Soil Giant Phage: Genome and Biological Characteristics of Sinorhizobium Jumbo Phage
Abstract
1. Introduction
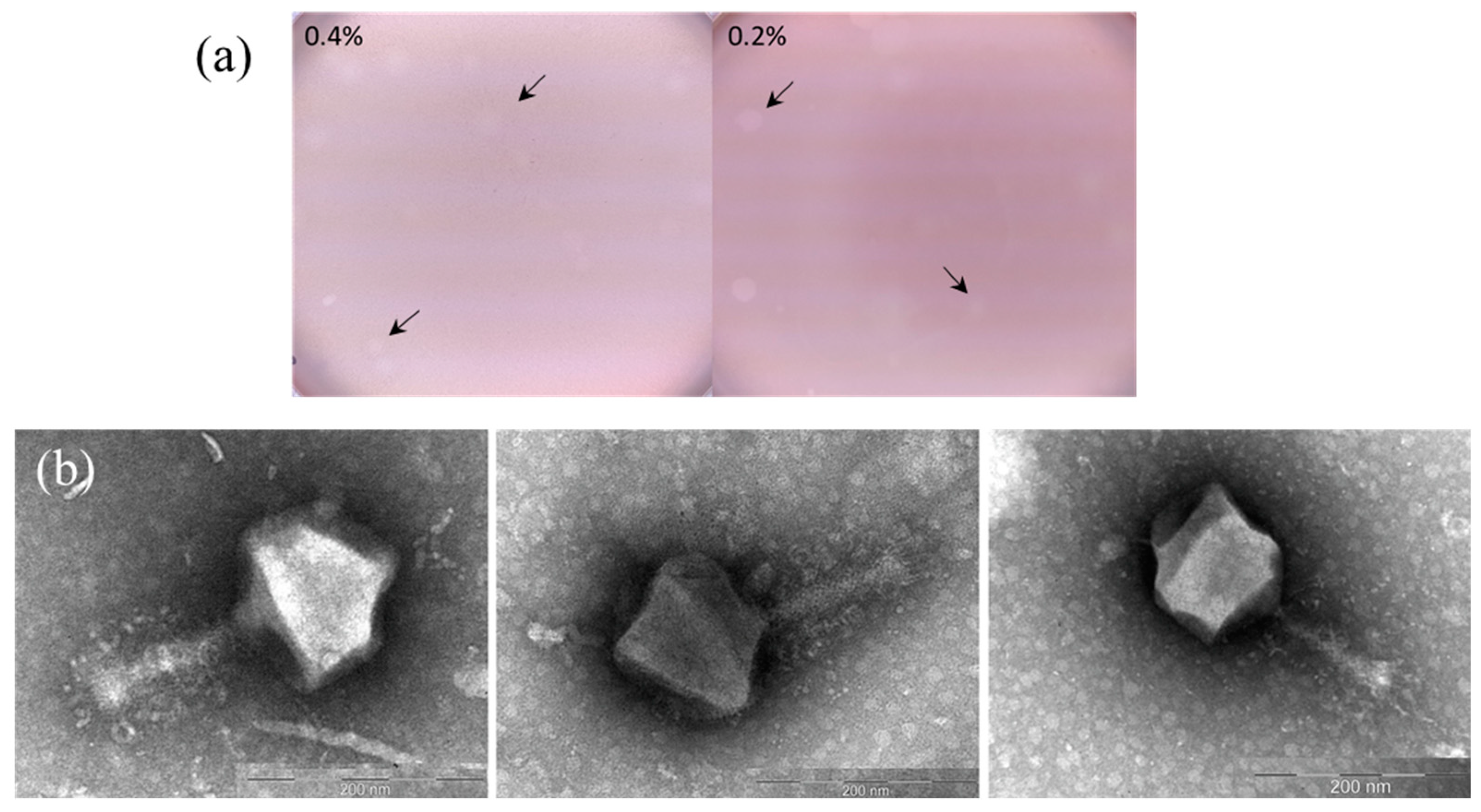
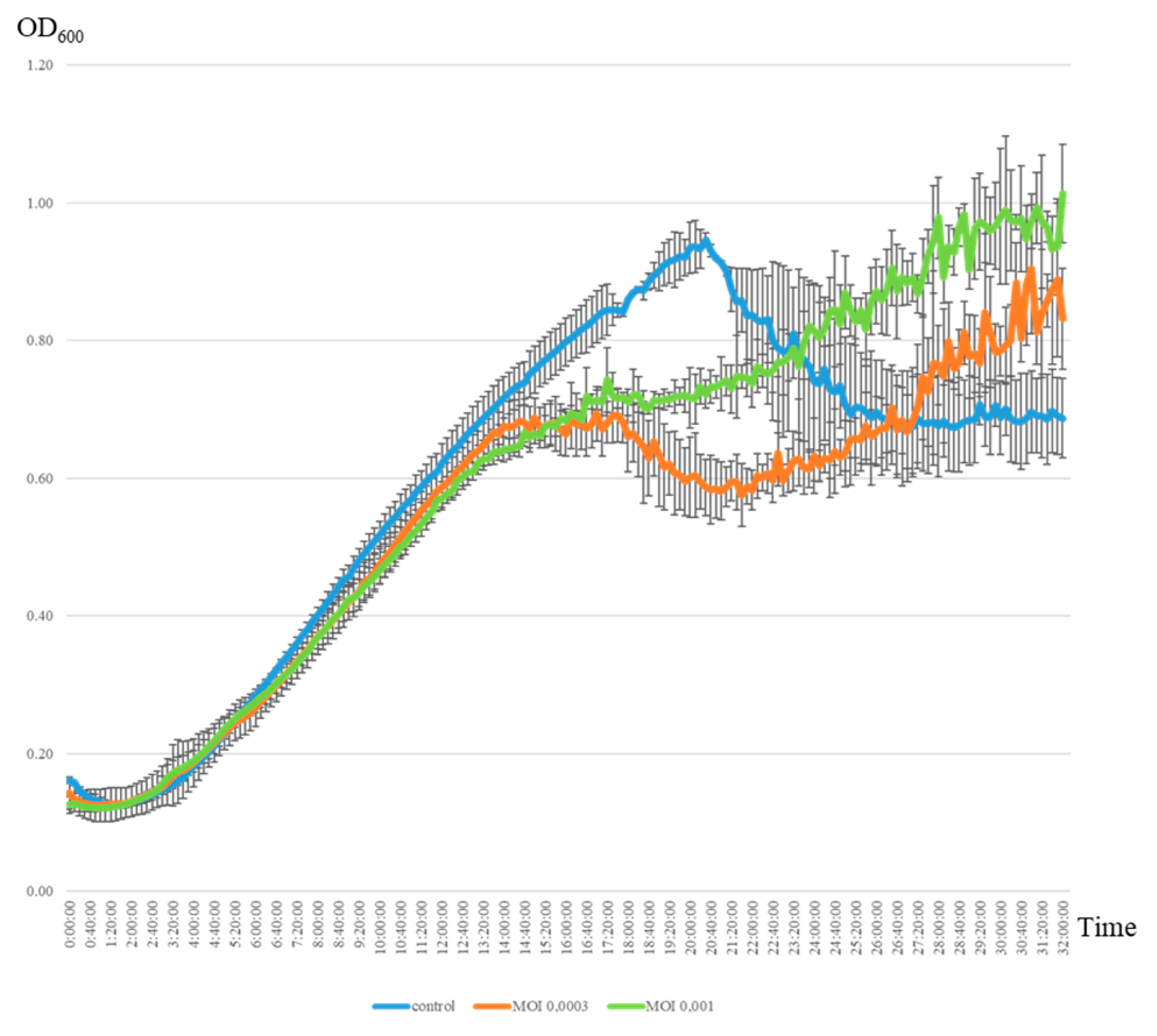
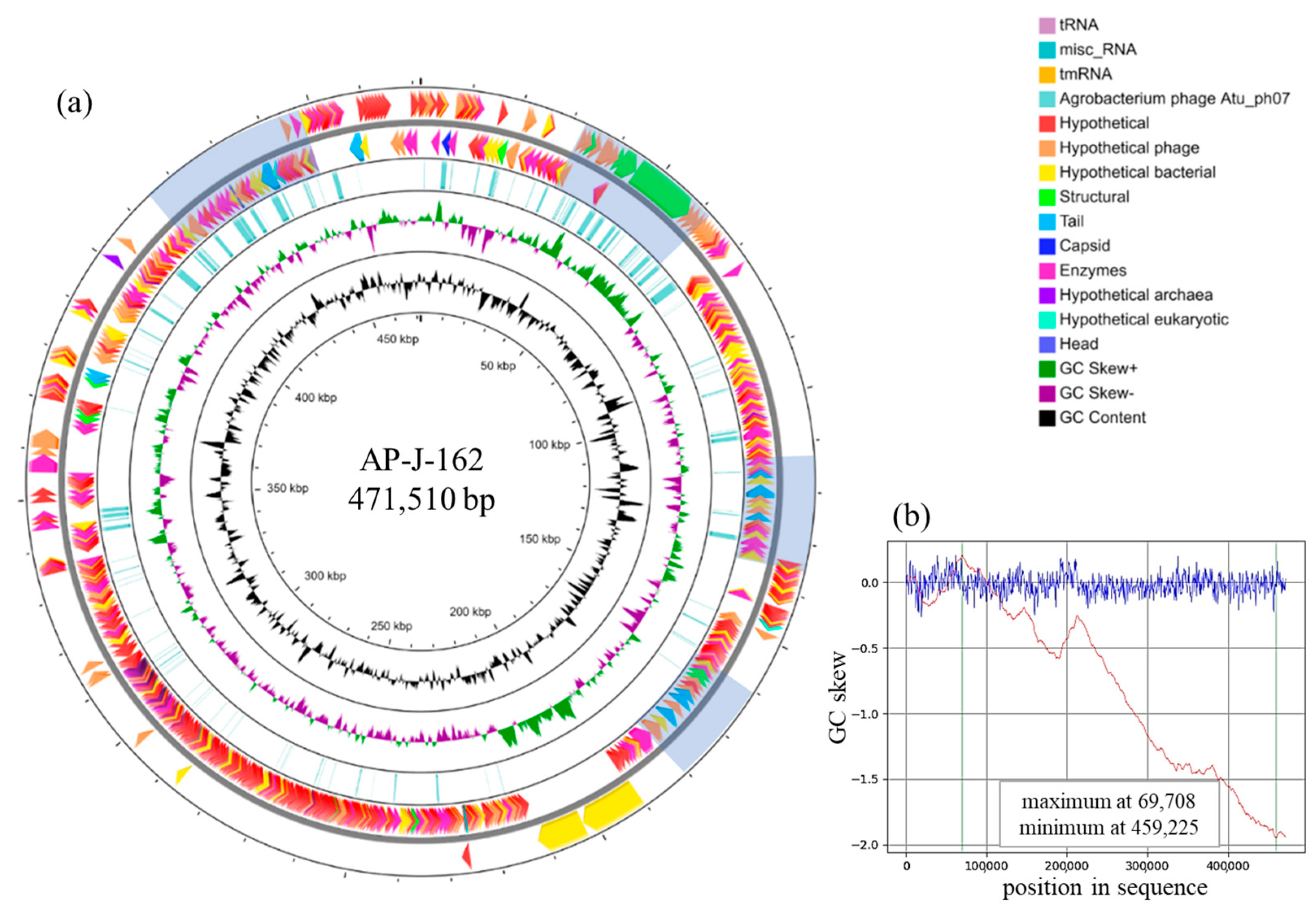
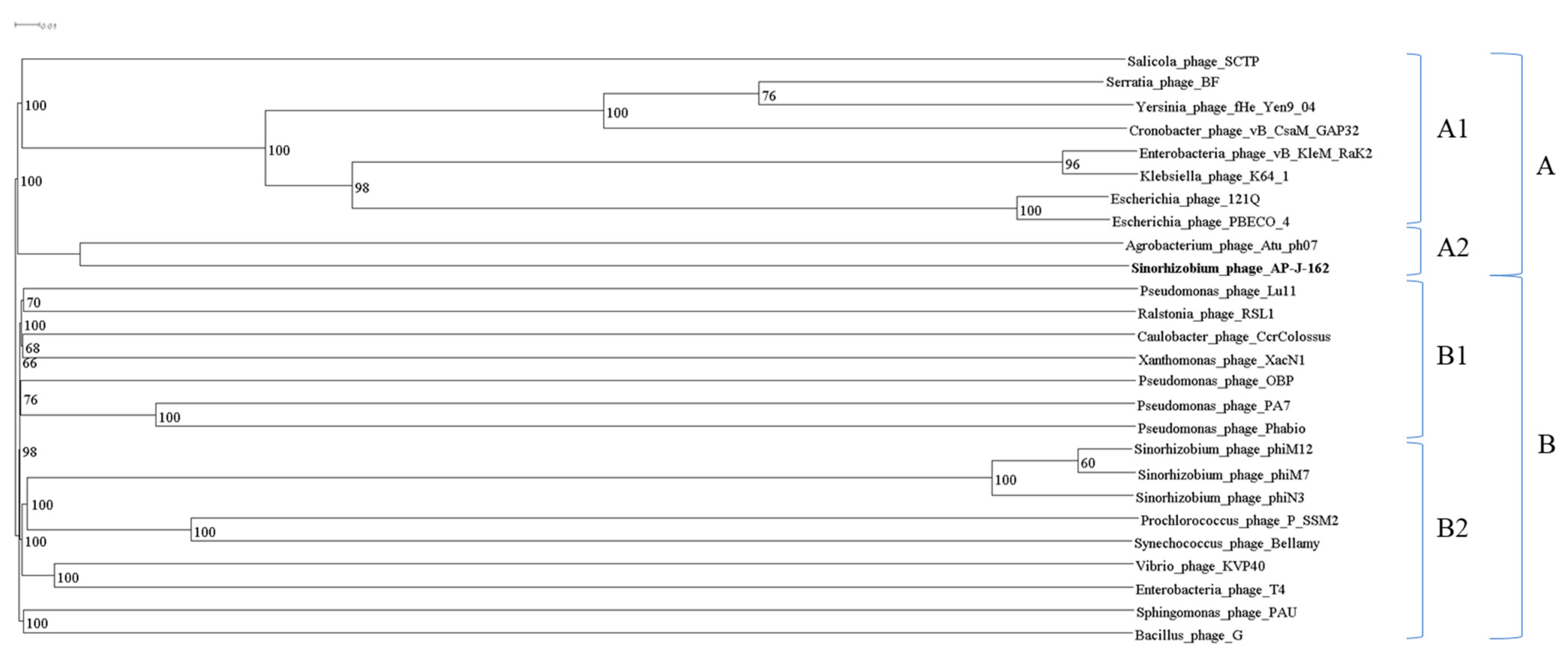
2. Results
3. Discussion
4. Materials and Methods
4.1. Phage Isolation and Purification
4.2. Bacterial Strains and Grows Conditions
4.3. Evaluation of Phage Lytic Activity (Host Range)
4.4. Transmission Electron Microscopy
4.5. One-Step Growth Curve of Phage AP-J-162
4.6. DNA Isolation, Sequencing, and Annotation of the Bacteriophage Genome
4.7. Phylogenetic Analysis
4.8. Analysis of Nucleotide and Amino Acid Sequences
4.9. Nucleotide Accession
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chevallereau, A.; Pons, B.J.; Van Houte, S.; Westra, E.R. Interactions between Bacterial and Phage Communities in Natural Environments. Nat. Rev. Microbiol. 2022, 20, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Fokine, A.; Rossmann, M.G. Molecular Architecture of Tailed Double-Stranded DNA Phages. Bacteriophage 2014, 4, e28281. [Google Scholar] [CrossRef] [PubMed]
- Giri, N. Bacteriophage Structure, Classification, Assembly and Phage Therapy. Biosci. Biotechnol. Res. Asia 2021, 18, 239–250. [Google Scholar] [CrossRef]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage Diversity, Genomics and Phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef]
- Elois, M.A.; Silva, R.D.; Pilati, G.V.T.; Rodríguez-Lázaro, D.; Fongaro, G. Bacteriophages as Biotechnological Tools. Viruses 2023, 15, 349. [Google Scholar] [CrossRef] [PubMed]
- Filippova, S.N.; Surgucheva, N.A.; Sorokin, V.V.; Akimov, V.N.; Karnysheva, E.A.; Brushkov, A.V.; Andersen, D.; Gal’chenko, V.F. Bacteriophages in arctic and antarctic low-temperature systems. Microbiology 2016, 85, 337–346. [Google Scholar] [CrossRef]
- Williamson, K.E.; Radosevich, M.; Wommack, K.E. Abundance and Diversity of Viruses in Six Delaware Soils. Appl. Environ. Microbiol. 2005, 71, 3119–3125. [Google Scholar] [CrossRef]
- Pratama, A.A.; Van Elsas, J.D. The ‘Neglected’ Soil Virome—Potential Role and Impact. Trends Microbiol. 2018, 26, 649–662. [Google Scholar] [CrossRef]
- Muscatt, G.; Hilton, S.; Raguideau, S.; Teakle, G.; Lidbury, I.D.E.A.; Wellington, E.M.H.; Quince, C.; Millard, A.; Bending, G.D.; Jameson, E. Crop Management Shapes the Diversity and Activity of DNA and RNA Viruses in the Rhizosphere. Microbiome 2022, 10, 181. [Google Scholar] [CrossRef]
