KL-Biome (Postbiotic Formulation of Lactiplantibacillus plantarum KM2) Improves Dexamethasone-Induced Muscle Atrophy in Mice
Abstract
1. Introduction
2. Results & Discussion
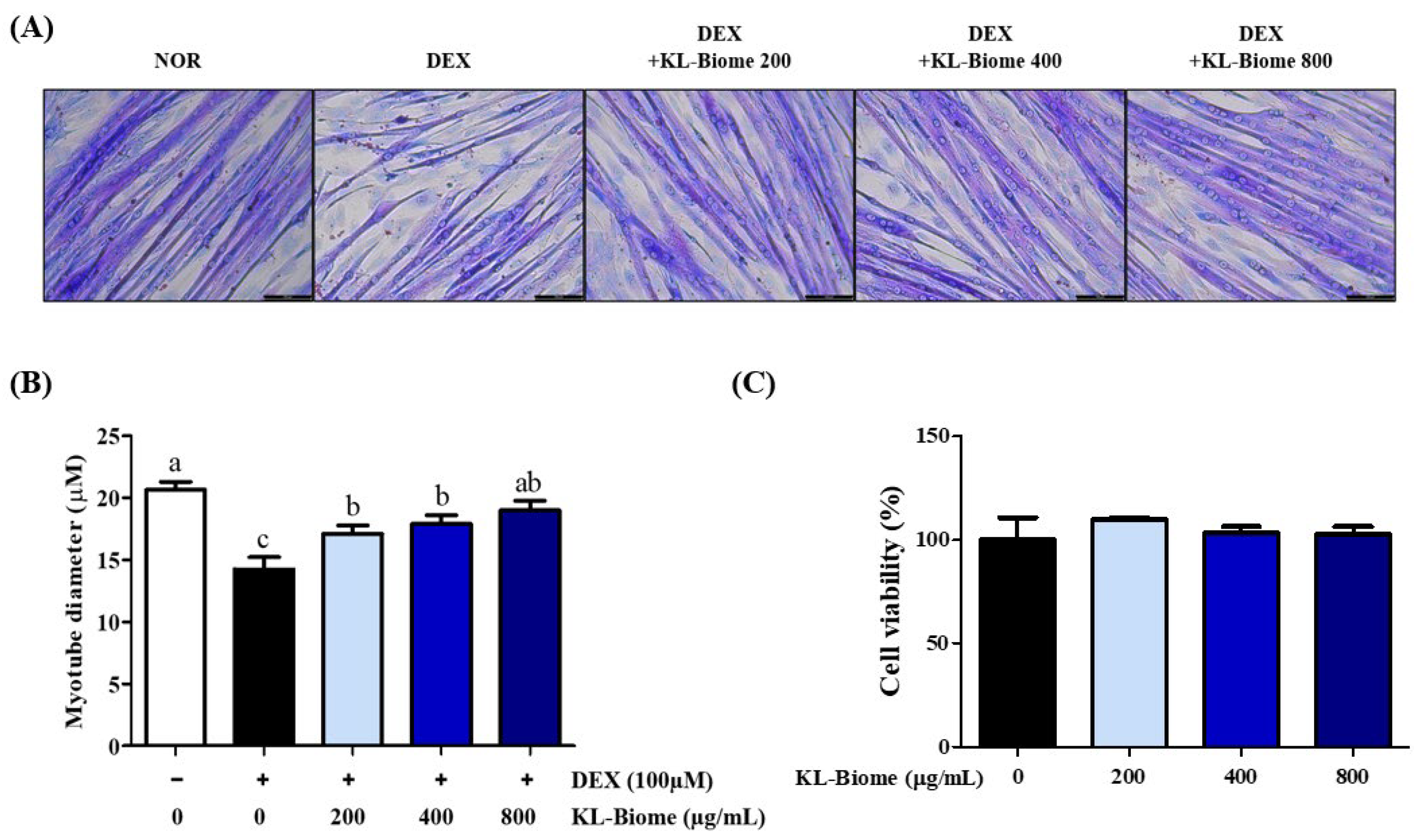
2.1. Effect of KL-Biome on Myotube Diameter in DEX-Induced Atrophic C2C12 Myotubes
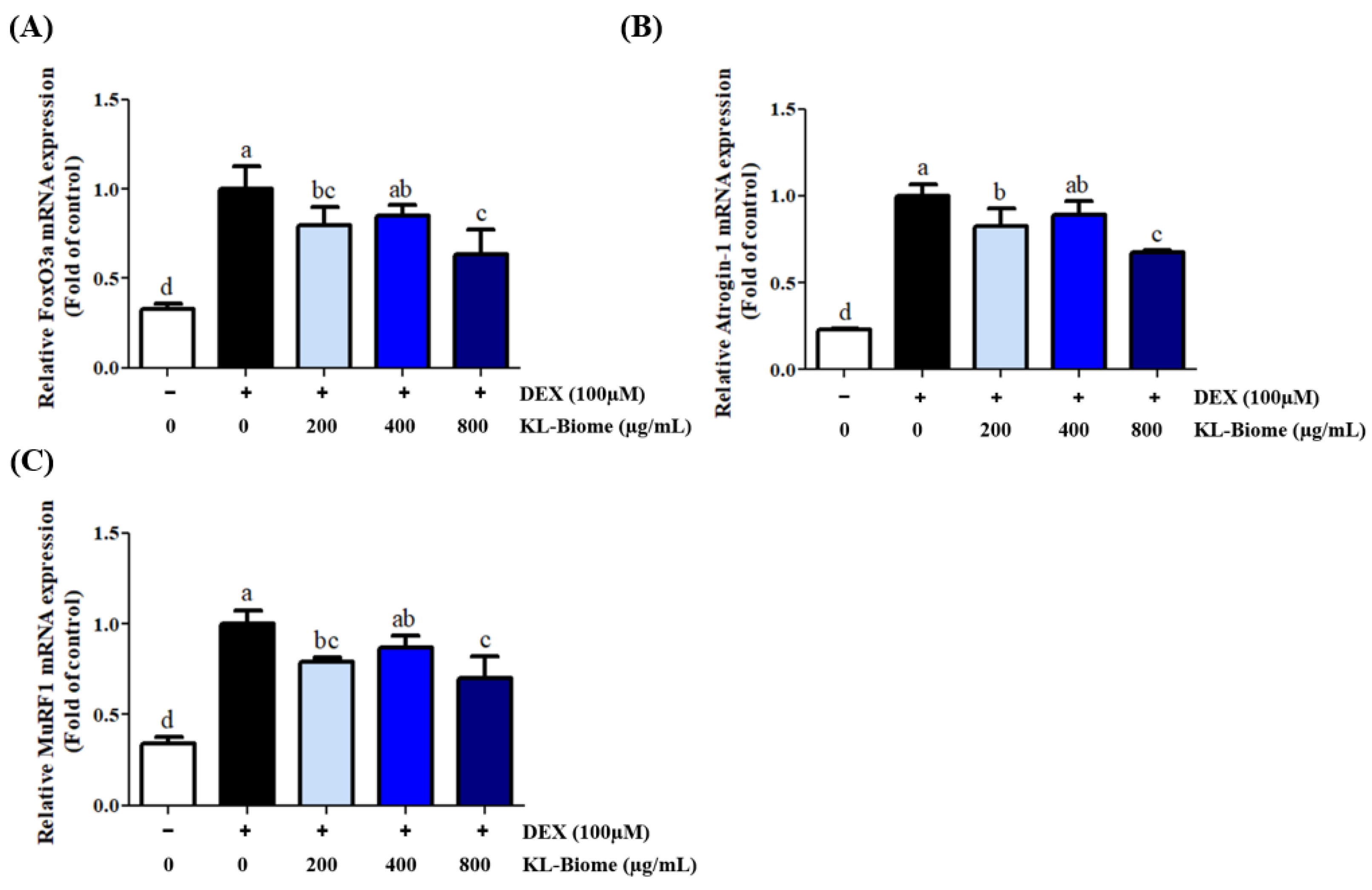
2.2. Effect of KL-Biome on Protein Degradation-Associated Gene Expression in DEX-Treated C2C12 Myotubes
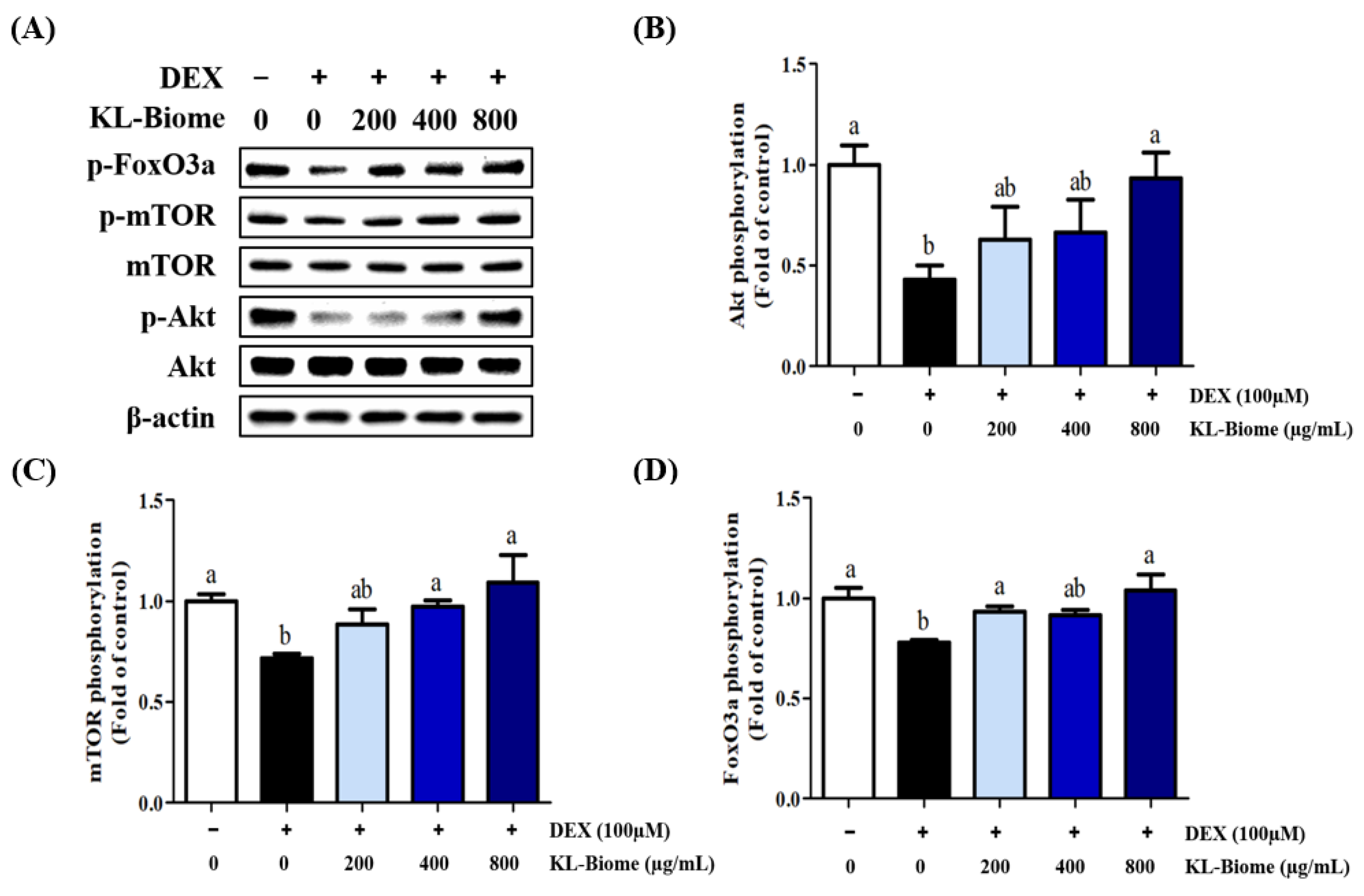
2.3. Effects of KL-Biome on the Protein Expression of Akt, mTOR, and FoxO3a in DEX-Treated C2C12 Myotubes
