Abstract
Chagas disease is caused by the intracellular protozoan parasite Trypanosoma cruzi. This disease affects mainly rural areas in Central and South America, where the insect vector is endemic. However, this disease has become a world health problem since migration has spread it to other continents. It is a complex disease with many reservoirs and vectors and high genetic variability. One of the host proteins involved in the pathogenesis is SLAMF1. This immune receptor acts during the infection of macrophages controlling parasite replication and thus affecting survival in mice but in a parasite strain-dependent manner. Therefore, we studied the role of SLAMF1 by quantitative proteomics in a macrophage in vitro infection and the different responses between Y and VFRA strains of Trypanosoma cruzi. We detected different significant up- or downregulated proteins involved in immune regulation processes, which are SLAMF1 and/or strain-dependent. Furthermore, independently of SLAMF1, this parasite induces different responses in macrophages to counteract the infection and kill the parasite, such as type I and II IFN responses, NLRP3 inflammasome activation, IL-18 production, TLR7 and TLR9 activation specifically with the Y strain, and IL-11 signaling specifically with the VFRA strain. These results have opened new research fields to elucidate the concrete role of SLAMF1 and discover new potential therapeutic approaches for Chagas disease.
1. Introduction
Chagas disease, also known as American trypanosomiasis, is a potentially life-threatening illness caused by the parasite Trypanosoma cruzi (T. cruzi), an intracellular protist that belongs to the Kinetoplastidae order and Trypanosomatidae family. This disease is named in honor of Carlos Ribeiro Justiniano Chagas, the researcher who discovered it in 1909, although the first evidence in human tissues dates to nearly 9 thousand years ago [1]. According to the World Health Organization, about 6–7 million people worldwide are estimated to be infected with T. cruzi, and 25 million are at risk of infection [2]. The disease is endemic to 21 countries in Central and South America, causing around 10–14 thousand deaths per year. However, Chagas disease has spread to other continents due to migration, becoming a world health problem [3,4,5]. More than 90% of patients live in the largest economies in the Western Hemisphere: Argentina, Brazil, Mexico, and the United States [6].
The immune response of mammal hosts is characterized by three principal mechanisms: recognition and killing of the parasite by the innate immune system (macrophages and dendritic cells), activation of the antigen-presenting cells, and the detection and elimination of the infected cells by the CD8+ T cells [7]. At the beginning of infection, macrophages, natural killer (NK) cells, and Tγδ cells are the first line of defense. They recognize surface molecules of T. cruzi by TLRs and other receptors and regulate the parasite’s replication through the generation of anti-microbial agents, such as reactive oxygen species (ROS) and reactive nitrogen intermediates (RNI) [8,9]. Furthermore, this ROS and RNI production activates the release of cytokines such as interleukin (IL)-12, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ [10]. Later, in adaptive immunity, the T helper 1 lymphocytes (Th1) activate the phagocytic cells to reduce the levels of parasites. Also, Th1 cells are critical for CD8+ T cells activation and the generation of antibodies, and their inhibition can lead to uncontrolled levels of parasites and death [10].
The Signaling Lymphocytic Activation Receptor (SLAM) family is a group of type I transmembrane glycoproteins that are expressed in many hematopoietic cells such as B cells, T cells, macrophages, dendritic cells, and NK cells. The SLAM family has nine members in humans and mice [11]. Seven are self-ligand receptors, and SLAMF2 and SLAMF4 interact between them [12]. SLAMF1 (CD150) is one of the better-studied receptors of this family and is expressed in the surface of T cells, B cells, macrophages, and dendritic cells. In the adaptive and innate immune response, SLAMF1 has four principal functions: it leads to an increase in IL-4 secretion in an SAP-dependent manner following TCR activation of CD4+ T cells, while in the thymus the SLAMF1–SLAMF1 interactions between DP thymocytes promote the development of NKT cells; also, SLAMF1 interacts with the hemagglutinin of the measles virus in activated T cells, and it is a sensor of Gram-negative bacteria such as Escherichia coli. In this last case, SLAMF1 internalizes into the macrophage phagosome to enhance the phagocytosis of the bacteria and increase the formation and activation of the NADPH oxidase 2 (NOX2) complex [11].
More importantly, SLAMF1 is related to Chagas disease. Previous studies have identified the principal changes in infected macrophages and organs mainly at the mRNA level of several genes [13,14,15]. Slamf1-/- mice infected by the Y strain were completely protected from an acute lethal parasite challenge [13]. Also, the cardiac damage and the parasite load in the heart were reduced, suggesting that this receptor controls the susceptibility to T. cruzi infection, at least with the virulent Y strain. Furthermore, a posterior investigation studied the immunological effect of SLAMF1 deficiency in in vitro and in vivo models with different strains of different DTUs of T. cruzi [14]. All the strains, except VFRA (DTU VI), showed a decrease in parasite load in infected Slamf1-/- macrophages compared to BALB/c controls. Immunological studies in vivo by RT-qPCR with the Y strain displayed that, in the absence of SLAMF1, the immune response protected mice from the otherwise lethal Y infection. SLAMF1 deficiency favored a pro-inflammatory response involving CD4+ and CD8+ T cells, dendritic cells, and classically activated macrophages. However, in the case of the VFRA strain, no major changes were observed in the absence of SLAMF1. Thus, these results suggested that T. cruzi infection is SLAMF1-dependent, except for the VFRA strain. SLAMF1 would act during the infection of macrophages controlling parasite replication and therefore affecting survival in mice but in a strain-dependent manner.
Thus, to further characterize the role of SLAMF1, we performed a quantitative proteomic analysis to uncover the differences between BALB/c and Slamf1-/- macrophages in the in vitro infection by T. cruzi, identifying the different and common responses between Y and VFRA strains of this parasite.
2. Results
2.1. Quantitative Proteomics Analysis of BALB/c and Slamf1-/- Macrophages Infected with the Y Strain of T. cruzi
We infected peritoneal BALB/c and Slamf1-/- macrophages for 24 h with two different strains, Y and VFRA, and we extracted the proteins of each condition for analysis by TMT-6plex. We detected a total of 1849 proteins for the quantitative study in all the conditions, and in the posterior analysis we established a threshold of <−0.5 and >0.5 for the log2 fold change and >1.3 for the −log10 (FDR). We chose 24 h post-infection (hpi) because this time has been established as the interval in which the parasite replicates inside the macrophage [14].
The Y strain was selected for three reasons: (1) the parasite load decreased in the Slamf1-/- macrophages; (2) the relevant changes in the immune response in these macrophages; (3) the SLAMF1-dependent lethality in infected susceptible BALB/c mice [14].
First, we focused on the BALB/c macrophages infected by the Y strain. After the quantitative analysis comparing with the control without infection, we obtained 18 significant proteins, 12 upregulated proteins, and 6 downregulated proteins (Figure 1). Then, we performed a gene ontology (GO) enrichment analysis to study the principal biological processes implicated in the Y strain infection with the significant proteins by fold change and FDR. We used the enrichment analysis tool from the Enrichr website and the “reduce and visualize gene ontology” (REVIGO) web tool to correlate the results. Considering the upregulated proteins (Figure 2), the most significant ontologies were related to the response against pathogens such as viruses and the IFN system (types I and II). Also, there was a slight negative regulation of TLR2 and TLR4 signaling pathways, while there was a positive regulation of TLR7 and TLR9 signaling pathways. Other significant GO terms were related to the lipid import, the protein translation pathways, the positive regulation of IL-18 production and the NACHT, LRR, and PYD domains-containing protein 3 (NLRP3) inflammasome complex assembly, the biosynthesis of nitric oxide synthase, and the alpha–beta T cell activation, among others.
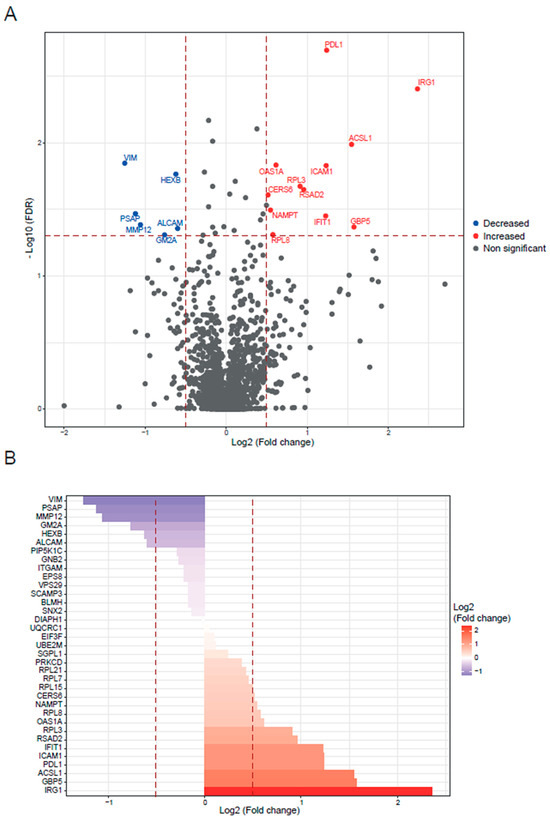
Figure 1.
BALB/c macrophages infected by Y strain vs. non-infected BALB/c macrophages. (A) Volcano plot of detected proteins with log2 fold change in mean intensities and −log10 FDR of each protein. Thresholds of significance are shown as dashed lines: <−0.5 and >0.5 for the log2 fold change and >1.3 for the −log10 FDR. Significant down- or upregulated proteins are represented in blue or red, respectively. Non-significant proteins are marked in grey. (B) Bar plot of the significant proteins (>1.3 for the −log10 FDR) classified by log2 fold change. Thresholds are shown as dashed lines: <−0.5 and >0.5.