- Amarillas, L.; Cháidez-Quiroz, C.; Sañudo-Barajas, A.; León-Félix, J. Complete Genome Sequence of a Polyvalent Bacteriophage, PhiKP26, Active on Salmonella and Escherichia coli. Arch. Virol. 2013, 158, 2395–2398. [Google Scholar] [CrossRef]
- Cai, L.; Tian, Y.; Li, Z.; Yang, Y.; Ai, C.; Zhang, R. A Broad-Host-Range Lytic Phage VB_VhaS-R18L as a Candidate against Vibriosis. Front. Microbiol. 2023, 14, 1191157. [Google Scholar] [CrossRef] [PubMed]
- Attai, H.; Boon, M.; Phillips, K.; Noben, J.-P.; Lavigne, R.; Brown, P.J.B. Larger Than Life: Isolation and Genomic Characterization of a Jumbo Phage That Infects the Bacterial Plant Pathogen, Agrobacterium tumefaciens. Front. Microbiol. 2018, 9, 1861. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, A.P.; Saksaganskaia, A.S.; Afonin, A.M.; Muntyan, V.S.; Vladimirova, M.E.; Dzyubenko, E.A.; Roumiantseva, M.L. A Temperate Sinorhizobium Phage, AP-16-3, Closely Related to Phage 16-3: Mosaic Genome and Prophage Analysis. Viruses 2023, 15, 1701. [Google Scholar] [CrossRef] [PubMed]
- Imam, M.; Alrashid, B.; Patel, F.; Dowah, A.S.A.; Brown, N.; Millard, A.; Clokie, M.R.J.; Galyov, E.E. VB_PaeM_MIJ3, a Novel Jumbo Phage Infecting Pseudomonas aeruginosa, Possesses Unusual Genomic Features. Front. Microbiol. 2019, 10, 2772. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Gao, M. Jumbo Bacteriophages: An Overview. Front. Microbiol. 2017, 8, 2772. [Google Scholar] [CrossRef]
- Al-Shayeb, B.; Sachdeva, R.; Chen, L.-X.; Ward, F.; Munk, P.; Devoto, A.; Castelle, C.J.; Olm, M.R.; Bouma-Gregson, K.; Amano, Y.; et al. Clades of Huge Phages from across Earth’s Ecosystems. Nature 2020, 578, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Devoto, A.E.; Santini, J.M.; Olm, M.R.; Anantharaman, K.; Munk, P.; Tung, J.; Archie, E.A.; Turnbaugh, P.J.; Seed, K.D.; Blekhman, R.; et al. Megaphages Infect Prevotella and Variants Are Widespread in Gut Microbiomes. Nat. Microbiol. 2019, 4, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Anantharaman, V.; Krishnan, A.; Burroughs, A.M.; Aravind, L. Jumbo Phages: A Comparative Genomic Overview of Core Functions and Adaptions for Biological Conflicts. Viruses 2021, 13, 63. [Google Scholar] [CrossRef]
- Aevarsson, A.; Kaczorowska, A.-K.; Adalsteinsson, B.T.; Ahlqvist, J.; Al-Karadaghi, S.; Altenbuchner, J.; Arsin, H.; Átlasson, Ú.Á.; Brandt, D.; Cichowicz-Cieślak, M.; et al. Going to Extremes—A Metagenomic Journey into the Dark Matter of Life. FEMS Microbiol. Lett. 2021, 368, fnab067. [Google Scholar] [CrossRef]
