Altered Inflammatory State and Mitochondrial Function Identified by Transcriptomics in Paediatric Congenital Heart Patients Prior to Surgical Repair
Abstract
:1. Introduction
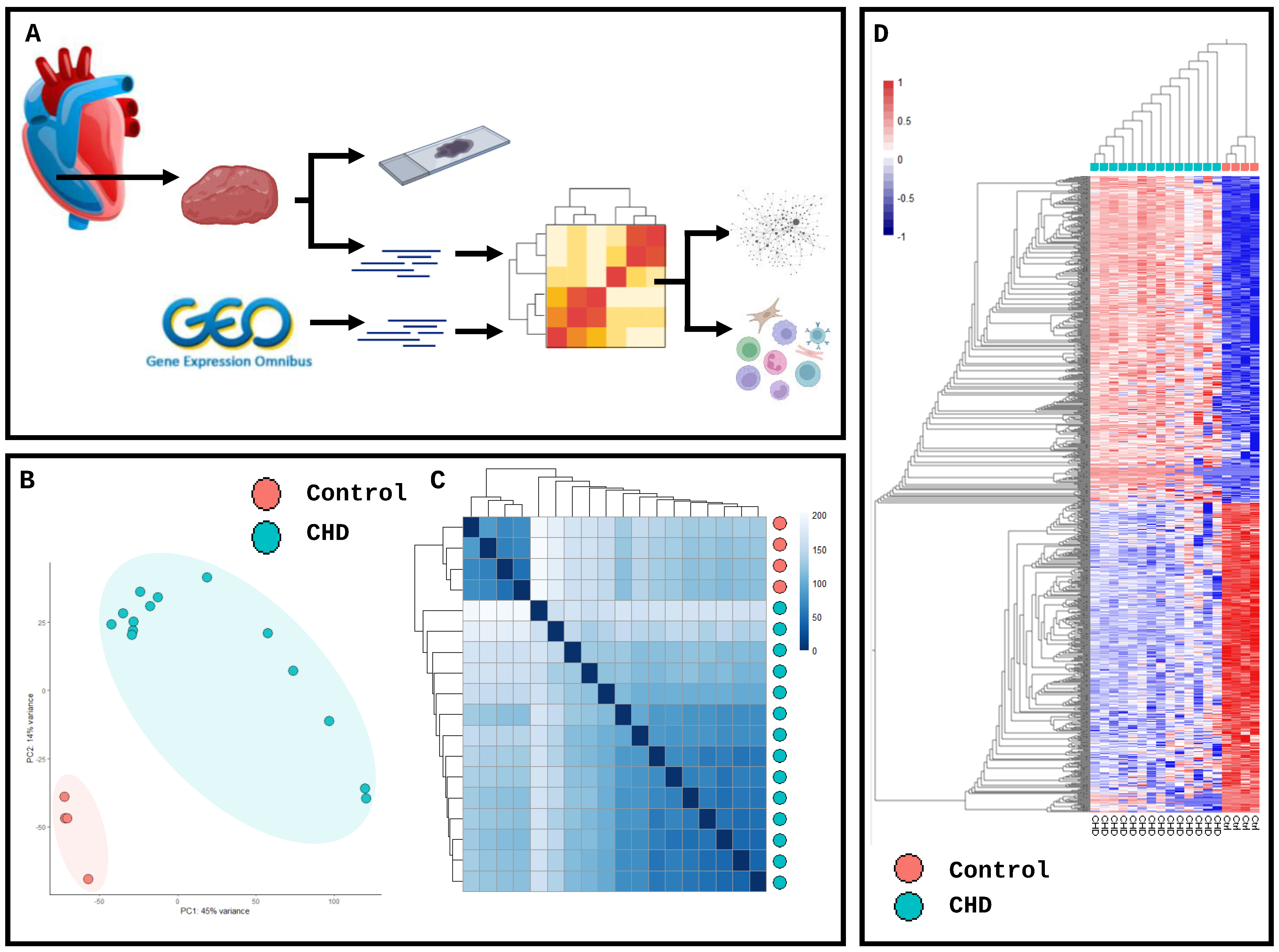
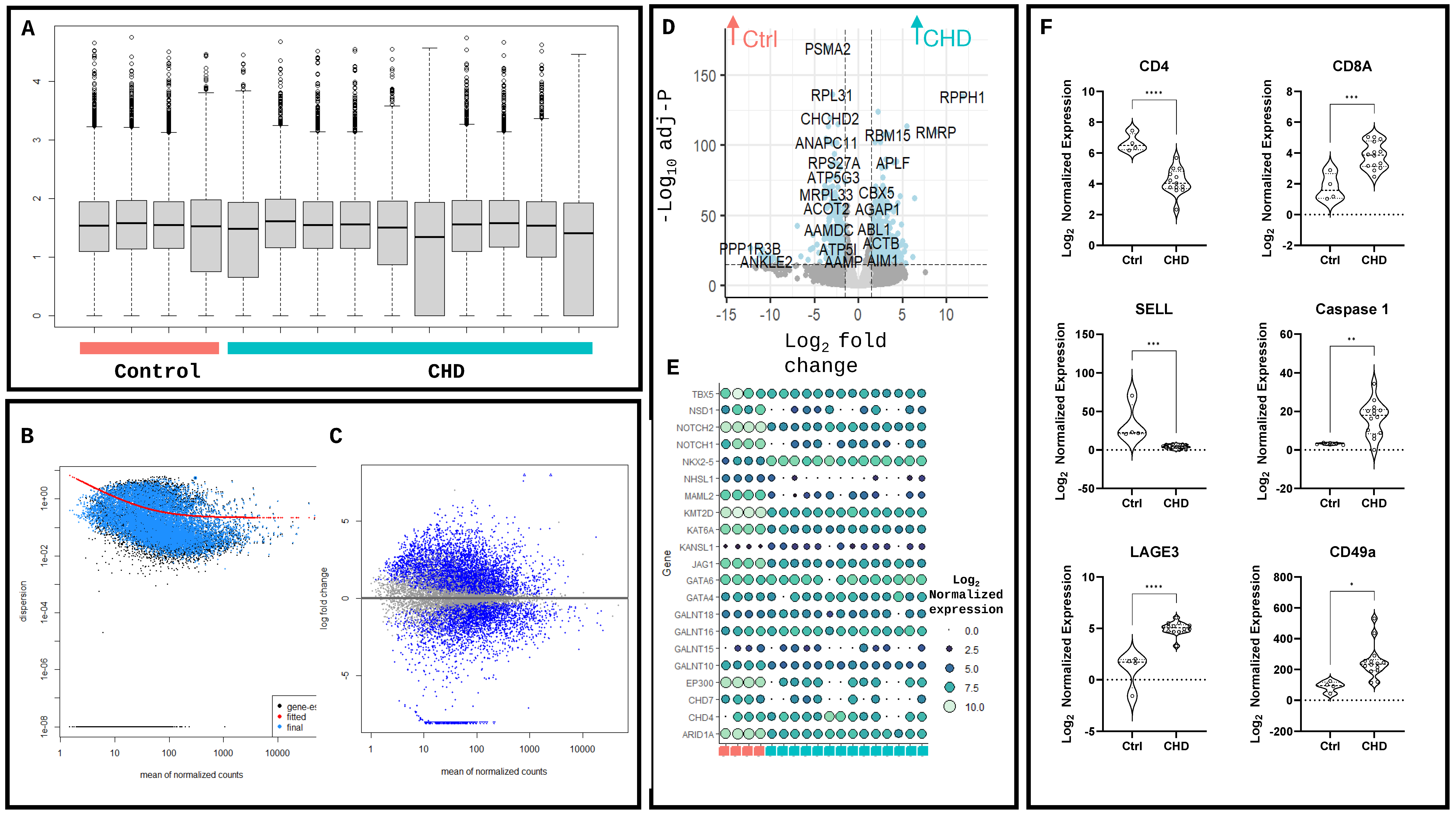
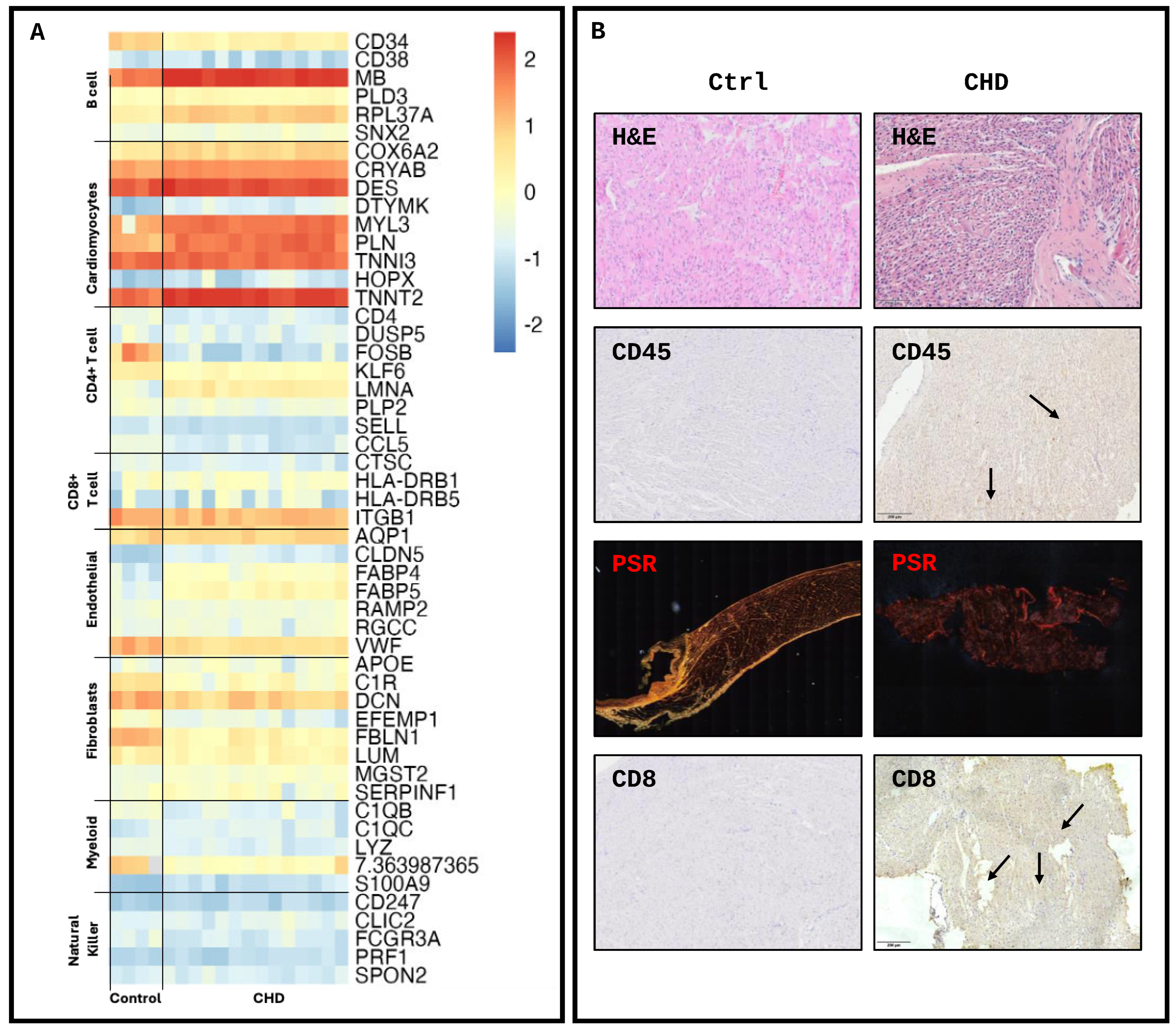
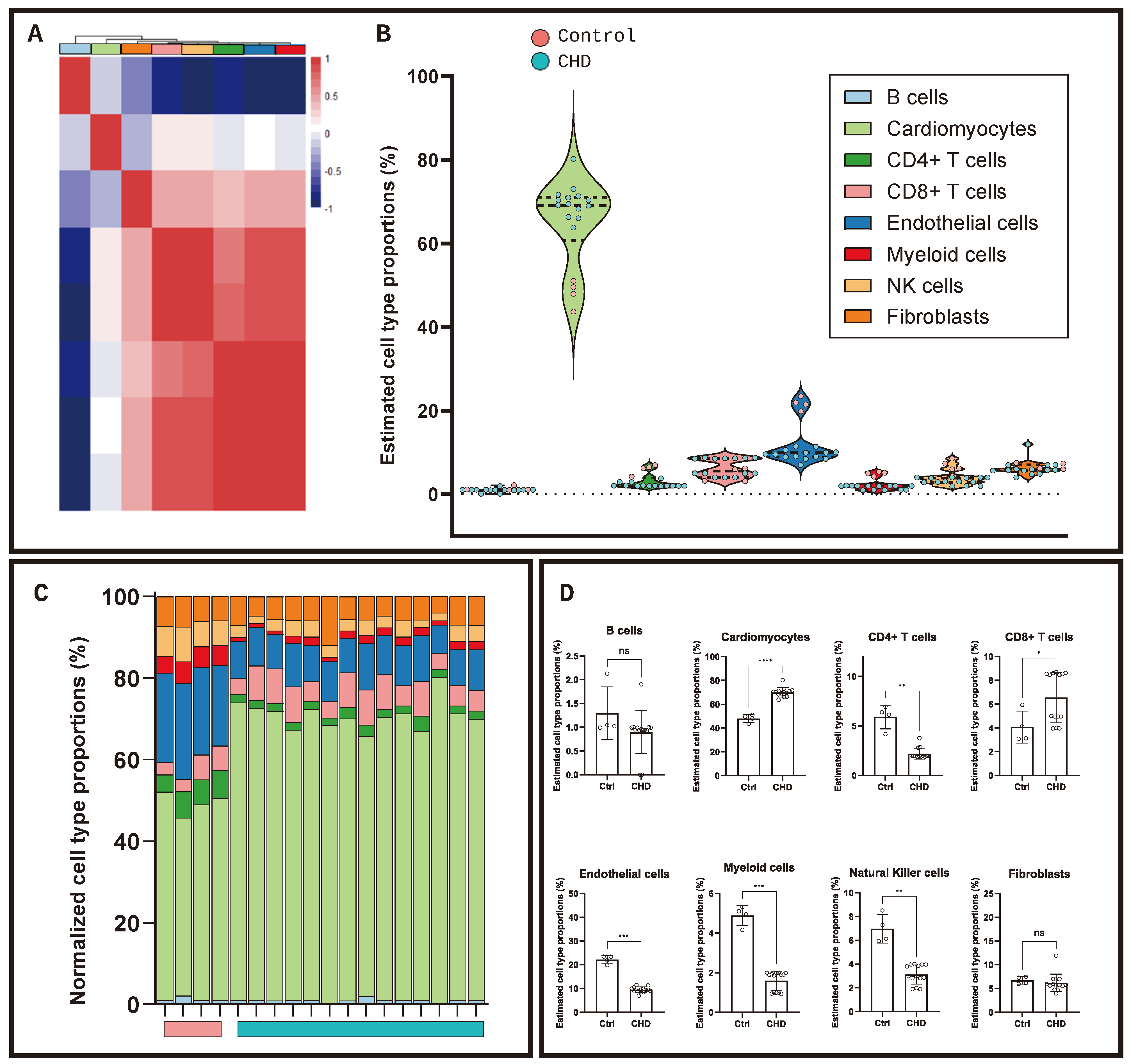
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Tissue Processing and Histological Analysis
4.3. Transcriptomic Analysis
4.4. Differential Enrichment and Network Analysis
4.5. Transcriptomic Deconvolution
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jacobs, J.P.; Mavroudis, C.; Quintessenza, J.A.; Chai, P.J.; Pasquali, S.K.; Hill, K.D.; Vricella, L.A.; Jacobs, M.L.; Dearani, J.A.; Cameron, D. Reoperations for pediatric and congenital heart disease: An analysis of the Society of Thoracic Surgeons (STS) congenital heart surgery database. Semin. Thorac. Cardiovasc. Surg. Pediatr. Card. Surg. Annu. 2014, 17, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Zomer, A.C.; Verheugt, C.L.; Vaartjes, I.; Uiterwaal, C.S.P.M.; Langemeijer, M.M.; Koolbergen, D.R.; Hazekamp, M.G.; Melle, J.P.v.; Konings, T.C.; Bellersen, L.; et al. Surgery in Adults with Congenital Heart Disease. Circulation 2011, 124, 2195–2201. [Google Scholar] [CrossRef]
- Ryo, K.; Goda, A.; Onishi, T.; Delgado-Montero, A.; Tayal, B.; Champion, H.C.; Simon, M.A.; Mathier, M.A.; Gladwin, M.T.; Gorcsan, J. Characterization of Right Ventricular Remodeling in Pulmonary Hypertension Associated With Patient Outcomes by 3-Dimensional Wall Motion Tracking Echocardiography. Circ. Cardiovasc. Imaging 2015, 8, e003176. [Google Scholar] [CrossRef]
- Bogaard, H.J.; Abe, K.; Vonk Noordegraaf, A.; Voelkel, N.F. The Right Ventricle Under Pressure: Cellular and Molecular Mechanisms of Right-Heart Failure in Pulmonary Hypertension. Chest 2009, 135, 794–804. [Google Scholar] [CrossRef]