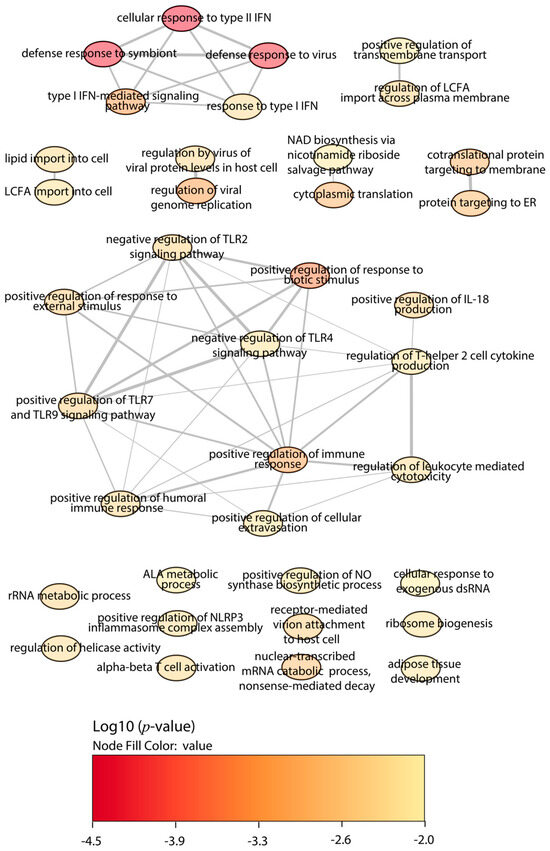
Figure 2.
GO enrichment analysis by Enrichr webtool of upregulated proteins in BALB/c macrophages infected with the Y strain. Significant biological processes’ GO terms are displayed in bubbles. Highly similar GO terms are linked by edges, and the line width indicates the degree of similarity. Color is determined by the log10 (p-value), with red being the most and beige being the least statistically significant, respectively. LCFA, long-chain fatty acid; ALA, alpha-linolenic acid; TLR, toll-like receptor; IFN, interferon; IL, interleukin.
Regarding the downregulated proteins in BALB/c macrophages infected with the Y strain, the enrichment of biological processes displayed less significant GO terms than the upregulated proteins (Figure 3). The most relevant were the glycoprotein catabolic process, the glycolipid metabolic process, and the neutrophil-mediated immunity and activation.
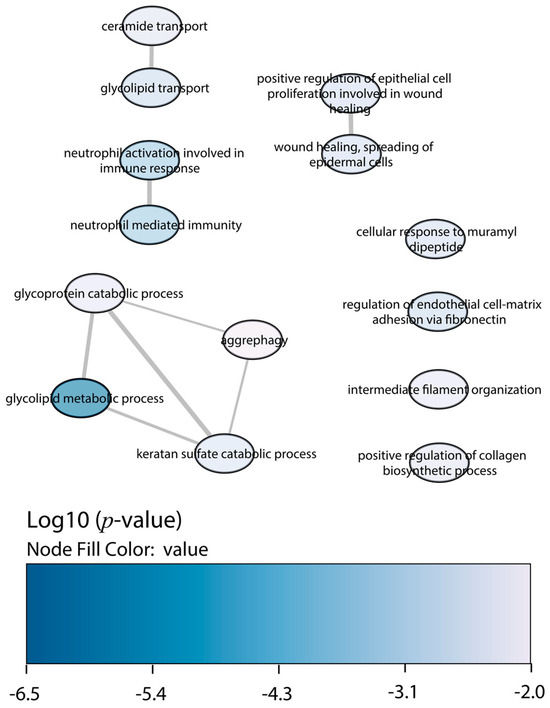
Figure 3.
GO enrichment analysis by Enrichr webtool of downregulated proteins in BALB/c macrophages infected with the Y strain. Significant biological processes’ GO terms are displayed in bubbles. Highly similar GO terms are linked by edges, and the line width indicates the degree of similarity. Color is determined by the log10 (p-value), with blue being the most and white being the least statistically significant, respectively.
The analysis of the Slamf1-/- macrophages infected with the Y strain revealed 25 significant proteins, 17 upregulated proteins, and 8 downregulated proteins (Figure 4). The GO enrichment analysis to study the principal biological processes implicated in the Y strain infection in the absence of the SLAMF1, considering the upregulated proteins (Figure 5), showed that the most significant GO terms were those related to the IFN types I and II, IL-1, TNF, IFN-β, NAD biosynthesis, cytokines signaling pathway, innate immune response, and the response against other pathogens as viruses. Also, other relevant GO terms were the alpha–beta T cell activation, the protein kinase C signaling, the negative regulation of TLR2 and TLR4, and the positive regulation of TLR7, TLR9, NLRP3 inflammasome complex assembly, nitric oxide synthase, and IL-18.
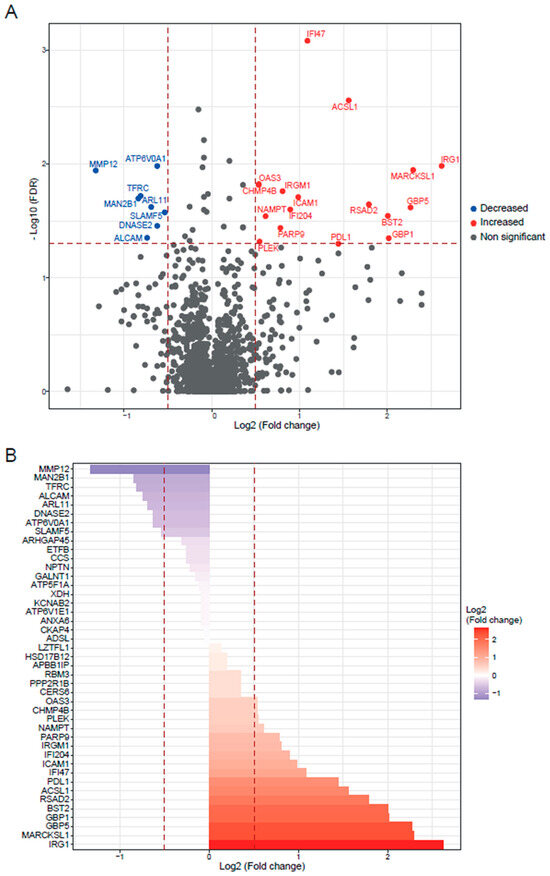
Figure 4.
Slamf1-/- macrophages infected by Y strain vs. non-infected Slamf1-/- macrophages. (A) Volcano plot of detected proteins with log2 fold change in mean intensities and −log10 FDR of each protein. Thresholds of significance are shown as dashed lines: <−0.5 and >0.5 for the log2 fold change and >1.3 for the −log10 FDR. Significant down- or upregulated proteins are represented in blue or red, respectively. Non-significant proteins are marked in grey. (B) Bar plot of the significant proteins (>1.3 for the −log10 FDR) classified by log2 fold change. Thresholds are shown as dashed lines: <−0.5 and >0.5.
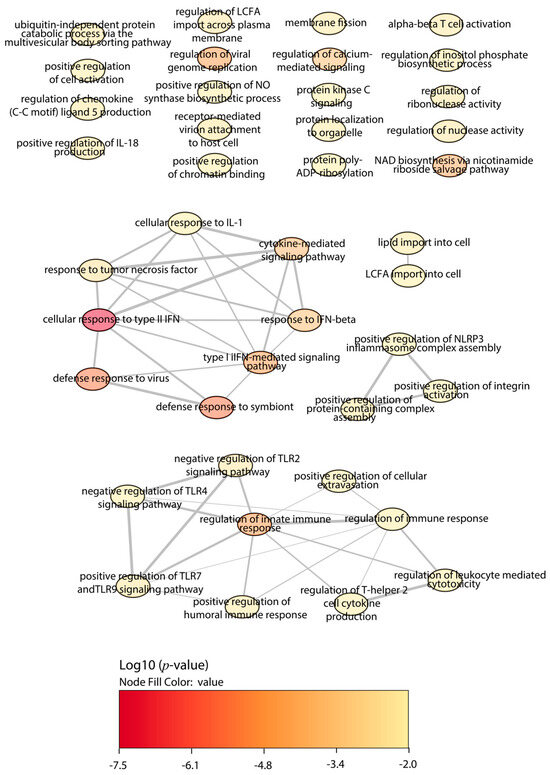
Figure 5.
GO enrichment analysis by Enrichr webtool of upregulated proteins in Slamf1-/- macrophages infected with the Y strain. Significant biological processes’ GO terms are displayed in bubbles. Highly similar GO terms are linked by edges, and the line width indicates the degree of similarity. Color is determined by the log10 (p-value), with red being the most and beige being the least statistically significant, respectively. LCFA, long-chain fatty acid; ALA, alpha-linolenic acid; TLR, toll-like receptor; IFN, interferon; IL, interleukin.
Regarding the downregulated proteins in Slamf1-/- macrophages infected with the Y strain, the GO enrichment also displayed less significant ontologies than the upregulated proteins (Figure 6). The most relevant GO terms were the iron ion transport, the negative regulation of mitochondrial fusion, the protein deglycosylation, and the glycoprotein and oligosaccharide (OS) catabolic processes, among others.
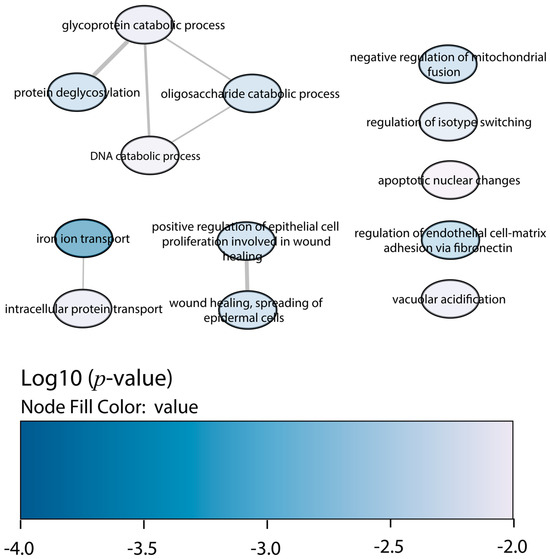
Figure 6.
