes Yes/ND Yes Gm +,Gm −,F,V | 0.25 a,b Cidal [20] Yes [11] Yes [11,13]/No [20] Yes Yes/ND Yes Gm +,Gm −,F,V | 0.125 a, 0.25 b Cidal [12,20] Yes [11] No [20]/Yes [11] Yes Yes/ND Yes Gm +,Gm −, F,V,P | 5 a, 1 b Cidal [34,35,64]/Static [65,66] Yes [43,67] No [31,34,42] Yes [31,42,43,44] Yes [35]/ND Yes Gm +,Gm −,F,V,P |
| Cell death inhibitors Extracellular inhibitors: K+ or Na+ Ca2+ or Mg2+ Tok1p inhibitors (TEA) Anoxia Intracellular inhibitors: CCCP or DNP Azide Antimycin A (+SHAM) Piericidin A Oligomycin A | Yes [68] Yes (Ca2+)/No (Mg2+) [24] Yes Yes [24] Yes [24] Yes [20,24] Yes [24] Yes Yes | Yes [69] ND Yes ND ND ND ND ND ND | Yes [20] Yes [20] Yes ND Yes Yes [20] ND Yes Yes | No [11,12]/Yes [20] No [20] Yes ND ND Yes [20] ND ND ND | Yes [31,36,43] Yes [43] Yes [36,44] Yes [34,35] Yes [34] No [34] ND Yes [34,35] Yes [35] |
| Cellular target | PM H+-ATPase | PM H+-ATPase | PM H+-ATPase Phospholipids [13] | PM H+-ATPase | PM H+-ATPase [34,35,36] PM (V) H+-ATPase [39] |
| Cell death type (yeast) | ND Permeabilization [14] | ND Permeabilization [23] | ND Permeabilization (No [20]/Yes [11,13]) | ND Permeabilization (No [20]/Yes [11,20]) | Apoptosis-like cell death [36,44] No permeabilization [31,42] |
4. Materials and Methods
4.1. Materials
4.2. Strains and Growth Conditions
4.3. Antimicrobial Assays
4.4. Permeability Assay
4.5. Extracellular Potassium Measurement
4.6. Oxygen Consumption Measurement
4.7. Measurement of ATP
4.8. Membrane Potential Monitoring
4.9. Measurements of Acidification of the External Medium
4.10. Statistics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Selsted, M.E.; Oullette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.; Barran, P.E.; Dorin, J.R. Structure-activity relationships in β-defensin peptides. Peptide Sci. 2007, 90, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Fruitwala, S.; El-Naccache, D.W.; Chang, T.L. Multifaceted immune functions of human defensins and underlying mechanisms. Semin. Cell Dev. Biol. 2019, 88, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zong, X.; Jin, M.; Min, J.; Wang, F.; Wang, Y. Mechanisms and regulation of defensins in host defense. Signal Transduct. Target Ther. 2023, 8, 300. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Lehrer, R.I. Defensins. Curr. Opin. Immunol. 1994, 6, 584–589. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef]
- Lehrer, R.I. Primate defensins. Nat. Rev. Microbiol. 2004, 2, 727–738. [Google Scholar] [CrossRef]
- Nguyen, T.X.; Cole, A.M.; Lehrer, R.I. Evolution of primate theta-defensins: A serpentine path to a sweet tooth. Peptides 2003, 24, 1647–1654. [Google Scholar] [CrossRef]
- Pazgier, M.; Hoover, D.M.; Yang, D.; Lu, W.; Lubkowski, J. Human β-defensins. Cell. Mol. Life Sci. 2006, 63, 1294–1313. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.J.A.; Kvansakul, M.; Lay, F.T.; Phan, T.K.; Hulett, M.D. Defensin-lipid interactions in membrane targeting: Mechanisms of action and opportunities for the development of antimicrobial and anticancer therapeutics. Biochem. Soc. Trans. 2022, 50, 423–437. [Google Scholar] [CrossRef]
- Ouhara, K.; Komatsuzawa, H.; Yamada, S.; Shiba, H.; Fujiwara, T.; Ohara, M.; Sayama, K.; Hashimoto, K.; Kurihara, H.; Sugai, M. Susceptibilities of periodontopathogenic and cariogenic bacteria β-defensin to antibacterial peptides, β-defensins and LL37, produced by human epithelial cells. J. Antimicrob. Chemother. 2005, 55, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Bartels, J.; Christophers, E.; Schroder, J.M. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 2001, 276, 5707–5713. [Google Scholar] [CrossRef] [PubMed]