- Rodela, M.L.; Sabet, S.; Peterson, A.; Dillon, J.G. Broad Environmental Tolerance for a Salicola Host-Phage Pair Isolated from the Cargill Solar Saltworks, Newark, CA, USA. Microorganisms 2019, 7, 106. [Google Scholar] [CrossRef]
- González, B.; Monroe, L.; Li, K.; Yan, R.; Wright, E.; Walter, T.; Kihara, D.; Weintraub, S.T.; Thomas, J.A.; Serwer, P.; et al. Phage G Structure at 6.1 Å Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus. J. Mol. Biol. 2020, 432, 4139–4153. [Google Scholar] [CrossRef]
- Krupovic, M.; Turner, D.; Morozova, V.; Dyall-Smith, M.; Oksanen, H.M.; Edwards, R.; Dutilh, B.E.; Lehman, S.M.; Reyes, A.; Baquero, D.P.; et al. Bacterial Viruses Subcommittee and Archaeal Viruses Subcommittee of the ICTV: Update of Taxonomy Changes in 2021. Arch. Virol. 2021, 166, 3239–3244. [Google Scholar] [CrossRef]
- Turner, D.; Adriaenssens, E.M.; Lehman, S.M.; Moraru, C.; Kropinski, A.M. Bacteriophage Taxonomy: A Continually Evolving Discipline. In Bacteriophage Therapy; Azeredo, J., Sillankorva, S., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2024; Volume 2734, pp. 27–45. ISBN 978-1-07-163522-3. [Google Scholar]
- Cazares, A.; Mendoza-Hernández, G.; Guarneros, G. Core and Accessory Genome Architecture in a Group of Pseudomonas Aeruginosa Mu-like Phages. BMC Genom. 2014, 15, 1146. [Google Scholar] [CrossRef] [PubMed]
- Casey, A.; Coffey, A.; McAuliffe, O. Genetics and Genomics of Bacteriophages: The Evolution of Bacteriophage Genomes and Genomic Research. In Bacteriophages; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 193–218. ISBN 978-3-319-41985-5. [Google Scholar]
- Chan, J.Z.-M.; Millard, A.D.; Mann, N.H.; Schäfer, H. Comparative Genomics Defines the Core Genome of the Growing N4-like Phage Genus and Identifies N4-like Roseophage Specific Genes. Front. Microbiol. 2014, 5, 506. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Li, Z.; Miller, E.S. Vibrio Phage KVP40 Encodes a Functional NAD+ Salvage Pathway. J. Bacteriol. 2017, 199, e00855-16. [Google Scholar] [CrossRef]
- Van Cauwenberghe, J.; Santamaría, R.I.; Bustos, P.; Juárez, S.; Ducci, M.A.; Figueroa Fleming, T.; Etcheverry, A.V.; González, V. Spatial Patterns in Phage-Rhizobium Coevolutionary Interactions across Regions of Common Bean Domestication. ISME J. 2021, 15, 2092–2106. [Google Scholar] [CrossRef]