- Wienecke, L.M.; Cohen, S.; Bauersachs, J.; Mebazaa, A.; Chousterman, B.G. Immunity and inflammation: The neglected key players in congenital heart disease? Heart Fail. Rev. 2022, 27, 1957–1971. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Sager, H.B.; Roh, J.D.; Valero-Muñoz, M.; Houstis, N.E.; Iwamoto, Y.; Sun, Y.; Wilson, R.M.; Wojtkiewicz, G.; Tricot, B.; et al. Cardiac macrophages promote diastolic dysfunction. J. Exp. Med. 2018, 215, 423–440. [Google Scholar] [CrossRef]
- Nevers, T.; Salvador, A.M.; Velazquez, F.; Ngwenyama, N.; Carrillo-Salinas, F.J.; Aronovitz, M.; Blanton, R.M.; Alcaide, P. Th1 effector T cells selectively orchestrate cardiac fibrosis in nonischemic heart failure. J. Exp. Med. 2017, 214, 3311–3329. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Wang, X.; Guo, G.; Wang, L.; Chen, S.; Yin, P.; Chen, K.; Chen, L.; Zhang, Z.; Chen, X.; et al. Resolving the intertwining of inflammation and fibrosis in human heart failure at single-cell level. Basic Res. Cardiol. 2021, 116, 55. [Google Scholar] [CrossRef]
- Garand, M.; Huang, S.S.Y.; Dineen, B.; Glass, I.A.; Eghtesady, P. Differential Regulation of Immune-Related Genes in the Developing Heart. Pediatr. Cardiol. 2024, 1–16. [Google Scholar] [CrossRef]
- Hwang, H.V.; Sandeep, N.; Nair, R.V.; Hu, D.Q.; Zhao, M.; Lan, I.S.; Fajardo, G.; Matkovich, S.J.; Bernstein, D.; Reddy, S. Transcriptomic and Functional Analyses of Mitochondrial Dysfunction in Pressure Overload-Induced Right Ventricular Failure. J. Am. Heart Assoc. 2021, 10, e017835. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Fahed, A.C.; Gelb, B.D.; Seidman, J.G.; Seidman, C.E. Genetics of Congenital Heart Disease. Circ. Res. 2013, 112, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Reichart, D.; Lindberg, E.L.; Maatz, H.; Miranda, A.M.A.; Viveiros, A.; Shvetsov, N.; Gärtner, A.; Nadelmann, E.R.; Lee, M.; Kanemaru, K.; et al. Pathogenic variants damage cell composition and single cell transcription in cardiomyopathies. Science 2022, 377, eabo1984. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, P.; Zhou, B.; Song, J.; Li, Z.; Zhang, M.; Guo, G.; Wang, Y.; Chen, X.; Han, L.; et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat. Cell Biol. 2020, 22, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Carreon, C.K.; Benini, A.; Baird, C.; Hoganson, D.; Borisuk, M.; Emani, S.; Hofferberth, S.; Padera, R.F., Jr.; Sanders, S.P. Pathology of valved venous homografts used as right ventricle-to-pulmonary artery conduits in congenital heart disease surgery. J. Thorac. Cardiovasc. Surg. 2019, 157, 342–350.e3. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.S.; Heider, A.; Si, M.S.; Ohye, R.G. Evaluation of Explanted CorMatrix Intracardiac Patches in Children With Congenital Heart Disease. Ann. Thorac. Surg. 2016, 102, 1329–1335. [Google Scholar] [CrossRef]
- Nordmeyer, S.; Kretzschmar, J.; Murin, P.; Cho, M.Y.; Foth, R.; Schlichting, U.; Berger, F.; Ovroutski, S.; Photiadis, J.; Sigler, M. ADAPT-treated pericardium for aortic valve reconstruction in congenital heart disease: Histological analysis of a series of human explants. Eur. J. Cardiothorac. Surg. 2019, 56, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Bolger, A.P.; Li, W.; Davlouros, P.A.; Volk, H.-D.; Poole-Wilson, P.A.; Coats, A.J.S.; Gatzoulis, M.A.; Anker, S.D. Elevated circulating levels of inflammatory cytokines and bacterial endotoxin in adults with congenital heart disease. Am. J. Cardiol. 