- Järva, M.; Phan, T.K.; Lay, F.T.; Caria, S.; Kvansakul, M.; Hulett, M.D. Human β-defensin 2 kills Candida albicans through phophatidylinositol 4,5-biphosphate-mediated membrane permeabilization. Sci. Adv. 2018, 4, eaat0979. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Barton, A.; Daher, K.A.; Harwig, S.S.; Ganz, T.; Selsted, M.E. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Invest. 1989, 84, 553–561. [Google Scholar] [CrossRef]
- Hill, C.P.; Yee, J.; Selsted, M.E.; Eisenberg, D. Crystal structure of defensin HNP-3, an amphiphilic dimer: Mechanisms of membrane permeabilization. Science 1991, 251, 1481–1485. [Google Scholar] [CrossRef]
- Hu, H.; Di, B.; Tolbert, W.D.; Gohain, N.; Yuan, W.; Gao, P.; Ma, B.; He, Q.; Pazgier, M.; Zhao, L.; et al. Systematic mutational analysis of human neutrophil α-defensin HNP4. Biochim. Biophys. Acta Biomembr. 2019, 1861, 835–844. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Lu, W. α-Defensins in human innate immunity. Immun. Rev. 2012, 245, 84–112. [Google Scholar] [CrossRef]
- Bonucci, A.; Balducci, E.; Pistolesi, S.; Pogni, R. The defensin-lipid interaction: Insights on the binding states of the human antimicrobial peptide HNP-1 to model bacterial membranes. Biochim. Biophys. Acta 2013, 1828, 758–764. [Google Scholar] [CrossRef]
- Feng, Z.; Jiang, B.; Chandra, J.; Ghannoum, M.; Nelson, S.; Weinberg, A. Human beta-defensins: Differential activity against candidal species and regulation by Candida albicans. J. Dent. Res. 2005, 84, 445–450. [Google Scholar] [CrossRef]
- Vylkova, S.; Nayyar, N.; Li, W.; Edgerton, M. Human beta-defensins kill Candida albicans in an energy-dependent and salt-sensitive manner without causing membrane disruption. Antimicrob. Agents Chemother. 2007, 51, 154–161. [Google Scholar] [CrossRef]
- De Leuw, E.; Li, C.; Zeng, P.; Li, C.; Diepeveen-de-Buin, M.; Lu, W.Y.; Breukink, E.; Lu, W. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 2010, 584, 1543–1548. [Google Scholar] [CrossRef]
- Munch, D.; Sahl, H.G. Structural variations of the cell wall precursor lipid II in Gram positive bacteria-Impact on binding and efficacy of antimicrobial peptides. Biochim. Biophys. Acta 2015, 1848, 3062–3071. [Google Scholar] [CrossRef] [PubMed]
- Mathew, B.; Ramakrishnan, N. Variations in the interaction of human defensins with Escherichia coli: Possible implications in bacterial killing. PLoS ONE 2017, 12, e0175858. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Ganz, T.; Szklarek, D.; Selsted, M.E. Modulation of the in vitro candidacidal activity of human neutrophil defensins by target cell metabolism and divalent cations. J. Clin. Invest. 1988, 81, 1829–1835. [Google Scholar] [CrossRef] [PubMed]
- Thevissen, K.; Terras, F.R.; Broekaert, W.F. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl. Environ. Microbiol. 1999, 65, 5451–5458. [Google Scholar] [CrossRef] [PubMed]
- Kjellerup, L.; Gordon, S.; Cohrt, K.O.; Brown, W.D.; Fuglsang, A.T.; Winther, A.L. Identification of antifungal H+-ATPase inhibitors with effect on plasma membrane potential. Antimicrob. Agents Chemother. 2017, 61, e00032-17. [Google Scholar] [CrossRef] [PubMed]
- Kagan, B.L.; Selsted, M.E.; Ganz, T.; Lehrer, R.I. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc. Natl. Acad. Sci. USA 1990, 87, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Lu, W. Defensins: A double-edged sword in host immunity. Front. Immunol. 2020, 11, 764. [Google Scholar] [CrossRef]
- Nigro, E.; Colavita, I.; Sarnataro, D.; Scudiero, O.; Zambrano, G.; Granata, V.; Daniele, A.; Carotenuto, A.; Galdiero, S.; Folliero, V.; et al. An ancestral host defence peptide within human β-defensin 3 recapitulates the antibacterial and antiviral activity of the full-length molecule. Sci. Rep. 2015, 5, 18450. [Google Scholar] [CrossRef]