- Cubo, M.T.; Alías-Villegas, C.; Balsanelli, E.; Mesa, D.; De Souza, E.; Espuny, M.R. Diversity of Sinorhizobium (Ensifer) Meliloti Bacteriophages in the Rhizosphere of Medicago Marina: Myoviruses, Filamentous and N4-Like Podovirus. Front. Microbiol. 2020, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.S.; Mohmmed, A.; Babu, C.R. Diversity among Rhizobiophages from Rhizospheres of Legumes Inhabiting Three Ecogeographical Regions of India. Soil Biol. Biochem. 2002, 34, 965–973. [Google Scholar] [CrossRef]
- Hodson, T.S.; Hyde, J.R.; Schouten, J.T.; Crockett, J.T.; Smith, T.A.; Merrill, B.D. Sinorhizobium Phage PhiN3, Complete Genome, KR052482. Microbiol. Mol. Biol. 2015. [Google Scholar]
- Maynard, N.D.; Birch, E.W.; Sanghvi, J.C.; Chen, L.; Gutschow, M.V.; Covert, M.W. A Forward-Genetic Screen and Dynamic Analysis of Lambda Phage Host-Dependencies Reveals an Extensive Interaction Network and a New Anti-Viral Strategy. PLoS Genet. 2010, 6, e1001017. [Google Scholar] [CrossRef]
- Almpanis, A.; Swain, M.; Gatherer, D.; McEwan, N. Correlation between Bacterial G+C Content, Genome Size and the G+C Content of Associated Plasmids and Bacteriophages. Microb. Genomics 2018, 4, e000168. [Google Scholar] [CrossRef]
- Zhang, Y.; Baranov, P.V.; Atkins, J.F.; Gladyshev, V.N. Pyrrolysine and Selenocysteine Use Dissimilar Decoding Strategies. J. Biol. Chem. 2005, 280, 20740–20751. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.-A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of Nearly 8000 Metagenome-Assembled Genomes Substantially Expands the Tree of Life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef]
- Young, J.P.W.; Moeskjær, S.; Afonin, A.; Rahi, P.; Maluk, M.; James, E.K.; Cavassim, M.I.A.; Rashid, M.H.; Aserse, A.A.; Perry, B.J.; et al. Defining the Rhizobium Leguminosarum Species Complex. Genes 2021, 12, 111. [Google Scholar] [CrossRef]
- Crick, F.H.C. Codon—Anticodon Pairing: The Wobble Hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Ayan, G.B.; Park, H.J.; Gallie, J. The Birth of a Bacterial TRNA Gene by Large-Scale, Tandem Duplication Events. eLife 2020, 9, e57947. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, J.; Ibba, M. Bacterial Transfer RNAs. FEMS Microbiol. Rev. 2015, 39, 280–300. [Google Scholar] [CrossRef]
- Lawrence, J.G.; Ochman, H. Molecular Archaeology of the Escherichia coli Genome. Proc. Natl. Acad. Sci. USA 1998, 95, 9413–9417. [Google Scholar] [CrossRef]