GO enrichment analysis by Enrichr webtool of downregulated proteins in Slamf1-/- macrophages infected with the Y strain. Significant biological processes’ GO terms are displayed in bubbles. Highly similar GO terms are linked by edges, and the line width indicates the degree of similarity. Color is determined by the log10 (p-value), with blue being the most and white being the least statistically significant, respectively.
2.2. Quantitative Proteomics Analysis of BALB/c and Slamf1-/- Macrophages Infected with the VFRA Strain of T. cruzi
The VFRA was selected since it infects the macrophages independent of SLAMF1 and exhibits an opposite behavior than the Y strain. While Y strain parasite loads decrease in the Slamf1-/- macrophages, the VFRA strain parasite loads remain unchanged. Furthermore, VFRA produces a chronic infection in mice, and the immune response associated with the VFRA infection is different from the Y strain response [14].
We performed the same analysis with the macrophages infected with the VFRA strain of T. cruzi. We obtained 12 significant proteins in the BALB/c infected macrophages: 6 upregulated and 6 downregulated (Figure S1); in the Slamf1-/- macrophages 19 significant proteins, 9 upregulated and 10 downregulated (Figure S2).
Regarding the GO enrichment with the upregulated proteins in the BALB/c macrophages infected with the VFRA strain, the most significant GO terms were the following: IFN type I and II responses, cellular response to IFN-β and IL-1, negative regulation of TLR2 and TLR4, autophagy of peroxisome and pexophagy, lipid import, response to mitochondrial depolarization and positive regulation of NLRP3 inflammasome complex assembly, and IL-18 and transmembrane transport, among others (Figure S3).
Considering the downregulated proteins in BALB/c macrophages infected with the VFRA strain, the enrichment of biological processes showed that the most relevant were glycoprotein, glycosphingolipid (GSL), and OS catabolic processes, negative regulation of mitochondrial fusion, protein deglycosylation, and neutrophil activation (Figure S4).
In the GO enrichment analysis in the Slamf1-/- macrophages infected with the VFRA strain, with the upregulated proteins, the most significant GO terms were the following: IFN type I and II responses, NAD biosynthesis, lipid import, the negative regulation of TLR2 and TLR4, and the positive regulation of NLRP3 inflammasome complex assembly, nitric oxide synthase, and IL-18 (Figure S5).
Finally, considering the downregulated proteins in Slamf1-/- macrophages infected with the VFRA strain, the enrichment of biological processes showed that the most relevant GO terms were the protein deglycosylation and the glycoprotein, GSL, and OS catabolic processes, among others (Figure S6).
2.3. Comparison between the Experimental Conditions and Protein–Protein Interaction Networks
To study the common and specific traits between the conditions with significant quantified proteins, we represented the data in a Venn diagram (Figure 7). Five upregulated proteins were common to all the conditions: immune-response gene 1 (IRG1), guanylate-binding protein (GBP) 5, intercellular adhesion molecule 1 (ICAM1), acyl-CoA synthetase long-chain family member 1 (ACSL1), and programmed cell death protein ligand 1 (PDL1); and one downregulated: metalloproteinase 12 (MMP12). Independently of SLAMF1, with the Y strain infection, there was one common upregulated protein, the radical S-adenosyl methionine domain containing 2 (RSAD2), and with the VFRA strain there was one common downregulated protein, the phospholipase D3 (PLD3). Moreover, independently of the strain, with the Slamf1-/- macrophages there were two common upregulated proteins, interferon-activable protein 204 (IFI204) and bone marrow stromal antigen 2 (BST2), and two common downregulated proteins, SLAMF5 and ADP ribosylation factor-like GTPase 11 (ARL11). Interestingly, Slamf1-/- macrophages infected by the Y strain displayed 10 unique significant proteins, 8 of them upregulated, while BALB/c macrophages infected by the Y strain had 7 unique proteins, and the other 2 conditions (Slamf1-/- and BALB/c macrophages infected by VFRA strain) 3 and 2, respectively.
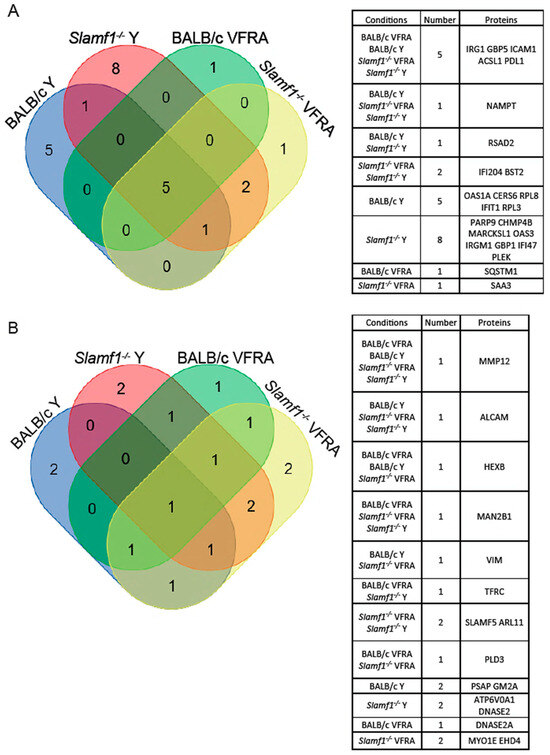
Figure 7.
Venn diagrams of shared proteins between macrophage conditions. (A) Upregulated proteins. (B) Downregulated proteins. Tables display the shared proteins in each combination.
Then, STRING was used to study the relationship between the distinct proteins detected in our experimental conditions (Figure S7). The analysis showed more relationships in upregulated proteins than in the downregulated ones. Our results displayed some proteins (IRG1, GBP5, PDL1, and ICAM1) correlated in all the conditions forming a unique section, in combination with other proteins in each specific condition, such as RSAD2, presented only in the Y infections.
Also, the association of SLAMF5 and MMP12 in Slamf1-/- macrophages was remarkable, whereas MMP12 was alone in the BALB/c macrophages, and ICAM1 was associated with ALCAM, an upregulated protein, and/or TFRC, a downregulated protein.
Furthermore, we represented in Venn diagrams the upregulated and downregulated enriched GO terms of each condition. Our objective was to see which biological processes were common or unique considering the absence of SLAMF1 and the two different strains of T. cruzi.
Considering the upregulated GO terms (Figure 8A and Table S1), all the conditions shared 11 GO terms, such as the negative regulation of TLR2 and TLR4 signaling pathways and the positive regulation of IL-18 production and NLRP3 inflammasome complex assembly. Remarkably, all the conditions displayed different GO terms related to IFN types I and II. Although the distinct GO terms are slightly different from each other, the activation of the IFN system could be added as a shared enriched pathway in all the infections in this study.
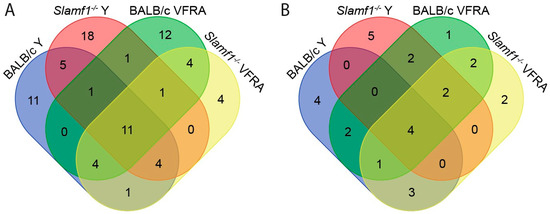
Figure 8.
Venn diagrams of enriched GO terms shared between macrophage conditions. (A) Enriched upregulated GO terms. (B) Enriched downregulated GO terms.
Regarding the Y strain infections, independently of the presence/absence of SLAMF1, there are five GO terms shared, like the positive regulation of TLR7 and TLR9 signaling pathways and the alpha–beta T cell activation. Interestingly, both Slamf1-/- macrophages Y and VFRA infections and BALB/c macrophages infected by VFRA shared the response to IFN-β, suggesting that in the BALB/c with the Y strain, this protective immune response is not so effective, as in VFRA the absence of SLAMF1 would allow the correct activation of this pathway.
Notably, there was a great quantity of unique upregulated GO terms for each condition (Table S2). BALB/c macrophages infected by the Y strain displayed 11 unique GO terms, Slamf1-/- macrophages infected by the Y strain 18 GO terms, BALB/c macrophages infected by the VFRA strain 12 GO terms, and Slamf1-/- macrophages infected by VFRA strain only 4 GO terms. In BALB/c macrophages infected by the Y strain, we found many unique GO terms related to the transcription–translation processes of the cell. However, in the other conditions, the unique GO terms were varied, and it was impossible to establish patrons or specific profiles. Only in the BALB/c infected by VFRA, 3 out of 12 GO terms were related to autophagy, suggesting that this process could be enriched in a normal VFRA infection.
The Venn diagram with the downregulated GO terms is shown in Figure 8B, and Table S3 lists the shared enriched GO terms of this analysis. All the conditions shared four enriched GO terms, although two were related to wound healing, not Chagas disease. However, all shared the glycoprotein catabolic process, suggesting that T. cruzi may inhibit glycoprotein degradation in the cell host. All the conditions except BALB/c infected by the Y strain displayed downregulation of the protein deglycosylation and OS catabolism. Interestingly, only in the BALB/c infections, independently of the strain, did we find markers of activation of neutrophils. This means that the neutrophil’s role in the innate immune response against T. cruzi is downregulated in a normal infection, but the depletion of SLAMF1 avoids it. Also, in the VFRA infections, independently on SLAMF1, GSL catabolism was detected, which would be a specific molecular process decreased only by this strain.
Finally, Table S4 compiles unique enriched downregulated GO terms in each condition. In BALB/c macrophages infected with the Y strain, there were four unique GO terms: ceramide transport, glycolipid transport, metabolism, and aggrephagy. Although this condition was the only one without deglycosylation or OS catabolism, the specific response of Y infection seems to be more correlated with the glycolipids and ceramides, two important molecules of the cell–host membrane. Also, this infection specifically produces the process known as aggrephagy, a type of macroautophagy of protein aggregates. Among the other unique GO terms, Slamf1-/- macrophages infected by the Y strain were the most relevant. It presented five unique GO terms, without relation between them. It displayed downregulation of iron transport, intracellular protein transport, the catabolism of DNA, nuclear apoptosis, and vacuolar acidification.