- Yount, N.Y.; Yeaman, M.R. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 7363–7368. [Google Scholar] [CrossRef]
- Viejo-Diaz, M.; Andrés, M.T.; Pérez-Gil, J.; Sánchez, M.; Fierro, J.F. Potassium efflux induced by a new lactoferrin-derived peptide mimicking the effect of native human lactoferrin on the bacterial cytoplasmic membrane. Biochemistry 2003, 68, 217–227. [Google Scholar] [CrossRef]
- Viejo-Diaz, M.; Andrés, M.T.; Fierro, J.F. Different anti-Candida activities of two human lactoferrin-derived peptides, Lfpep and kaliocin-1. Antimicrob. Agents Chemother. 2005, 49, 2583–2588. [Google Scholar] [CrossRef]
- Yount, N.Y.; Andrés, M.T.; Fierro, J.F.; Yeaman, M.R. The gamma-core motif correlates with antimicrobial activity in cysteine-containing kaliocin-1 originating from transferrins. Biochim. Biophys. Acta 2007, 1768, 2862–2872. [Google Scholar] [CrossRef]
- Andrés, M.T.; Fierro, J.F. Antimicrobial mechanism of action of transferrins: Selective inhibition of H+-ATPase. Antimicrob. Agents Chemother. 2010, 54, 4335–4342. [Google Scholar] [CrossRef] [PubMed]
- Andrés, M.T.; Acosta-Zaldívar, M.; Fierro, J.F. Antifungal mechanism of action of lactoferrin: Identification of H+-ATPase (P3A-Type) as a new apoptotic-cell membrane receptor. Antimicrob. Agents Chemother. 2016, 60, 4206–4216. [Google Scholar] [CrossRef] [PubMed]
- Andrés, M.T.; Acosta-Zaldívar, M.; González-Seisdedos, J.; Fierro, J.F. Cytosolic acidification is the first transduction signal of lactoferrin-induced regulated cell death pathway. Int. J. Mol. Sci. 2019, 20, 5838. [Google Scholar] [CrossRef]
- Santos-Pereira, C.; Andrés, M.T.; Chaves, S.R.; Fierro, J.F.; Gerós, H.; Manon, S.; Rodrigues, L.R.; Côrte-Real, M. Lactoferrin perturbs lipid rafts and requires integrity of Pma1p-lipid rafts association to exert its antifungal activity against Saccharomyces cerevisiae. Int. J. Biol. Macromol. 2021, 171, 343–357. [Google Scholar] [CrossRef]
- Santos-Pereira, C.; Rocha, J.F.; Fernandes, H.; Rodrigues, L.R.; Côrte-Real, M.; Sousa, S.F. The milk-derived lactoferrin inhibits V-ATPase activity by targeting its V1 domain. Int. J. Biol. Macromol. 2021, 186, 54–70. [Google Scholar] [CrossRef] [PubMed]
- Santos-Pereira, C.; Andrés, M.T.; Fierro, J.F.; Rodrigues, L.R.; Côrte-Real, M. A review on lactoferrin as a proton pump inhibitor. Int. J. Biol. Macromol. 2022, 202, 309–317. [Google Scholar] [CrossRef]
- Monk, B.C.; Niimi, K.; Lin, S.; Knight, A.; Kardos, T.B.; Cannon, R.D.; Parshot, R.; King, A.; Lun, D.; Harding, D.R. Surface-active fungicidal D-peptide inhibitors of the plasma membrane proton pump that block azole resistance. Antimicrob. Agents Chemother. 2005, 49, 57–70. [Google Scholar] [CrossRef]
- Clausen, J.D.; Kjellerup, L.; Cohrt, K.O.; Hansen, J.B.; Dalby-Brown, W.; Winther, A.L. Elucidation of antimicrobial activity and mechanism of action by N-substituted carbazole derivatives. Bioorg. Med. Chem. Lett. 2017, 27, 4564–4570. [Google Scholar] [CrossRef] [PubMed]
- Viejo-Diaz, M.; Andrés, M.T.; Fierro, J.F. Effects of human lactoferrin on the cytoplasmic membrane of Candida albicans cells related with its candidacidal activity. FEMS Immunol. Med. Microbiol. 2004, 42, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Viejo-Diaz, M.; Andrés, M.T.; Fierro, J.F. Modulation of in vitro fungicidal activity of human lactoferrin against Candida albicans by extracellular cation concentration and target cell metabolic activity. Antimicrob. Agents Chemother. 2004, 48, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- Andrés, M.T.; Viejo-Diaz, M.; Fierro, J.F. Human lactoferrin induces apoptosis-like cell death in Candida albicans: Critical role of K+-channel-mediated K+-efflux. Antimicrob. Agents Chemother. 2008, 52, 4081–4088. [Google Scholar] [CrossRef]