- Marri, P.R.; Golding, G.B. Gene Amelioration Demonstrated: The Journey of Nascent Genes in Bacteria. Genome 2008, 51, 164–168. [Google Scholar] [CrossRef]
- Sickmier, E.A.; Kreuzer, K.N.; White, S.W. The Crystal Structure of the UvsW Helicase from Bacteriophage T4. Structure 2004, 12, 583–592. [Google Scholar] [CrossRef][Green Version]
- Torres-Miranda, A.; Vega-Sagardía, M.; Garrido, D. Probiotics, Microbiome and the Concept of Cross-Feeding. In Comprehensive Gut Microbiota; Elsevier: Amsterdam, The Netherlands, 2022; pp. 199–220. ISBN 978-0-12-822036-8. [Google Scholar]
- Weigel, C.; Seitz, H. Bacteriophage Replication Modules. FEMS Microbiol. Rev. 2006, 30, 321–381. [Google Scholar] [CrossRef] [PubMed]
- Węgrzyn, A.; Czyż, A.; Gabig, M.; Węgrzyn, G. ClpP/ClpX-Mediated Degradation of the Bacteriophage λ O Protein and Regulation of λ Phage and λ Plasmid Replication. Arch. Microbiol. 2000, 174, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y. Phage Lytic Enzymes. Viruses 2019, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, M.M.; Stanley, E.; Fitzgerald, G.F.; Van Sinderen, D. Identification and Characterization of a Lysis Module Present in a Large Proportion of Bacteriophages Infecting Streptococcus thermophilus. Appl. Environ. Microbiol. 1999, 65, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; São-José, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanović, N.; Fagorzi, C.; Mengoni, A.; Lassalle, F.; di Cenzo, G.C. Taxonomy of Rhizobiaceae Revisited: Proposal of a New Framework for Genus Delimitation. Int. J. Syst. Evol. Microbiol. 2022, 72, 005243. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, E.; Rocha, E.P.C. Phage-Plasmids Promote Recombination and Emergence of Phages and Plasmids. Nat. Commun. 2024, 15, 1545. [Google Scholar] [CrossRef]
- Barnet, Y.M. Bacteriophages of Rhizobium Trifolii I. Morphology and Host Range. J. Gen. Virol. 1972, 15, 1–15. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. In Bacteriophages; Clokie, M.R.J., Kropinski, A.M., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 1, pp. 69–76. ISBN 978-1-58829-682-5. [Google Scholar]
- Roumiantseva, M.L.; Vladimirova, M.E.; Saksaganskaia, A.S.; Muntyan, V.S.; Kozlova, A.P.; Afonin, A.M.; Baturina, O.A.; Simarov, B.V. Ensifer meliloti L6-AK89, an Effective Inoculant of Medicago lupulina Varieties: Phenotypic and Deep-Genome Screening. Agronomy 2022, 12, 766. [Google Scholar] [CrossRef]