2003, 92, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Qing, M.; Schumacher, K.; Heise, R.; Wöltje, M.; Vazquez-Jimenez, J.F.; Richter, T.; Arranda-Carrero, M.; Hess, J.; von Bernuth, G.; Seghaye, M.-C. Intramyocardial synthesis of pro- and anti-inflammatory cytokines in infants with congenital cardiac defects. J. Am. Coll. Cardiol. 2003, 41, 2266–2274. [Google Scholar] [CrossRef]
- Elder, R.W.; George, R.P.; McCabe, N.M.; Rodriguez, F.H., III; Book, W.M.; Mahle, W.T.; Kirk, A.D. Immunologic Aging in Adults with Congenital Heart Disease: Does Infant Sternotomy Matter? Pediatr. Cardiol. 2015, 36, 1411–1416. [Google Scholar] [CrossRef]
- Cancrini, C.; Romiti, M.L.; Finocchi, A.; Di Cesare, S.; Ciaffi, P.; Capponi, C.; Pahwa, S.; Rossi, P. Post-natal ontogenesis of the T-cell receptor CD4 and CD8 Vbeta repertoire and immune function in children with DiGeorge syndrome. J. Clin. Immunol. 2005, 25, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Ashrafian, H.; Frenneaux, M.P.; Opie, L.H. Metabolic Mechanisms in Heart Failure. Circulation 2007, 116, 434–448. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.; Gehmlich, K.; Denning, C.; Pavlovic, D. Complex Relationship Between Cardiac Fibroblasts and Cardiomyocytes in Health and Disease. J. Am. Heart Assoc. 2021, 10, e019338. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, D.; Cappabianca, G.; Rotunno, C.; Castellaneta, F.; Quagliara, T.; Carrozzo, A.; Mastro, F.; Charitos, I.A.; Beghi, C. The Preoperative Inflammatory Status Affects the Clinical Outcome in Cardiac Surgery. Antibiotics 2019, 8, 176. [Google Scholar] [CrossRef] [PubMed]
- Urbanowicz, T.; Olasińska-Wiśniewska, A.; Michalak, M.; Perek, B.; Al-Imam, A.; Rodzki, M.; Witkowska, A.; Straburzyńska-Migaj, E.; Bociański, M.; Misterski, M.; et al. Pre-operative systemic inflammatory response index influences long-term survival rate in off-pump surgical revascularization. PLoS ONE 2022, 17, e0276138. [Google Scholar] [CrossRef]