2.4. Functional Enrichment Analysis
Our last analysis aimed to predict which specific cellular pathways could be affected by the proteins with significant changes. We performed a functional enrichment analysis with the Enrichr web tool using the BioPlanet database (Figure 9 and Figure 10).
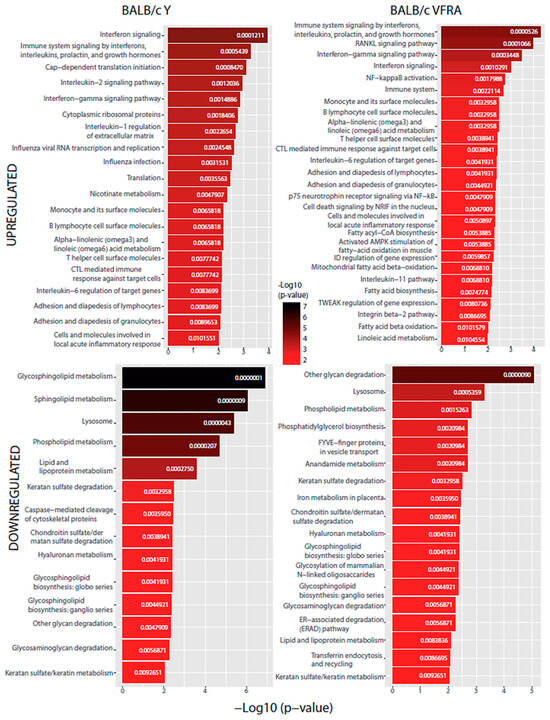
Figure 9.
BioPlanet database functional enrichment analysis of BALB/c macrophages. Significant pathways of BALB/c macrophages infected by the Y strain and the VFRA strain according to the significantly upregulated and downregulated proteins.
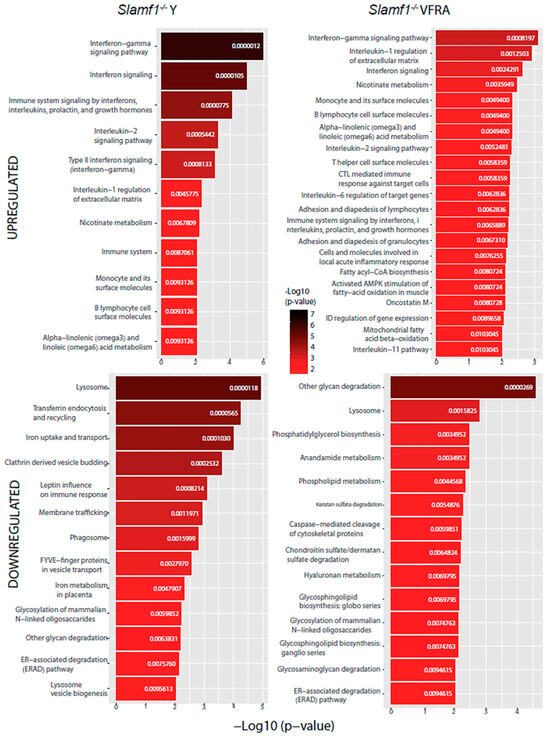
Figure 10.
BioPlanet database functional enrichment analysis of Slamf1-/- macrophages. Significant pathways of Slamf1-/- macrophages infected by the Y strain and the VFRA strain according to the significantly upregulated and downregulated proteins.
All the conditions shared the IFN signaling pathway, as we had already seen in the enrichment GO analysis, confirming that, as in virus infections, T. cruzi activates IFN signaling in the host. Specifically, the IFN-γ pathway was increased in all of them in this analysis. Also, we found in every condition, except in the Slamf1-/- macrophages infected by the Y strain, the upregulation of IL-6, while in all of them except the BALB/c macrophages infected by the VFRA strain, we had upregulated the IL-1β regulation of the extracellular matrix and IL-2 signaling pathway. Furthermore, in all the VFRA infections, independently of SLAMF1 expression, we had an upregulation of the IL-11 pathway. In BALB/c macrophages infected by the Y strain, we obtained pathways related to the translation process and the replication and infection of viruses, among others, while in BALB/c macrophages infected by the VFRA strain we found mostly pathways related to the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) activation.
Regarding the downregulated enriched pathways, all the conditions shared other glycan degradation and lysosome pathways. Also, we found in every condition, except in the Slamf1-/- macrophages infected by the Y strain, the downregulation of GSL biosynthesis and glycosaminoglycan degradation, while in all of them, except the BALB/c macrophages infected by the Y strain, we had the downregulation of the endoplasmic-reticulum-associated degradation pathway. Furthermore, in all the VFRA infections, independently of SLAMF1 expression, there was a downregulation of phosphatidylglycerol (PG) biosynthesis. Finally, specifically in the Slamf1-/- macrophages infected by the Y strain, we obtained a downregulation of pathways related to membrane trafficking, clathrin vesicles, iron uptake, lysosome biogenesis, or phagosome.
3. Discussion
Previously to this work, a decreased parasite replication in Slamf1-/- macrophages infected with Dm28, Y, 10R26, M6421, and Bug2148 strains was detected compared to BALB/c macrophages at 24 hpi, indicating that SLAMF1 is necessary for the internalization and intracellular replication of these strains [14]. However, VFRA displayed the opposite behavior. Thus, we performed a proteomic study on the effect of the absence of SLAMF1 in macrophages after Y and VFRA strains of in vitro infection.
We detected some significantly upregulated proteins upon infection: IRG1, GBP5, ICAM1, ACSL1, and PDL1 independently on the presence or absence of SLAMF1. IRG1 is a novel M1 macrophage marker and produces itaconic acid that inhibits the isocitrate lyase, the key enzyme of the glyoxylate shunt [16], necessary in T. cruzi to survive on limited carbon conditions [17]. Hearts of BALB/c mice infected with the Y strain displayed high levels of itaconic acid production [18], and increased levels of Irg1 were detected in the heart tissue and intestine of Y and VFRA-infected BALB/c and Slamf1-/- mice [14]. Thus, our results support further research into itaconic acid and its therapeutic potential for treating T. cruzi infection.
GBP5 and other GBPs are strongly induced by IFN-γ, a cytokine increased in cells and organs infected by T. cruzi [14,19,20]. GBPs facilitate cell-autonomous immunity through inflammasome activation and/or microbe elimination [21,22]. We have described for the first time the relationship between GBP5 and this parasite. T. cruzi infections also displayed upregulation of ICAM1 (CD54) [23,24], which deficiency generates a higher infection susceptibility and a decrease in T cell recruitment [25]. Our results support the role of ICAM1 as a defense mechanism against T. cruzi. On the other hand, our study is the first that correlates the increase in ACSL1 with Chagas disease. Interestingly, ACSL1 increases in acute myocardial infarction patients [26], which is one of the symptoms of chronic Chagas disease.
The PD1/PDL1 pathway inhibits the proliferation and activation of macrophages leading to T cell anergy [27], while its blockade reinvigorates exhausted CD8+ T cells, reducing the pathogen burden [28,29]. However, there are some controversies considering its role in T. cruzi infections. Some researchers have shown that PD1 inhibition leads to reduced parasitemia [30], but others have described that PDL1 inhibition favors a higher parasitemia [31]. On the other hand, PD1–PDL1 interaction protected against heart damage in chronic T. cruzi infection, reducing the exacerbated immune response without modifying the parasite load [32], and increased PDL1 levels were detected in BALB/c macrophages as in our proteomic study [33]. Altogether, the PD1/PDL1 pathway may regulate the activation of infected macrophages to prevent immune system exacerbation.
Regarding the downregulated protein found in all infections, MMP12, an increase was detected in T. cruzi infections at 24 hpi in mouse macrophages [34], contrary to our proteomic analysis. However, the same report analyzed the expression at 48 hpi too, revealing a significant reduction in MMP12. These different results could be due to the different parasite-infecting strains used. Moreover, MMP12 was identified as a resolution-promoting factor in myocardial infarction, a symptom of chronic Chagas disease, inducing M2 macrophages and the secretion of anti-inflammatory cytokines [35]. Thus, the decreased expression of MMP12 would enhance the cardiac disease preventing inflammation resolution.
Considering the strain-specific proteins without a dependency on SLAMF1, Y strain infections displayed one common upregulated protein (RSAD2), and VFRA strain infections had one common downregulated protein (PLD3). RSAD2 is an IFN-stimulated protein that restricts various families of viruses [36,37], while PLD3 digests ssRNA and ssDNA in lysosomes/endosomes in viral and bacterial infections [38,39]. This is the first time both proteins have been related to T. cruzi infections.
Addressing the effect of SLAMF1 using the Slamf1-/- macrophages, we detected two common upregulated proteins (IFI204 and BST2) and two common downregulated proteins (SLAMF5 and ARL11), independently on the infecting strain. IFI204 is orthologous to the human IFN-γ inducible protein 16 (IFI16), which was increased in infected dendritic cells [40], while BST2 is induced by type I IFN and traps viral particles onto the cell surface [41]. Our study points to IFI204/IFI16 and BST2 as potential Chagas disease targets and SLAMF1 as their inhibitor through some unknown mechanism. On the other hand, SLAMF5 produces IL-6 and TNF-α [42], while ARL11 regulates pro-inflammatory effectors [43] and is associated with M2 macrophages [44]. Lower Tnf levels were described in Slamf1-/- macrophages infected with Y and VFRA and the switch to M1 phenotype [14]. Decreased levels of both SLAMF5 and ARL11 agree with those results, although more studies must be conducted to unravel the mechanisms that connect SLAMF1 to them.
Slamf1-/- macrophages infected by the lethal Y strain displayed the biggest number of altered unique proteins. These proteins either blocked or induced by SLAMF1 may be involved in the disappearance of this strain’s lethality in the absence of SLAMF1, making them attractive molecules to understand and avoid pathogenicity. In contrast, Slamf1-/- macrophages infected by the VFRA strain only displayed three significant unique proteins. This agrees with previous observations that VFRA barely changes the cytokine production in Slamf1-/- macrophages [14].