- Beck, N.K.; Callahan, K.; Nappier, S.P.; Kim, H.; Sobsey, M.D.; Meschke, J.S. DEVELOPMENT OF A SPOT-TITER CULTURE ASSAY FOR QUANTIFYING BACTERIA AND VIRAL INDICATORS. J. Rapid Methods Autom. Microbiol. 2009, 17, 455–464. [Google Scholar] [CrossRef]
- Khalatbari-Limaki, S.; Hosseinzadeh, S.; Shekarforoush, S.S.; Berizi, E. The Morphological and Biological Characteristics of a Virulent PI Phage Isolated from Slaughterhouse Sewage in Shiraz, Iran. Iran. J. Microbiol. 2020, 12, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.A.; Tsoungui Obama, H.C.J.; Kamanga, G.; Kayanula, L.; Adil Mahmoud Yousif, N. The Many Definitions of Multiplicity of Infection. Front. Epidemiol. 2022, 2, 961593. [Google Scholar] [CrossRef] [PubMed]
- Frage, B.; Döhlemann, J.; Robledo, M.; Lucena, D.; Sobetzko, P.; Graumann, P.L.; Becker, A. Spatiotemporal Choreography of Chromosome and Megaplasmids in the Sinorhizobium Meliloti Cell Cycle. Mol. Microbiol. 2016, 100, 808–823. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M. Practical Advice on the One-Step Growth Curve. In Bacteriophages; Clokie, M.R.J., Kropinski, A.M., Lavigne, R., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1681, pp. 41–47. ISBN 978-1-4939-7341-5. [Google Scholar]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-Based Phylogeny and Classification of Prokaryotic Viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program: Table 1. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S. Estimating Phylogenetic Trees from Distance Matrices. Estim. Phylogenetic Trees Distance Matrices 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
| Genome Annotation | Sinorhizobium Phage AP-J-162 | Agrobacterium Phage Atu_ph07 |
|---|---|---|
| Genome size, kb | 471.5 | 490.4 |
| G + C, % | 47.13 | 37.1 |
| ORFs: | 711 | 714 |
| Unique | 304 | 390 |
| Hypothetical proteins | 191 | 214 |
| Predicted function | 148 | 110 |
| tRNA | 66 | 33 |
| tmRNA | 1 | 0 |
| Misc. R.N.A. | 1 | 0 |
| ORF similar | 152 | |
| ORFs encoding: | 21 | |
| (i) structural elements of phage particle | ||
| (ii) enzymes | 54 | |
| (iii) hypothetical proteins | 77 | |
| № ORF (gp) | Length, a.a. | Predicted Function | Best Match (Identity/Cover, %) | The Closer Structural Analog Predicted with I-TASSER Web Server (AlphaFold2 Algorithm) (PDB Accession) | |
|---|---|---|---|---|---|