- Soto-Heredero, G.; Desdín-Micó, G.; Mittelbrunn, M. Mitochondrial dysfunction defines T cell exhaustion. Cell Metab. 2021, 33, 470–472. [Google Scholar] [CrossRef]
- Scharping, N.E.; Rivadeneira, D.B.; Menk, A.V.; Vignali, P.D.A.; Ford, B.R.; Rittenhouse, N.L.; Peralta, R.; Wang, Y.; Wang, Y.; DePeaux, K.; et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 2021, 22, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Noly, P.-E.; Piquereau, J.; Coblence, M.; Ataam, J.A.; Guihaire, J.; Rucker-Martin, C.; Decante, B.; Haddad, F.; Fadel, E.; Mercier, O. Right ventricular mitochondrial respiratory function in a piglet model of chronic pulmonary hypertension. J. Thorac. Cardiovasc. Surg. 2020, 159, 129–140. [Google Scholar] [CrossRef]
- Laroumanie, F.; Douin-Echinard, V.; Pozzo, J.; Lairez, O.; Tortosa, F.; Vinel, C.; Delage, C.; Calise, D.; Dutaur, M.; Parini, A.; et al. CD4+ T Cells Promote the Transition From Hypertrophy to Heart Failure During Chronic Pressure Overload. Circulation 2014, 129, 2111–2124. [Google Scholar] [CrossRef]
- Cui, Q.; Sun, S.; Zhu, H.; Xiao, Y.; Jiang, C.; Zhang, H.; Liu, J.; Ye, L.; Shen, J. Volume Overload Initiates an Immune Response in the Right Ventricle at the Neonatal Stage. Front. Cardiovasc. Med. 2021, 8, 772336. [Google Scholar] [CrossRef]
- Ruan, Y.; Hu, J.; Che, Y.; Liu, Y.; Luo, Z.; Cheng, J.; Han, Q.; He, H.; Zhou, Q. CHCHD2 and CHCHD10 regulate mitochondrial dynamics and integrated stress response. Cell Death Dis. 2022, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Alkhaja, A.K.; Jans, D.C.; Nikolov, M.; Vukotic, M.; Lytovchenko, O.; Ludewig, F.; Schliebs, W.; Riedel, D.; Urlaub, H.; Jakobs, S.; et al. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell 2012, 23, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Karamanlidis, G.; Bautista-Hernandez, V.; Fynn-Thompson, F.; Nido, P.d.; Tian, R. Impaired Mitochondrial Biogenesis Precedes Heart Failure in Right Ventricular Hypertrophy in Congenital Heart Disease. Circ. Heart Fail. 2011, 4, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Fedak, P.W.M.; Weisel, R.D.; Butany, J.; Rao, V.; Maitland, A.; Li, R.-K.; Dhillon, B.; Yau, T.M. Fundamentals of Reperfusion Injury for the Clinical Cardiologist. Circulation 2002, 105, 2332–2336. [Google Scholar] [CrossRef]
- Di Lisa, F.; Bernardi, P. Mitochondria and ischemia–reperfusion injury of the heart: Fixing a hole. Cardiovasc. Res. 2006, 70, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Schoenmann, N.; Tannenbaum, N.; Hodgeman, R.M.; Raju, R.P. Regulating mitochondrial metabolism by targeting pyruvate dehydrogenase with dichloroacetate, a metabolic messenger. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166769. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, W.; Devarakonda, T.; Zhou, H.; Wang, X.Y.; Salloum, F.N.; Spiegel, S.; Fang, X. Functional analysis of molecular and pharmacological modulators of mitochondrial fatty acid oxidation. Sci. Rep. 2020, 10, 1450. [Google Scholar] [CrossRef] [PubMed]
- Bartoli-Leonard, F.; Wilkinson, F.L.; Schiro, A.; Serracino Inglott, F.; Alexander, M.Y.; Weston, R. Loss of SIRT1 in diabetes accelerates DNA damage-induced vascular calcification. Cardiovasc. Res. 2021, 117, 836–849. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Bartoli-Leonard, F.; Chelvanambi, S.; Pham, T.; Turner, M.E.; Blaser, M.C.; Caputo, M.; Aikawa, M.; Pang, A.; Muehlschlegel, J.; Aikawa, E. Single-cell T cell receptor sequencing of paired tissue and blood samples reveals clonal expansion of CD8+ effector T cells in patients with calcific aortic valve disease. bioRxiv 2023. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartoli-Leonard, F.; Harris, A.G.; Saunders, K.; Madden, J.; Cherrington, C.; Sheehan, K.; Baquedano, M.; Parolari, G.; Bamber, A.; Caputo, M. Altered Inflammatory State and Mitochondrial Function Identified by Transcriptomics in Paediatric Congenital Heart Patients Prior to Surgical Repair. Int. J. Mol. Sci. 2024, 25, 7487. https://doi.org/10.3390/ijms25137487
Bartoli-Leonard F, Harris AG, Saunders K, Madden J, Cherrington C, Sheehan K, Baquedano M, Parolari G, Bamber A, Caputo M. Altered Inflammatory State and Mitochondrial Function Identified by Transcriptomics in Paediatric Congenital Heart Patients Prior to Surgical Repair. International Journal of Molecular Sciences. 2024; 25(13):7487. https://doi.org/10.3390/ijms25137487
Chicago/Turabian StyleBartoli-Leonard, Francesca, Amy G. Harris, Kelly Saunders, Julie Madden, Carrie Cherrington, Karen Sheehan, Mai Baquedano, Giulia Parolari, Andrew Bamber, and Massimo Caputo. 2024. "Altered Inflammatory State and Mitochondrial Function Identified by Transcriptomics in Paediatric Congenital Heart Patients Prior to Surgical Repair" International Journal of Molecular Sciences 25, no. 13: 7487. https://doi.org/10.3390/ijms25137487
APA StyleBartoli-Leonard, F., Harris, A. G., Saunders, K., Madden, J., Cherrington, C., Sheehan, K., Baquedano, M., Parolari, G., Bamber, A., & Caputo, M. (2024). Altered Inflammatory State and Mitochondrial Function Identified by Transcriptomics in Paediatric Congenital Heart Patients Prior to Surgical Repair. International Journal of Molecular Sciences, 25(13), 7487. https://doi.org/10.3390/ijms25137487