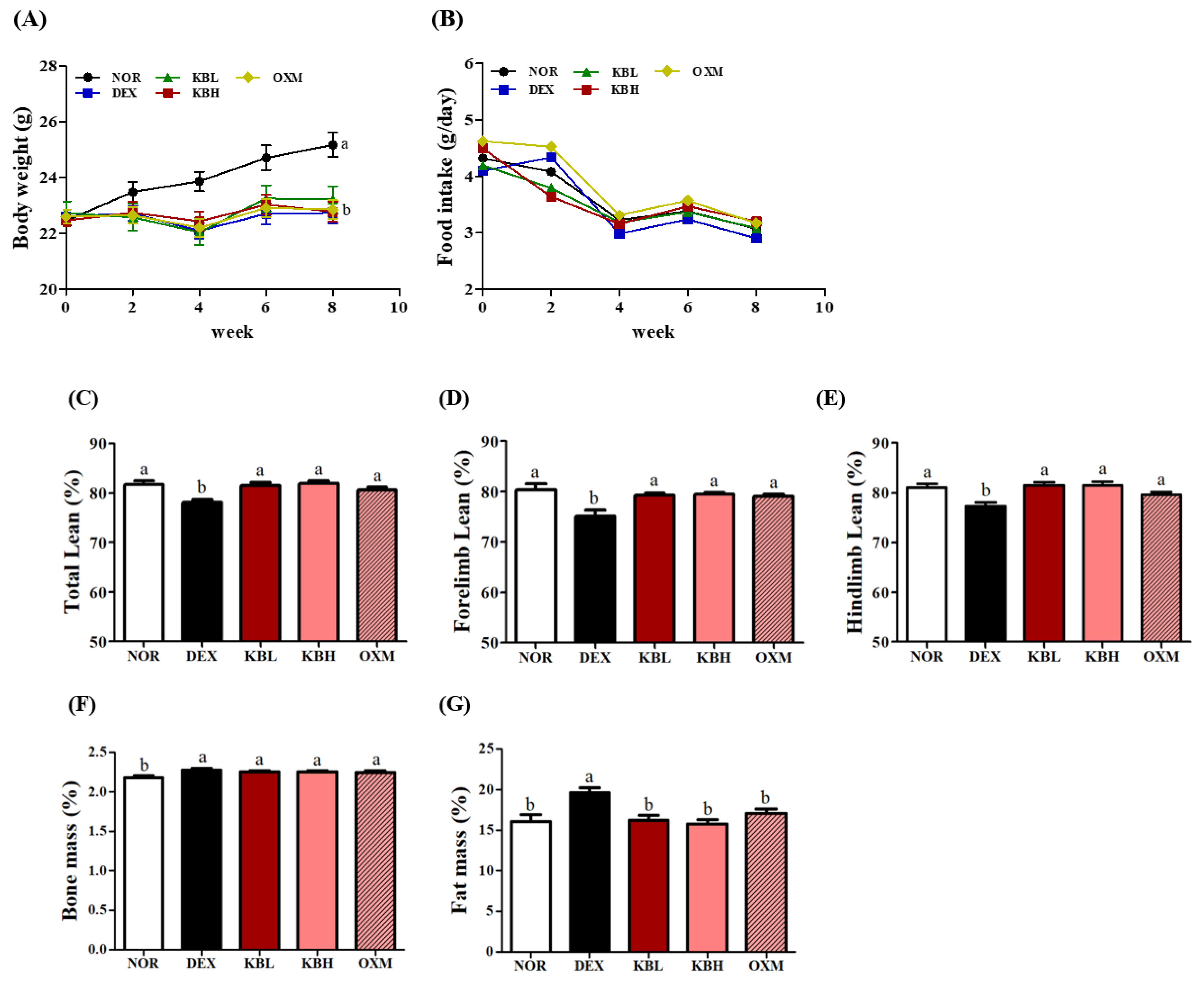
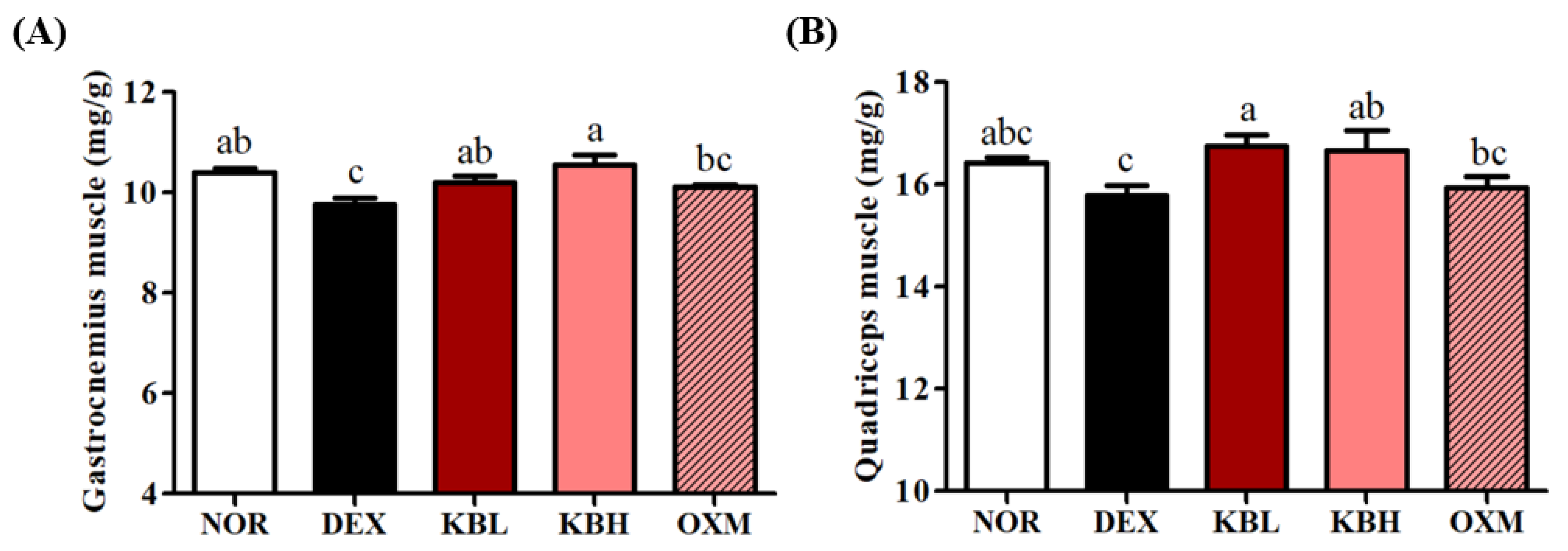
2.4. Effects of KL-Biome on Body Composition and Muscle Weight in DEX-Treated Mice
2.5. Effect of KL-Biome on Biochemical Markers in DEX-Treated Mice
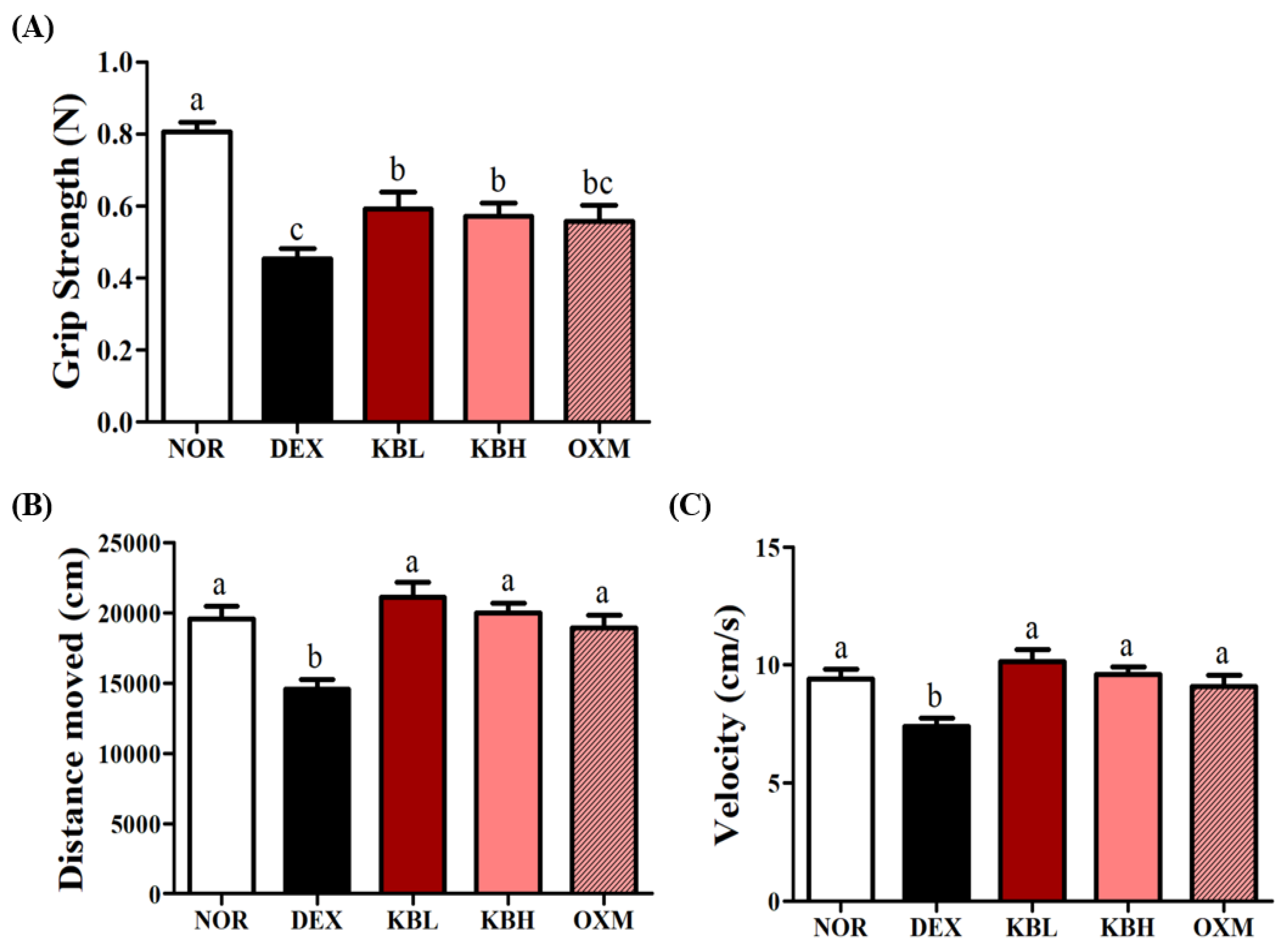
2.6. Effects of KL-Biome on Muscle Function in DEX-Treated Mice
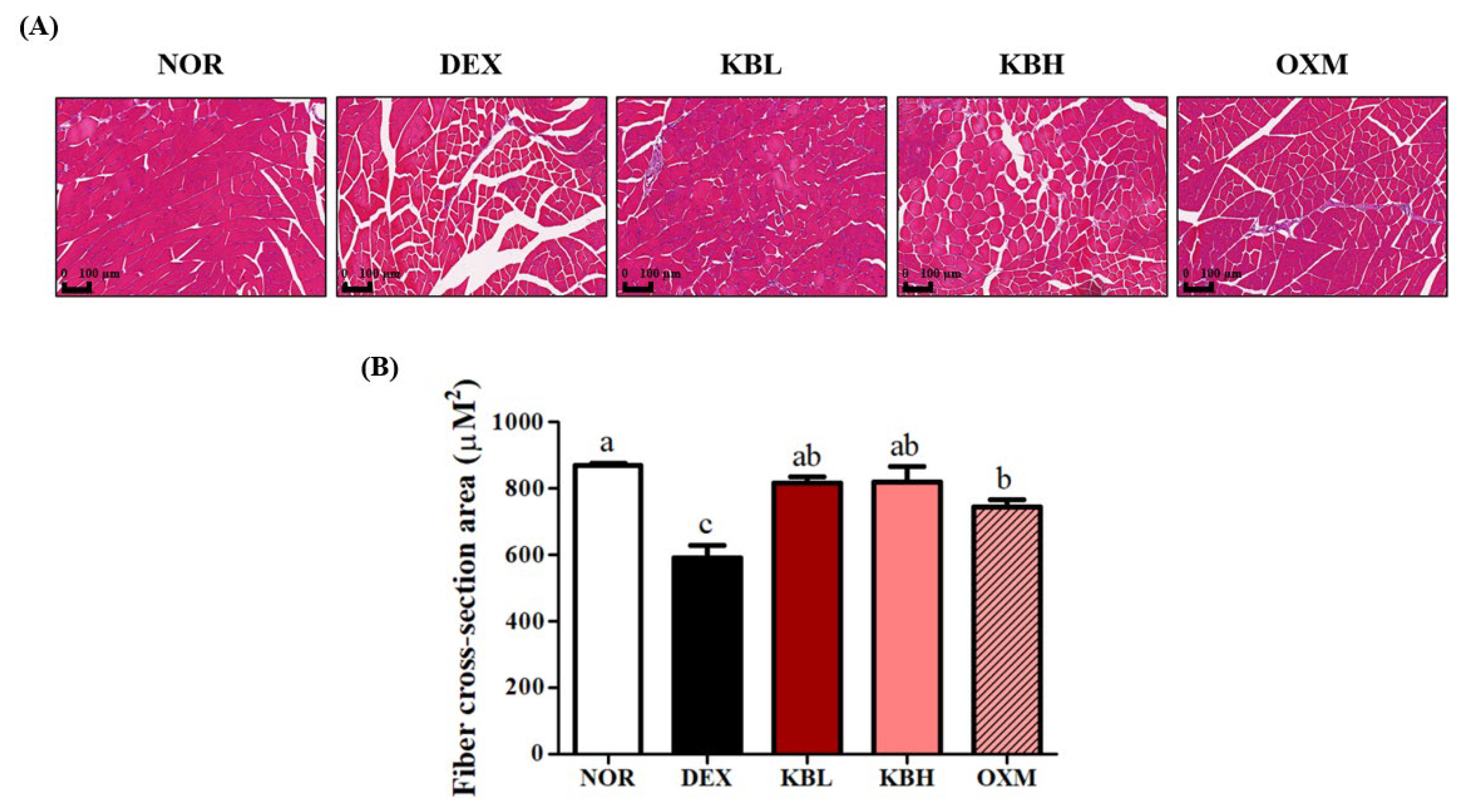
2.7. Effect of KL-Biome on Structural Damage of Muscle Tissues in DEX-Treated Mice
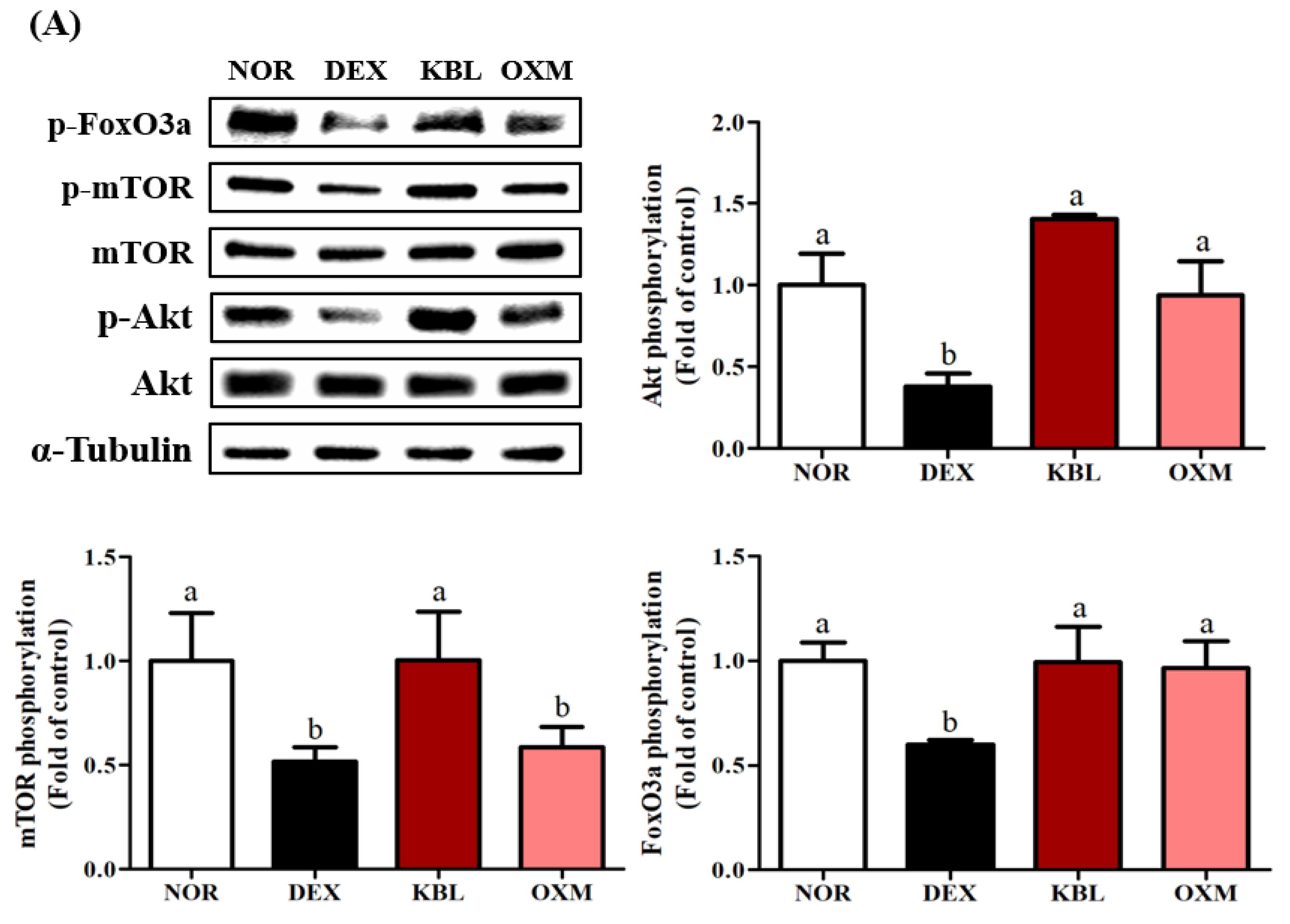