Among these unique significant proteins in Slamf1-/- macrophages infected by the Y strain, several have been described as macrophage response regulators: PARP9, MARCKSL1, IRGM1, ATP6V0A1, GBP1, and IFI47 [45,46,47,48,49,50,51]. They would contribute to the innate immune response promoting macrophage activation and the killing of the parasite. This agrees with the reduced parasite load in Slamf1-/- macrophages infected by the Y strain [14], suggesting that SLAMF1 signaling after the T. cruzi interaction affects these critical proteins to resolve the infection. Alternatively, the Y strain may promote changes in them to persist in the cell, but the decrease in its internalization by the absence of SLAMF1 reduces the effect. This points out the relevance of SLAMF1 and these proteins as possible therapeutic targets.
To further understand the relevance of our results, we performed GO and functional enrichment analysis. Table 1 summarizes the most relevant macrophage pathways suffering a positive or negative regulation. We detected nine common pathways in all the conditions without an SLAMF1 effect, some implicated in innate immunity. IFN responses activate parasite killing through microbicidal molecules and ROS in the early stages of T. cruzi infection [52]. Also, NLRP3 inflammasome induces the secretion of the inflammatory cytokines IL-1β and IL-18, which guide pyroptosis and nitric oxide production, favoring T. cruzi elimination [53]. Our study supports the key role of these processes in dealing with the T. cruzi infection in macrophages independently of the strain.

Table 1.
Most remarkable upregulated or downregulated macrophage pathways in our proteomic analysis. Text color legend for pathways shared by all the conditions (red); three conditions (green); two conditions (blue); unique to a condition (black). Abbreviations: LCFA, long-chain fatty acid; ALA, alpha-linolenic acid; LA, linoleic acid; GSL, glycosphingolipid; Cer, ceramide; GL, glycolipid; AEA, anandamide; PG, phosphatidylglycerol; GP, glycoprotein; GAG, glycosaminoglycan; OS, oligosaccharide; ERAD, endoplasmic-reticulum-associated protein degradation; PKC, protein kinase C; RANKL, receptor activator of nuclear factor kappa-B ligand; TWEAK, TNF-related weak inducer of apoptosis. Arrows up indicate upregulation and arrows down indicate downregulation.
Also, we found an increase in the alpha-linolenic acid (ALA) and linoleic acid (LA) metabolism. LA is a precursor of omega-6 fatty acids with predominantly pro-inflammatory actions, while ALA is a precursor of omega-3 fatty acids with mostly anti-inflammatory effects [54]. Their increment may be due to the homeostatic mechanisms of inflammation. Other changes in lipids included downregulation of the GSL catabolism and PG biosynthesis in VFRA-infected macrophages. GSLs are critical for the immune response and the parasite phagocytosis [55] while PG displays anti-inflammatory effects [56]. Thus, these downregulations would prevent parasite proliferation in macrophages and contribute to the persistence of the pro-inflammatory response against the VFRA strain.
Finally, other responses were also specific to parasite strain or SLAMF1-dependent strain. IL-11 activation was VFRA-specific, being upregulated in infections by Tulahuen strain [57], which belongs to the DTU VI as VFRA, suggesting that IL-11 activation would be DTU-specific. TLR7 and TLR9 signaling was specifically increased with Y strain, while the downregulation of the TLR2 and TLR4 signaling was found in all the conditions. TLR2 and TLR4 sense molecules of the trypomastigote surface inducing the nitric oxide and pro-inflammatory cytokine production [52], and their downregulation could be negative feedback signaling to avoid an over-enhanced immune response or a regulation mechanism induced by the parasite. On the other hand, TLR9 induces cytokine production and activation of Th1 responses [52], and a triple defect in TLR3, 7, and 9 made mice more susceptible to the infection [58]. Thus, we have pointed out that the Y strain, which causes mice mortality, enhances TLR signaling, while VFRA, a non-lethal strain in mice, does not produce this effect in macrophages. Moreover, as TLR9 recognized CpG DNA and TLR7 ssRNA in the lysosomes, this suggests a bigger exposure to parasite intracellular content due to better T. cruzi degradation. This would correlate with a previous proteomic study in which VFRA displayed more antioxidant molecules than the Y strain [59]. As VFRA is more resistant to these killing processes, it would release less DNA and ssRNA in the lysosome, explaining the increase in TLR7 and TLR9 activation only with the Y strain infection.
In summary, we have identified different down- or upregulated proteins in Y and VFRA infections in macrophages. Some are SLAMF1-dependent and/or strain-dependent, whereas the others are common to all T. cruzi infections. We have highlighted the relevance of most of them as critical innate immune regulators and displayed the significant functional pathways in which they participate, opening new avenues of research in Chagas disease. Independently on SLAMF1, T. cruzi induces different responses in macrophages to deal with the infection and kill the parasite, such as the type I and II IFN responses, NLRP3 inflammasome activation, IL-18 production, TLR7 and TLR9 activation specifically in the Y strain infection, and IL-11 signaling specifically in the VFRA strain infection.
4. Materials and Methods
4.1. Parasite Culture
T. cruzi VFRA strain was obtained from Dr. M. Miles (London School of Hygiene and Tropical Medicine, London, UK) through the European program ChagasEpiNet. Y strain was obtained from Harvard Medical School (Boston, MA, USA). This strain was isolated originally from a human patient in Brazil [60].
Trypomastigotes of T. cruzi were co-cultured in vitro with Vero cells (ATCC number CCL-81). Vero cells were cultured with complete Roswell Park Memorial Institute (RPMI, Thermo Fisher Scientific, Waltham, MA USA) medium containing 2 mM L-glutamine, 100 U/mL penicillin, 1000 U/mL gentamicin, 10 μg/mL streptomycin, and 0.1 mM non-essential amino acids and supplemented with 5% fetal bovine serum (FBS, Gibco Life Technologies, Grand Island, NY, USA). Cells were maintained at 37 °C in an atmosphere of 5% CO2 until the cells reached 80% confluence in the biosafety level 3 (BSL3) cell culture laboratories of the Centro de Biología Molecular Severo Ochoa (Madrid, Spain). Then, Vero cells were infected with trypomastigotes following a 10:1 parasite–cell ratio. After 4 days, the supernatant of the infected cells with the trypomastigotes was collected, dead cells and amastigotes were removed by centrifugation at 1000× g for 5 min, and trypomastigotes were collected by centrifugation at 1600× g for 10 min. These trypomastigotes were used for macrophage infections.
4.2. Mice and Ethic Statement
Female mice of 10–12 weeks old were used. Slamf1-/- mice were obtained from Dr. Cox Terhost (Harvard Medical School, Boston, MA, USA), while BALB/c mice were obtained from Charles River Laboratories. Mice were maintained under pathogen-free conditions at Centro de Biología Molecular Severo Ochoa (Madrid, Spain) animal facility.
This study was carried out in strict accordance with the European Commission’s legislation for the protection of animal use purposes (2010/63/EU). The protocol for the treatment of the animals was approved by the “Comité de Etica de Investigación de la Comunidad de Madrid” (Spain, permits PROEX 148/15, and PROEX 213.6/21). Animals had unlimited access to food and water. They were euthanized in a CO2 chamber and all efforts were made to minimize their suffering.
4.3. Isolation of Peritoneal Macrophages and In Vitro Infection
BALB/c and Slamf1-/- 10–12 weeks-old female mice were injected intraperitoneally with 10% thioglycollate (Gibco, Grand Island, NY, USA) (1 mL per mouse). After 4 days, mice were euthanized by CO2 inhalation. Peritoneal cells were collected by lavage with 10 mL of cold phosphate-buffered saline (PBS), and they were pelleted by centrifugation (260× g, for 10 min, at 4 °C). Then, they were seeded in p100 plates (107 cells/plate) in RPMI medium supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 1000 U/mL gentamicin, 10 μg/mL streptomycin, 0.1 mM non-essential amino acids, and 5% FBS overnight at 37 °C in an atmosphere of 5% CO2. Non-adherent cells were removed by gently washing three times with warm PBS. These peritoneal macrophages were infected with trypomastigotes of Y or VFRA strains at a ratio of 10 parasites per cell in the BSL3 cell culture laboratories of the Centro de Biología Molecular Severo Ochoa (Madrid, Spain). At 24 hpi, supernatant with the trypomastigotes was eliminated and the macrophages, including the control macrophages without infection, were washed three times with PBS to remove the unbound parasites.
4.4. Protein Extraction and Digestion for the Proteomic Analysis
For protein extraction, three cell replicates (n = 3) for each condition were incubated in RIPA buffer (150 mM NaCl, 20 mM Tris, pH = 7.6, 0.5% sodium deoxycholate, 1 mM EDTA, 0.1% SDS, 1% Triton-X100) with protease (cOmplete tablets Easypack, Roche, Basel, Switzerland) and phosphatase (PhosSTOP, Roche, Basel, Switzerland) inhibitors for 30 min at 4 °C and centrifuged at 20,800× g for 15 min, and the supernatants were kept. For protein quantification, bicinchoninic acid assay (BCA Protein Assay, Thermo Fisher Scientific, Waltham, MA USA) was utilized using supernatants, and extrapolation was based on a standard curve of known concentrations of bovine serum albumin.