| 9 | 408 | Major capsid protein | Myoviridae sp. (77.5/97) | 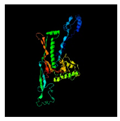 | Capsid protein bacteriophage T4 (5VF3) |
| 29 | 574 | Portal protein | Agrobacterium phage Atu_ph07 (57.58/100) |  | Portal protein bacteriophage T4 (3JA7) |
| 31 | 279 | Baseplate | Agrobacterium phage Atu_ph07 (50.18/98) | 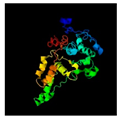 | Transporter protein Chlamydomonas reinhardtii (5ZME) |
| 50 | 103 | Baseplate wedge protein | Agrobacterium phage Atu_ph07 (62.14/100) | ||
| 56 | 131 | Baseplate wedge protein | Agrobacterium phage Atu_ph07 (51.56/97) | 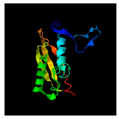 | Hexagonal pre-attachment T4 baseplate–tail tube complex (5IV5) |
| 57 | 1198 | Baseplate wedge protein | Agrobacterium phage Atu_ph07 (62.35/100) | ||
| 58 | 4455 | Structural protein | Agrobacterium phage Atu_ph07 (49.61/99) | ||
| 139 | 166 | Tail protein | Caulobacter phage CcrColossus (33.77/87) | ||
| 141 | 959 | Tail fiber protein | Myoviridae sp. (28.88/59) | 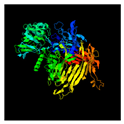 | Tc toxin Photorhabdus luminescens (6SUF) |
| 145 | 602 | Tail fiber protein | Agrobacterium phage Atu_ph07 (31.34/98) |  | Apoptosome Drosophila melanogaster (4V4L) |
| 184 | 248 | Outer membrane protein | Verticillium longisporum (65.71/42) | ||
| 213 | 375 | Hemagglutinin | Agrobacterium phage Atu_ph07 (31.77/99) | 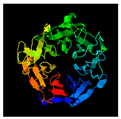 | Human integrin (3VI3) |
| 214 | 394 | Hemagglutinin | Agrobacterium phage Atu_ph07 (34.91/94) | ||
| 218 | 311 | Tail protein | Agrobacterium phage Atu_ph07 (45.16/99) | 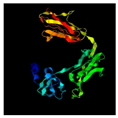 | IFN-alpha/beta-binding protein C12R Ectromelia virus Moscow (3OQ3) |
| 219 | 726 | Tail protein | Agrobacterium phage Atu_ph07 (40.06/99) | 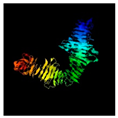 | S-layer protein RsaA Caulobacter crescentus (8BQE) |
| 220 | 679 | Tail protein | Agrobacterium phage Atu_ph07 (34.2/98) | ||
| 221 | 788 | Tail fiber protein | Agrobacterium phage Atu_ph07 (30.41/59) |  | ATPase Sus scrofa (4HQJ) |
| 282 | 171 | Outer membrane protein | Agrobacterium phage Atu_ph07 (35.09/100) | ||
| 569 | 1280 | Tail fiber protein | Agrobacterium phage Atu_ph07 (32.52/42) |  | Toxin B Clostridioides difficile (2BVL) |
| 572 | 564 | Potential structural protein | Thiobacillus sp. (44.07/93) | ||
| 573 | 210 | Potential structural protein | Ectothiorhodospiraceae bacterium 2226 (40.0/93) | ||