2.8. Effect of KL-Biome on Major Signaling Factors Affecting Protein Synthesis and Degradation in DEX-Treated Mice
2.9. Effect of KL-Biome on Gut Microbiome Composition
3. Materials and Methods
3.1. Materials
3.2. Preparation of KL-Biome
3.3. C2C12 Myoblasts Cell Culture
3.4. Myotube Diameter
3.5. DEX-Induced Atrophy Mouse Model Microsystems Ltd.
3.6. Body Composition
3.7. Blood and Tissue Collection and Serum Biochemical Analysis
3.8. Grip Strength Test
3.9. Exercise Performance Test
3.10. Histological Analysis
3.11. RNA Extraction and Quantitative Real-Time Polymerize Chain Reaction (qRT-PCR)
3.12. Western Blot Analysis
3.13. Cecal Gut Microbiota Analysis via Shotgun Sequencing
3.14. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larsson, L.; Degens, H.; Li, M.; Salviati, L.; Lee, Y.I.; Thompson, W.; Kirkland, J.L.; Sandri, M. Sarcopenia: Aging-related loss of muscle mass and function. Physiol. Rev. 2019, 99, 427–511. [Google Scholar] [CrossRef] [PubMed]
- von Haehling, S.; Morley, J.E.; Anker, S.D. An overview of sarcopenia: Facts and numbers on prevalence and clinical impact. J. Cachexia Sarcopenia Muscle 2010, 1, 129–133. [Google Scholar] [CrossRef] [PubMed]