Protein extracts were prepared in up to 50 µL of sample reduction buffer and then applied onto 1.2 cm wide wells of a conventional SDS-PAGE gel (0.75 mm thick, 4% stacking, and 10% resolving). The run was stopped when the front entered 3 mm into the resolving gel so that the whole proteome became concentrated in the stacking/resolving gel interface. The unseparated protein bands were visualized by Coomassie staining, excised, cut into cubes (2 × 2 mm), and placed in 0.5 mL microcentrifuge tubes [61]. The gel pieces were destained in acetonitrile/water (1:1), were reduced and alkylated (disulfide bonds from cysteinyl residues were reduced with 10 mM DTT for 1 h at 56 °C, and then thiol groups were alkylated with 10 mM iodoacetamide for 30 min at room temperature in darkness), and digested in situ with sequencing grade trypsin (Promega, Madison, WI, USA) as described by Shevchenko et al. [62], with minor modifications. The gel pieces were shrunk by removing all liquid using sufficient acetonitrile. Acetonitrile was pipetted out, and the gel pieces were dried in a speed vac. The dried gel pieces were re-swollen in 100 mM Tris-HCl pH 8, 10 mM CaCl2 with 60 ng/µL trypsin at 5:1 protein/enzyme (w/w) ratio. Digestion was performed in the presence of 0.2% RapiGest (Waters, Milford, MA, USA). The tubes were kept on ice for 2 h and incubated at 37 °C for 12 h. Digestion was stopped by the addition of 1% trifluoroacetic acid. Whole supernatants were dried down and then desalted onto OMIX Pipette tips C18 (Agilent Technologies, Santa Clara, CA, USA) until the mass spectrometric analysis was performed.
4.5. TMT Labeling and Fractionation
The resultant peptide mixture from desalted proteins tryptic digest (50 µg) was labeled using chemicals from the TMT sixplex Isobaric Mass Tagging Kit (Thermo Fisher Scientific, Waltham, MA, USA) as described by the manufacturer. Briefly, peptides were dissolved in 50 μL of 100 mM triethylammonium bicarbonate, adjusted to pH 8. For labeling, each TMT reagent was dissolved in 41 μL of acetonitrile and added to the respective peptide mixture and then incubated at room temperature for one hour. Labeling was stopped by the addition of 8 μL 5% hydroxylamine. Whole supernatants were dried down, and the six samples were mixed to obtain the “6plex-labeled mixture”. The mixture was analyzed by Reverse-Phase Liquid Chromatography–Tandem Mass Spectroscopy (RP-LC-MS/MS) to check the efficiency of the labeling.
The sample was then fractionated using the Pierce High pH Reversed-Phase Peptide Fractionation Kit (Thermo Fisher Scientific, Waltham, MA, USA) as described, with minor modifications. The sample was re-swollen in 0.1% trifluoroacetic acid and then loaded onto an equilibrated, high-pH, reversed-phase fractionation spin column. A step gradient of increasing acetonitrile concentrations (5–50%) in a volatile high pH (Triethylamine (0.1%)) was then applied to the columns to elute bound peptides into nine different fractions collected by centrifugation. The fractions obtained from a high-pH, reversed-phase 6plex-labeled mixture were dried and stored until analysis by mass spectrometry for quantification.
4.6. Analysis by RP-LC-MS/MS
The fractions were resuspended in 10 µL of 0.1% FA and analyzed by RP-LC-MS/MS in an Easy-nLC II system coupled to an ion trap LTQ-Orbitrap-Velos-Pro hybrid mass spectrometer (Thermo Scientific, Waltham, MA, USA). The peptides were concentrated by reverse-phase chromatography using a 0.1 mm × 20 mm C18 RP precolumn (Thermo Fisher Scientific, Waltham, MA, USA) and then separated using a 0.075 mm × 250 mm bioZen 2.6 µm Peptide XB-C18 RP column (Phenomenex, Alcobendas, Spain) operating at 0.25 μL/min. Peptides were eluted using a 90 min dual gradient. The gradient profile was set as follows: 5–25% solvent B for 68 min, 25–40% solvent B for 22 min, 40–100% solvent B for 2 min, and 100% solvent B for 18 min (solvent A: 0.1% FA in water; solvent B: 0.1% FA, 80% acetonitrile in water). ESI ionization was performed using a Nano-bore emitters Stainless Steel ID 30 μm (Proxeon, Waltam MA, USA) interface at 2.1 kV spray voltage with S-Lens of 60% [63].
The instrument method consisted of a data-dependent top-20 experiment with an Orbitrap MS1 scan at a resolution (m/Δm) of 30,000 followed by twenty higher-energy collision dissociation (HCD) MS/MS mass-analyzed in the Orbitrap at 7500 (Δm/m) resolution. MS2 experiments were performed using HCD to generate high-resolution and high-mass-accuracy MS2 spectra. The minimum MS signal for triggering MS/MS was set to 500. The lock mass option was enabled for both MS and MS/MS mode, and the polydimethylcyclosiloxane ions (protonated (Si(CH3)2O))6; m/z 445.120025) were used for internal recalibration of the mass spectra.
Peptides were detected in survey scans from 400 to 1600 amu (1 μscan) using an isolation width of 1.3 u (in mass-to-charge ratio units), normalized collision energy of 40% for HCD fragmentation, and dynamic exclusion applied for 60 s periods. Charge-state screening was enabled to reject unassigned and singly charged protonated ions.
4.7. Quantitative Proteomic Data Analysis and Representation
Peptide identification from raw data was carried out using PEAKS Studio X+ search engine (Bioinformatics Solutions Inc., Waterloo, ON, Canada). Database search was performed against uniprot-mus-musculus.fasta (55,462 entries; UniProt release 03/2020) merged to uniprot-trypanosoma-cruzi (19,242 entries; UniProt release 03/2020). The following constraints were used for the searches: tryptic cleavage after Arg and Lys (semi-specific), up to two missed cleavage sites, and tolerances of 20 ppm for precursor ions and 0.05 Da for MS/MS fragment ions, and the searches were performed allowing optional Met oxidation and Cys carbamidomethylation and fixed TMT 6plex reagent labeling at the N-terminus and lysine residues. FDR for peptide spectrum matches and proteins was limited to 0.01. Only those proteins with at least two unique peptides discovered from LC/MS/MS analyses were considered reliably identified and sent to be quantified [63,64,65].
Quantitation of TMT labeled peptides was performed with PEAKS Studio X+ search engine, selected “Reporter Ion Quantification iTRAQ/TMT” under the “Quantifications” options. We used auto-normalization mode, which calculates a global ratio from the total intensity of all labels in all quantifiable peptides. The −10LgP, quality, and reporter ion intensity were used for the spectrum filter, and significance (PEAKSQ method) was used for peptide and protein abundance calculation. For the protein quantification, we considered protein groups for peptide uniqueness, used unique peptides for protein quantification, and the modified peptides were excluded.
For data representation, volcano plots and bar plots of each analysis were created using R programming language (v.4.2.2) with the package ggplot2. Enrichment GO terms study was carried out with Enrichr webtool, posterior REVIGO [66] analysis, and represented by Cytoscape (v.3.10.1). Protein–protein interaction networks were performed by STRING webtool (v.12.0) and represented by Cytoscape (version 3.10.1). Enrichment pathways analysis was performed in the Enrichr web tool using the BioPlanet database. Venn diagrams were created with the following online tool: https://bioinformatics.psb.ugent.be/webtools/Venn/ accessed on 4 July 2024. Figure representations were prepared in Adobe Illustrator CS5 (2010).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25137493/s1.
Author Contributions
Conceptualization, M.F. and N.G.; methodology, A.H.-C., B.B., E.M. and A.M.; software, A.H.-C., J.d.M.-S., E.M. and A.M.; validation, A.H.-C. and J.d.M.-S.; formal analysis, A.H.-C. and J.d.M.-S.; investigation, A.H.-C.; resources, A.H.-C., B.B., E.M. and A.M.; data curation, A.H.-C., J.d.M.-S., E.M. and A.M.; writing—original draft preparation, A.H.-C.; writing—review and editing, A.H.-C., M.F. and N.G.; visualization, A.H.-C. and N.G.; supervision, M.F. and N.G.; project administration, M.F. and N.G.; funding acquisition, M.F. and N.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Ministerio de Economía y competitividad and Fondo Europeo de Desarrollo Regional (SAF2016-75988-R (MINECO/FEDER), SAF2015-63868-R (MINECO/FEDER) to M.F.), Ministerio de Ciencia, Innovación y Universidades-Agencia Estatal de Investigación and Fondo Europeo de Desarrollo Regional (PGC2018-096132-B-I00 and PID2021-123389OB-I00 to N.G.). A.H-C. was a recipient of a FPU contract from the Ministerio de Ciencia, Innovación y Universidades; J.M-S. was a recipient of a Garantía Juvenil predoctoral fellowship (Comunidad de Madrid). Grant PT17/0019, of the PE I + D + i 2013-2016, funded by ISCIII and ERDF.
Institutional Review Board Statement
The animal study protocol was approved by the Ethics Committee of Centro de Biología Molecular Severo Ochoa (CSIC-UAM, Madrid, Spain) and the Comité de Etica de Investigación de la Comunidad de Madrid (Madrid, Spain), for studies involving animals: PROEX 148/15 on date 2015/06/29, and PROEX 213.6/21 on date 2021/07/02) for studies involving animals.
Informed Consent Statement
Not applicable for studies involving animals.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We thank the personnel of the Proteomics Service of the Centro de Biología Molecular Severo Ochoa (CBMSO) Consejo Superior the Investigaciones Científicas-Universidad Autónoma de Madrid (CSIC-UAM) (Madrid, Spain) for their technical assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Guhl, F.; Jaramillo, C.; Vallejo, G.A.; Yockteng, R.; Cárdenas-Arroyo, F.; Fornaciari, G.; Arriaza, B.; Aufderheide, A.C. Isolation of Trypanosoma Cruzi DNA in 4000-Year-Old Mummified Human Tissue from Northern Chile. Am. J. Phys. Anthropol. 1999, 108, 401–407. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Chagas Disease (American Trypanosomiasis). Available online: https://www.who.int/health-topics/chagas-disease (accessed on 6 May 2024).