- Sijpesteijn, A.K. Induction of cytochrome formation and stimulation of oxidative dissimilation by hemin in Streptococcus lactis and Leuconostoc mesenteroides. Antonie Van Leeuwenhoek 1970, 36, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Duwat, P.; Sourice, S.; Cesselin, B.; Lamberet, G.; Vido, K.; Gaudu, P.; Le Loir, Y.; Violet, F.; Loubière, P.; Gruss, A. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. 2001, 183, 4509–4516. [Google Scholar] [CrossRef] [PubMed]
- Edgerton, M.; Koshlukova, S.E.; Araujo, M.W.; Patel, R.C.; Dong, J.; Bruenn, J.A. Salivary histatin 5 and human neutrophil defensin 1 kill Candida albicans via shared pathways. Antimicrob. Agents Chemother. 2000, 44, 3310–3316. [Google Scholar] [CrossRef]
- Serrano, R. Transport across yeast vacuolar and plasma membranes. In The Molecular Biology of the Yeast Saccharomyces. Genome, Dynamics, Protein Synthesis and Energetics; Strathern, J.N., Jones, E.W., Broach, J.R., Eds.; Cold Spring Harbour Laboratory Press: Cold Spring Harbor, NY, USA, 1991; pp. 523–585. [Google Scholar]
- Thevissen, K.; Marchand, A.; Chaltin, P.; Meert, M.K.; Cammue, B.P.A. Antifungal carbazoles. Curr. Med. Chem. 2009, 16, 2205–2211. [Google Scholar] [CrossRef]
- Bublitz, M.; Kjellerup, L.; Cohrt, K.O.; Gordon, S.; Mortensen, A.L.; Clausen, J.D.; Pallin, T.D.; Hansen, J.B.; Fuglsang, A.T.; Dalby-Brown, W.; et al. Tetrahydrocarbazoles are a novel class of potent P-type ATPase inhibitors with antifungal activity. PLoS ONE 2018, 13, e0188620. [Google Scholar] [CrossRef]
- Ihi, T.; Nakazato, M.; Mukae, H.; Matsukura, S. Elevated concentrations of human neutrophil peptides in plasma, blood, and body fluids from patients with infections. Clin. Infect. Dis. 1997, 25, 1134–1140. [Google Scholar] [CrossRef]
- Lehrer, R.I. Questions and answers about defensins. Clin. Infect. Dis. 1997, 25, 1141–1142. [Google Scholar] [CrossRef][Green Version]
- Berkestedt, I.; Herwald, H.; Ljunggren, L.; Nelson, A.; Bodelsson, M. Elevated plasma levels of antimicrobial polypeptides in patients with severe sepsis. J. Innate Immun. 2010, 2, 478–482. [Google Scholar] [CrossRef]
- Chileveru, H.R.; Lim, S.A.; Chairatana, P.; Wommack, A.J.; Chiang, I.L.; Nolan, E.M. Visualizing attack of Escherichia coli by the antimicrobial peptide human defensin 5. Biochemistry 2015, 54, 1767–1777. [Google Scholar] [CrossRef]
- Olson, V.L.; Hansing, R.L.; McClary, D.O. The role of metabolic energy in the lethal action of basic proteins on Candida albicans. Can. J. Microbiol. 1977, 23, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Zaldívar, M.; Andrés, M.T.; Rego, A.; Pereira, C.S.; Fierro, J.F.; Côrte-Real, M. Human lactoferrin triggers a mitochondrial and caspase-dependent regulated cell death in Saccharomyces cerevisiae. Apoptosis 2016, 21, 163–173. [Google Scholar] [CrossRef]
- Stevens, H.C.; Nichols, J.W. The proton electrochemical gradient across the plasma membrane of yeast is necessary for phospholipid flip. J. Biol. Chem. 2007, 282, 17563–17567. [Google Scholar] [CrossRef]
- Bracey, D.; Holyoak, C.D.; Nebe-von Caron, G.; Coote, P.J. Determination of the intracellular pH (pHi) of growing cells of Saccharomyces cerevisiae: The effect of reduced expression of the membrane H+-ATPase. J. Microbiol. Methods 1998, 31, 113–125. [Google Scholar] [CrossRef]
- Olsen, L.F.; Andersen, A.Z.; Lunding, A.; Brasen, J.C.; Poulsen, A.K. Regulation of glycolytic oscillations by mitochondrial and plasma membrane H+-ATPases. Biophys. J. 2009, 96, 3850–3861. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S. Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of the CSalphabeta defensins. Mol. Immunol. 2008, 45, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, I.; O’Connor, P.M.; Hill, C.; Stanton, C.; Ross, R.P. Actinomyces produces defensin-like bacteriocins (Actifensins) with a highly degenerate structure and broad antimicrobial activity. J. Bacteriol. 2020, 202, e00529-19. [Google Scholar] [CrossRef] [PubMed]