| 589 | 411 | Tail protein | Pseudomonas sp. Fl4BN1 (38.83/69) | 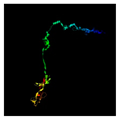 | Subunit of pre-attachment T4 baseplate–tail tube complex (5IV5) |
| 590 | 303 | Tail protein | Agrobacterium phage Atu_ph07 (36.31/99) | ||
| 647 | 957 | Potential structural protein | Myoviridae sp. (41.8/39) | ||
| 657 | 143 | Head completion protein | Agrobacterium phage Atu_ph07 (61.76/95) | 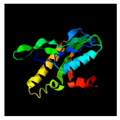 | TnsA/TnsC transposition complex Escherichia coli (1T0F) |
| 658 | 331 | Potential structural protein | Agrobacterium phage Atu_ph07 (46.71/98) | ||
| 659 | 194 | Potential structural protein | Agrobacterium phage Atu_ph07 (69.63/98) | ||
| 665 | 1092 | Tail sheath monomer | Agrobacterium phage Atu_ph07 (59.85/99) |  | Secreted extracellular protein A (SepA) Shigella flexneri (5J44) |
| 696 | 967 | Tail fiber protein | Flammeovirga sp. EKP202 (42.61/39) | ||
| 708 | 671 | Potential structural protein | Agrobacterium phage Atu_ph07 (38.03/90) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozlova, A.P.; Muntyan, V.S.; Vladimirova, M.E.; Saksaganskaia, A.S.; Kabilov, M.R.; Gorbunova, M.K.; Gorshkov, A.N.; Grudinin, M.P.; Simarov, B.V.; Roumiantseva, M.L. Soil Giant Phage: Genome and Biological Characteristics of Sinorhizobium Jumbo Phage. Int. J. Mol. Sci. 2024, 25, 7388. https://doi.org/10.3390/ijms25137388
Kozlova AP, Muntyan VS, Vladimirova ME, Saksaganskaia AS, Kabilov MR, Gorbunova MK, Gorshkov AN, Grudinin MP, Simarov BV, Roumiantseva ML. Soil Giant Phage: Genome and Biological Characteristics of Sinorhizobium Jumbo Phage. International Journal of Molecular Sciences. 2024; 25(13):7388. https://doi.org/10.3390/ijms25137388
Chicago/Turabian StyleKozlova, Alexandra P., Victoria S. Muntyan, Maria E. Vladimirova, Alla S. Saksaganskaia, Marsel R. Kabilov, Maria K. Gorbunova, Andrey N. Gorshkov, Mikhail P. Grudinin, Boris V. Simarov, and Marina L. Roumiantseva. 2024. "Soil Giant Phage: Genome and Biological Characteristics of Sinorhizobium Jumbo Phage" International Journal of Molecular Sciences 25, no. 13: 7388. https://doi.org/10.3390/ijms25137388
APA StyleKozlova, A. P., Muntyan, V. S., Vladimirova, M. E., Saksaganskaia, A. S., Kabilov, M. R., Gorbunova, M. K., Gorshkov, A. N., Grudinin, M. P., Simarov, B. V., & Roumiantseva, M. L. (2024). Soil Giant Phage: Genome and Biological Characteristics of Sinorhizobium Jumbo Phage. International Journal of Molecular Sciences, 25(13), 7388. https://doi.org/10.3390/ijms25137388