- Pérez-Molina, J.A.; Molina, I. Chagas Disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Rassi, A., Jr.; Rassi, A.; Marcondes de Rezende, J. American Trypanosomiasis (Chagas Disease). Infect. Dis. Clin. N. Am. 2012, 26, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Schmunis, G.A.; Yadon, Z.E. Chagas Disease: A Latin American Health Problem Becoming a World Health Problem. Acta Trop. 2010, 115, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.; Bottazzi, M.E.; Strub-Wourgaft, N.; Sosa-Estani, S.; Torrico, F.; Pajín, L.; Abril, M.; Sancho, J. A New Patient Registry for Chagas Disease. PLoS Negl. Trop. Dis. 2020, 14, e0008418. [Google Scholar] [CrossRef] [PubMed]
- Tarleton, R.L. Immune System Recognition of Trypanosoma Cruzi. Curr. Opin. Immunol. 2007, 19, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, M.; Garg, N.J. P47phox-/- Mice Are Compromised in Expansion and Activation of CD8+ T Cells and Susceptible to Trypanosoma Cruzi Infection. PLoS Pathog. 2014, 10, e1004516. [Google Scholar] [CrossRef]
- Muñoz-Fernández, M.A.; Fernández, M.A.; Fresno, M. Synergism between Tumor Necrosis Factor-Alpha and Interferon-Gamma on Macrophage Activation for the Killing of Intracellular Trypanosoma Cruzi through a Nitric Oxide-Dependent Mechanism. Eur. J. Immunol. 1992, 22, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.M.; Oliveira, A.C.; Bellio, M. The Immune Response to Trypanosoma Cruzi: Role of Toll-Like Receptors and Perspectives for Vaccine Development. J. Parasitol. Res. 2012, 2012, 507874. [Google Scholar] [CrossRef]
- Sintes, J.; Engel, P. SLAM (CD150) Is a Multitasking Immunoreceptor: From Cosignalling to Bacterial Recognition. Immunol. Cell Biol. 2011, 89, 161–163. [Google Scholar] [CrossRef]
- van Driel, B.J.; Liao, G.; Engel, P.; Terhorst, C. Responses to Microbial Challenges by SLAMF Receptors. Front. Immunol. 2016, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Calderón, J.; Maganto-Garcia, E.; Punzón, C.; Carrión, J.; Terhorst, C.; Fresno, M. The Receptor Slamf1 on the Surface of Myeloid Lineage Cells Controls Susceptibility to Infection by Trypanosoma Cruzi. PLoS Pathog. 2012, 8, e1002799. [Google Scholar] [CrossRef]
- Poveda, C.; Herreros-Cabello, A.; Callejas-Hernández, F.; Osuna-Pérez, J.; Maza, M.C.; Chillón-Marinas, C.; Calderón, J.; Stamatakis, K.; Fresno, M.; Gironès, N. Interaction of Signaling Lymphocytic Activation Molecule Family 1 (SLAMF1) Receptor with Trypanosoma Cruzi Is Strain-Dependent and Affects NADPH Oxidase Expression and Activity. PLoS Negl. Trop. Dis. 2020, 14, e0008608. [Google Scholar] [CrossRef]
- Rodriguez, H.O.; Guerrero, N.A.; Fortes, A.; Santi-Rocca, J.; Gironès, N.; Fresno, M. Trypanosoma Cruzi Strains Cause Different Myocarditis Patterns in Infected Mice. Acta Trop. 2014, 139, 57–66. [Google Scholar] [CrossRef]
- Michelucci, A.; Cordes, T.; Ghelfi, J.; Pailot, A.; Reiling, N.; Goldmann, O.; Binz, T.; Wegner, A.; Tallam, A.; Rausell, A.; et al. Immune-Responsive Gene 1 Protein Links Metabolism to Immunity by Catalyzing Itaconic Acid Production. Proc. Natl. Acad. Sci. USA 2013, 110, 7820–7825. [Google Scholar] [CrossRef] [PubMed]
- Tallam, A.; Perumal, T.M.; Antony, P.M.; Jäger, C.; Fritz, J.V.; Vallar, L.; Balling, R.; Del Sol, A.; Michelucci, A. Gene Regulatory Network Inference of Immunoresponsive Gene 1 (IRG1) Identifies Interferon Regulatory Factor 1 (IRF1) as Its Transcriptional Regulator in Mammalian Macrophages. PLoS ONE 2016, 11, e0149050. [Google Scholar] [CrossRef]
- Gironès, N.; Carbajosa, S.; Guerrero, N.A.; Poveda, C.; Chillón-Marinas, C.; Fresno, M. Global Metabolomic Profiling of Acute Myocarditis Caused by Trypanosoma Cruzi Infection. PLoS Negl. Trop. Dis. 2014, 8, e3337. [Google Scholar] [CrossRef]
- Dutra, W.O.; Menezes, C.a.S.; Magalhães, L.M.D.; Gollob, K.J. Immunoregulatory Networks in Human Chagas Disease. Parasite Immunol. 2014, 36, 377–387. [Google Scholar] [CrossRef]
- Ferreira, L.R.P.; Frade, A.F.; Baron, M.A.; Navarro, I.C.; Kalil, J.; Chevillard, C.; Cunha-Neto, E. Interferon-γ and Other Inflammatory Mediators in Cardiomyocyte Signaling during Chagas Disease Cardiomyopathy. World J. Cardiol. 2014, 6, 782–790. [Google Scholar] [CrossRef]
- Kirkby, M.; Enosi Tuipulotu, D.; Feng, S.; Lo Pilato, J.; Man, S.M. Guanylate-Binding Proteins: Mechanisms of Pattern Recognition and Antimicrobial Functions. Trends Biochem. Sci. 2023, 48, 883–893. [Google Scholar] [CrossRef]
- Kutsch, M.; Coers, J. Human Guanylate Binding Proteins: Nanomachines Orchestrating Host Defense. FEBS J. 2021, 288, 5826–5849. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Calderon, T.M.; Berman, J.W.; Braunstein, V.L.; Weiss, L.M.; Wittner, M.; Tanowitz, H.B. Infection of Endothelial Cells with Trypanosoma Cruzi Activates NF-kappaB and Induces Vascular Adhesion Molecule Expression. Infect. Immun. 1999, 67, 5434–5440. [Google Scholar] [CrossRef] [PubMed]
- Laucella, S.; Salcedo, R.; Castaños-Velez, E.; Riarte, A.; De Titto, E.H.; Patarroyo, M.; Orn, A.; Rottenberg, M.E. Increased Expression and Secretion of ICAM-1 during Experimental Infection with Trypanosoma Cruzi. Parasite Immunol. 1996, 18, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Michailowsky, V.; Celes, M.R.N.; Marino, A.P.; Silva, A.A.; Vieira, L.Q.; Rossi, M.A.; Gazzinelli, R.T.; Lannes-Vieira, J.; Silva, J.S. Intercellular Adhesion Molecule 1 Deficiency Leads to Impaired Recruitment of T Lymphocytes and Enhanced Host Susceptibility to Infection with Trypanosoma Cruzi. J. Immunol. 2004, 173, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cui, R.; Du, R.; Song, C.; Xie, F.; Ren, L.; Li, J. Platelet-Derived Microvesicles Mediate Cardiomyocyte Ferroptosis by Transferring ACSL1 During Acute Myocardial Infarction. Mol. Biotechnol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhang, Y.; Wang, J.; Gu, J. Defects in Macrophage Reprogramming in Cancer Therapy: The Negative Impact of PD-L1/PD-1. Front. Immunol. 2021, 12, 690869. [Google Scholar] [CrossRef] [PubMed]
- Attanasio, J.; Wherry, E.J. Costimulatory and Coinhibitory Receptor Pathways in Infectious Disease. Immunity 2016, 44, 1052–1068. [Google Scholar] [CrossRef] [PubMed]
- Gigley, J.P.; Bhadra, R.; Moretto, M.M.; Khan, I.A. T Cell Exhaustion in Protozoan Disease. Trends Parasitol. 2012, 28, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, F.R.S.; Mariano, F.S.; Oliveira, C.J.F.; Pavanelli, W.R.; Guedes, P.M.M.; Silva, G.K.; Campanelli, A.P.; Milanezi, C.M.; Azuma, M.; Honjo, T.; et al. Regulation of Trypanosoma Cruzi-Induced Myocarditis by Programmed Death Cell Receptor 1. Infect. Immun. 2011, 79, 1873–1881. [Google Scholar] [CrossRef]
- Arana, Y.; Gálvez, R.I.; Jacobs, T. Role of the PD-1/PD-L1 Pathway in Experimental Trypanosoma Cruzi Infection and Potential Therapeutic Options. Front. Immunol. 2022, 13, 866120. [Google Scholar] [CrossRef]
- Fonseca, R.; Salgado, R.M.; Borges da Silva, H.; do Nascimento, R.S.; D’Império-Lima, M.R.; Alvarez, J.M. Programmed Cell Death Protein 1-PDL1 Interaction Prevents Heart Damage in Chronic Trypanosoma Cruzi Infection. Front. Immunol. 2018, 9, 997. [Google Scholar] [CrossRef] [PubMed]
- Dulgerian, L.R.; Garrido, V.V.; Stempin, C.C.; Cerbán, F.M. Programmed Death Ligand 2 Regulates Arginase Induction and Modifies Trypanosoma Cruzi Survival in Macrophages during Murine Experimental Infection. Immunology 2011, 133, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, S.; Garg, N.J. Trypanosoma Cruzi Induces the PARP1/AP-1 Pathway for Upregulation of Metalloproteinases and Transforming Growth Factor β in Macrophages: Role in Cardiac Fibroblast Differentiation and Fibrosis in Chagas Disease. mBio 2020, 11, e01853-20. [Google Scholar] [CrossRef] [PubMed]