- Ericksen, B.; Wu, Z.; Lu, W.; Lehrer, R.I. Antibacterial activity and specificity of the six human α-defensins. Antimicrob. Agents Chemother. 2005, 49, 269–275. [Google Scholar] [CrossRef]
- Wilde, C.G.; Griffith, J.E.; Marra, M.N.; Snable, J.L.; Scott, R.W. Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J. Biol. Chem. 1989, 264, 11200–11203. [Google Scholar] [CrossRef] [PubMed]
- Arnold, R.R.; Cole, M.F.; McGhee, J.R. A bactericidal effect for human lactoferrin. Science 1977, 197, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Kong, W.; Nakayama, K. Human lactoferrin binds and removes the hemoglobin receptor protein of the periodontopathogen Porphyromonas gingivalis. J. Biol. Chem. 2000, 275, 30002–30008. [Google Scholar] [CrossRef] [PubMed]
- Aguila, A.; Herrera, A.G.; Morrison, D.; Cosgrove, B.; Perojo, A.; Montesinos, I.; Pérez, J.; Sierra, G.; Gemmell, C.G.; Brock, J.H. Bacteriostatic activity of human lactoferrin against Staphylococcus aureus is a function of its iron-binding properties and is not influenced by antibiotic resistance. FEMS Immunol. Med. Microbiol. 2001, 31, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Andrés, M.T.; Viejo-Diaz, M.; Pérez, F.; Fierro, J.F. Antibiotic tolerance induced by lactoferrin in clinical Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 2005, 49, 1613–1616. [Google Scholar] [CrossRef]
- Turner, J.; Cho, Y.; Dinh, N.N.; Waring, A.J.; Lehrer, R.I. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 1998, 42, 2206–2214. [Google Scholar] [CrossRef]
- García, J.R.; Krause, A.; Schulz, S.; Rodríguez-Jiménez, F.J.; Klüver, E.; Adermann, K.; Forssmann, U.; Frimpong-Boateng, A.; Bals, R.; Forssmann, W.G. Human beta-defensin 4: A novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 2001, 15, 1819–1821. [Google Scholar] [CrossRef] [PubMed]
| Amino Acid Sequence | Aa (No.) | mav | z | HR | |
|---|---|---|---|---|---|
| α-defensins |  | ||||
| HNP-1 | 30 | 3448 | +3 | 38 | |
| HNP-4 | 33 | 3802 | +4 | 39 | |
| β-defensins | |||||
| hBD-2 | 41 | 4328 | +6 | 29 | |
| hBD-3 | 45 | 5155 | +11 | 23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrés, M.T.; Fierro, P.; Antuña, V.; Fierro, J.F. The Antimicrobial Activity of Human Defensins at Physiological Non-Permeabilizing Concentrations Is Caused by the Inhibition of the Plasma Membrane H+-ATPases. Int. J. Mol. Sci. 2024, 25, 7335. https://doi.org/10.3390/ijms25137335
Andrés MT, Fierro P, Antuña V, Fierro JF. The Antimicrobial Activity of Human Defensins at Physiological Non-Permeabilizing Concentrations Is Caused by the Inhibition of the Plasma Membrane H+-ATPases. International Journal of Molecular Sciences. 2024; 25(13):7335. https://doi.org/10.3390/ijms25137335
Chicago/Turabian StyleAndrés, María T., Patricia Fierro, Victoria Antuña, and José F. Fierro. 2024. "The Antimicrobial Activity of Human Defensins at Physiological Non-Permeabilizing Concentrations Is Caused by the Inhibition of the Plasma Membrane H+-ATPases" International Journal of Molecular Sciences 25, no. 13: 7335. https://doi.org/10.3390/ijms25137335
APA StyleAndrés, M. T., Fierro, P., Antuña, V., & Fierro, J. F. (2024). The Antimicrobial Activity of Human Defensins at Physiological Non-Permeabilizing Concentrations Is Caused by the Inhibition of the Plasma Membrane H+-ATPases. International Journal of Molecular Sciences, 25(13), 7335. https://doi.org/10.3390/ijms25137335