- Mouton, A.J.; Rivera Gonzalez, O.J.; Kaminski, A.R.; Moore, E.T.; Lindsey, M.L. Matrix Metalloproteinase-12 as an Endogenous Resolution Promoting Factor Following Myocardial Infarction. Pharmacol. Res. 2018, 137, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; He, J.; Zhou, X.; Pan, H.; He, F.; Du, A.; Yu, B.; Jiang, N.; Li, X.; Yuan, K.; et al. Discovering Common Pathogenetic Processes between COVID-19 and Tuberculosis by Bioinformatics and System Biology Approach. Front. Cell. Infect. Microbiol. 2023, 13, 1280223. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Zhang, R.; Liu, L.-L.; Li, H.-J.; Zhu, H. Expression Analysis and Antiviral Activity of Koi Carp (Cyprinus Carpio) Viperin against Carp Edema Virus (CEV). Fish Shellfish Immunol. 2024, 148, 109519. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.L.; Huang, D.; Blane, T.R.; Thinnes, T.C.; Murakami, Y.; Fukui, R.; Miyake, K.; Nemazee, D. Cleavage of DNA and RNA by PLD3 and PLD4 Limits Autoinflammatory Triggering by Multiple Sensors. Nat. Commun. 2021, 12, 5874. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.L.; Huang, D.; Huber, C.; Mårtensson, A.; Tardif, V.; Skog, P.D.; Blane, T.R.; Thinnes, T.C.; Osborn, K.; Chong, H.S.; et al. PLD3 and PLD4 Are Single-Stranded Acid Exonucleases That Regulate Endosomal Nucleic-Acid Sensing. Nat. Immunol. 2018, 19, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Gil-Jaramillo, N.; Rocha, A.P.; Raiol, T.; Motta, F.N.; Favali, C.; Brigido, M.M.; Bastos, I.M.D.; Santana, J.M. The First Contact of Human Dendritic Cells With Trypanosoma Cruzi Reveals Response to Virus as an Unexplored Central Pathway. Front. Immunol. 2021, 12, 638020. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, K.; Wang, S.; Du, J. Multi-Functional BST2/Tetherin against HIV-1, Other Viruses and LINE-1. Front. Cell. Infect. Microbiol. 2022, 12, 979091. [Google Scholar] [CrossRef]
- Sintes, J.; Romero, X.; de Salort, J.; Terhorst, C.; Engel, P. Mouse CD84 Is a Pan-Leukocyte Cell-Surface Molecule That Modulates LPS-Induced Cytokine Secretion by Macrophages. J. Leukoc. Biol. 2010, 88, 687–697. [Google Scholar] [CrossRef]
- Arya, S.B.; Kumar, G.; Kaur, H.; Kaur, A.; Tuli, A. ARL11 Regulates Lipopolysaccharide-Stimulated Macrophage Activation by Promoting Mitogen-Activated Protein Kinase (MAPK) Signaling. J. Biol. Chem. 2018, 293, 9892–9909. [Google Scholar] [CrossRef] [PubMed]
- Brito, C.; Costa-Silva, B.; Barral, D.C.; Pojo, M. Unraveling the Relevance of ARL GTPases in Cutaneous Melanoma Prognosis through Integrated Bioinformatics Analysis. Int. J. Mol. Sci. 2021, 22, 9260. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Goettsch, C.; Sharma, A.; Ricchiuto, P.; Goh, W.W.B.; Halu, A.; Yamada, I.; Yoshida, H.; Hara, T.; Wei, M.; et al. PARP9 and PARP14 Cross-Regulate Macrophage Activation via STAT1 ADP-Ribosylation. Nat. Commun. 2016, 7, 12849. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukkarasu, S.; Ahmed, M.; Rosa, B.A.; Boothby, M.; Cho, S.H.; Rangel-Moreno, J.; Mbandi, S.K.; Schreiber, V.; Gupta, A.; Zuniga, J.; et al. Poly(ADP-Ribose) Polymerase 9 Mediates Early Protection against Mycobacterium Tuberculosis Infection by Regulating Type I IFN Production. J. Clin. Investig. 2023, 133, e158630. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Lu, S.; Garces, J.; Jin, T.; Li, J. Protein Kinase C-Regulated Dynamitin-Macrophage-Enriched Myristoylated Alanine-Rice C Kinase Substrate Interaction Is Involved in Macrophage Cell Spreading. J. Biol. Chem. 2000, 275, 23948–23956. [Google Scholar] [CrossRef] [PubMed]
- Hsin, I.-L.; Sheu, G.-T.; Jan, M.-S.; Sun, H.-L.; Wu, T.-C.; Chiu, L.-Y.; Lue, K.-H.; Ko, J.-L. Inhibition of Lysosome Degradation on Autophagosome Formation and Responses to GMI, an Immunomodulatory Protein from Ganoderma Microsporum. Br. J. Pharmacol. 2012, 167, 1287–1300. [Google Scholar] [CrossRef] [PubMed]
- Maric-Biresev, J.; Hunn, J.P.; Krut, O.; Helms, J.B.; Martens, S.; Howard, J.C. Loss of the Interferon-γ-Inducible Regulatory Immunity-Related GTPase (IRG), Irgm1, Causes Activation of Effector IRG Proteins on Lysosomes, Damaging Lysosomal Function and Predicting the Dramatic Susceptibility of Irgm1-Deficient Mice to Infection. BMC Biol. 2016, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Buijze, H.; Brinkmann, V.; Hurwitz, R.; Dorhoi, A.; Kaufmann, S.H.E.; Pei, G. Human GBP1 Is Involved in the Repair of Damaged Phagosomes/Endolysosomes. Int. J. Mol. Sci. 2023, 24, 9701. [Google Scholar] [CrossRef] [PubMed]
- Collazo, C.M.; Yap, G.S.; Sempowski, G.D.; Lusby, K.C.; Tessarollo, L.; Vande Woude, G.F.; Sher, A.; Taylor, G.A. Inactivation of LRG-47 and IRG-47 Reveals a Family of Interferon Gamma-Inducible Genes with Essential, Pathogen-Specific Roles in Resistance to Infection. J. Exp. Med. 2001, 194, 181–188. [Google Scholar] [CrossRef]
- Macaluso, G.; Grippi, F.; Di Bella, S.; Blanda, V.; Gucciardi, F.; Torina, A.; Guercio, A.; Cannella, V. A Review on the Immunological Response against Trypanosoma Cruzi. Pathogens 2023, 12, 282. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, R.V.H.; Zamboni, D.S. Inflammasome Activation in Response to Intracellular Protozoan Parasites. Trends Parasitol. 2020, 36, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Misheva, M.; Johnson, J.; McCullagh, J. Role of Oxylipins in the Inflammatory-Related Diseases NAFLD, Obesity, and Type 2 Diabetes. Metabolites 2022, 12, 1238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; de Waard, A.A.; Wuhrer, M.; Spaapen, R.M. The Role of Glycosphingolipids in Immune Cell Functions. Front. Immunol. 2019, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, V.; Uaratanawong, R.; Patel, R.R.; Patel, H.; Bao, W.; Hartney, B.; Cohen, E.; Chen, X.; Zhong, Q.; Isales, C.M.; et al. Phosphatidylglycerol Inhibits Toll-Like Receptor-Mediated Inflammation by Danger-Associated Molecular Patterns. J. Investig. Dermatol. 2019, 139, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Nisimura, L.M.; Coelho, L.L.; de Melo, T.G.; Vieira, P.d.C.; Victorino, P.H.; Garzoni, L.R.; Spray, D.C.; Iacobas, D.A.; Iacobas, S.; Tanowitz, H.B.; et al. Trypanosoma Cruzi Promotes Transcriptomic Remodeling of the JAK/STAT Signaling and Cell Cycle Pathways in Myoblasts. Front. Cell. Infect. Microbiol. 2020, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Caetano, B.C.; Carmo, B.B.; Melo, M.B.; Cerny, A.; dos Santos, S.L.; Bartholomeu, D.C.; Golenbock, D.T.; Gazzinelli, R.T. Requirement of UNC93B1 Reveals a Critical Role for TLR7 in Host Resistance to Primary Infection with Trypanosoma Cruzi. J. Immunol. 2011, 187, 1903–1911. [Google Scholar] [CrossRef] [PubMed]
- Herreros-Cabello, A.; Callejas-Hernández, F.; Fresno, M.; Gironès, N. Comparative Proteomic Analysis of Trypomastigotes from Trypanosoma Cruzi Strains with Different Pathogenicity. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 76, 104041. [Google Scholar] [CrossRef]
- Amato Neto, V. Origin of the “Y Strain” of Trypanosoma Cruzi. Rev. Inst. Med. Trop. Sao Paulo 2010, 52, 171. [Google Scholar] [CrossRef][Green Version]
- Sanchiz, Á.; Morato, E.; Rastrojo, A.; Camacho, E.; González-de la Fuente, S.; Marina, A.; Aguado, B.; Requena, J.M. The Experimental Proteome of Leishmania Infantum Promastigote and Its Usefulness for Improving Gene Annotations. Genes 2020, 11, 1036. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Suárez, H.; Andreu, Z.; Mazzeo, C.; Toribio, V.; Pérez-Rivera, A.E.; López-Martín, S.; García-Silva, S.; Hurtado, B.; Morato, E.; Peláez, L.; et al. CD9 Inhibition Reveals a Functional Connection of Extracellular Vesicle Secretion with Mitophagy in Melanoma Cells. J. Extracell. Vesicles 2021, 10, e12082. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xiao, W.; Su, Z.; Cheng, J.; Zheng, C.; Zhang, Z.; Wang, Y.; Wang, L.; Xu, B.; Li, S.; et al. Hippocampal Proteomic Alteration in Triple Transgenic Mouse Model of Alzheimer’s Disease and Implication of PINK 1 Regulation in Donepezil Treatment. J. Proteome Res. 2019, 18, 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Gong, P.; Zhao, W.; Zhang, J.; Wu, X.; Xin, C.; Xiong, Z.; Li, Z.; Wu, X.; Wan, Q.; et al. Quantitative iTRAQ-Based Proteomic Analysis of Piperine Protected Cerebral Ischemia/Reperfusion Injury in Rat Brain. Neurochem. Int. 2019, 124, 51–61. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).