- James, D.E.; Stockli, J.; Birnbaum, M.J. The aetiology and molecular landscape of insulin resistance. Nat. Rev. Mol. Cell. Biol. 2021, 22, 751–771. [Google Scholar] [CrossRef]
- Fanzani, A.; Conraads, V.M.; Penna, F.; Martinet, W. Molecular and cellular mechanism of skeletal muscle atrophy: An update. J. Cachexia Sarcopenia Muscle 2012, 3, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Li, M.; Wang, K.; Qi, G.; Liu, H.; Wang, W.; Ji, Y.; Chang, M.; Deng, C.; Xu, F.; et al. Diabetic muscular atrophy: Molecular mechanisms and promising therapies. Front. Endocrinol. 2022, 13, 917113. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, S.; Kim, H.; Garcia-Perez, I.; Reza, M.M.; Martin, K.A.; Kundu, P.; Cox, L.M.; Selkrig, J.; Posma, J.M.; Zhang, H.; et al. The gut microbiota influences skeletal muscle mass and function in mice. Sci. Transl. Med. 2019, 11, eaan5662. [Google Scholar] [CrossRef]
- Liu, C.; Cheung, W.H.; Li, J.; Chow, S.K.H.; Yu, J.; Wong, S.H.; Ip, M.; Sung, J.J.Y.; Wong, R.M.Y. Understanding the gut microbiota and sarcopenia: A systematic review. J. Cachexia Sarcopenia Muscle 2021, 12, 1393–1407. [Google Scholar] [CrossRef] [PubMed]
- Prokopidis, K.; Chambers, E.; Lochlainn, M.N.; Witard, O.C. Mechanism linking the gut-muscle axis with muscle protein metabolism and anabolic resistance: Implications for older adults at risk of sarcopenia. Front. Physiol. 2021, 12, 770455. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Huang, Y.; Yu, X. A narrative review of gut-muscle axis and sarcopenia: The potential role of gut microbiota. Int. J. Gen. Med. 2021, 14, 1263–1273. [Google Scholar] [CrossRef]
- De Vries, M.C.; Vaughan, E.E.; Kleerebezem, M.; de Vos, W.M. Lactobacillus plantarum—Survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 2006, 16, 1018–1028. [Google Scholar] [CrossRef]
- Liu, Y.W.; Liong, M.T.; Tsai, Y.C. New perspectives of Lactobacillus plantarum as a probiotic: The gut-heart-brain axis. J. Microbiol. 2018, 56, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, Y.; Zhang, L.; Zhang, X.; Huang, L.; Li, D.; Niu, C.; Yang, Z.; Wang, Q. Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. 2012, 135, 1914–1919. [Google Scholar] [CrossRef] [PubMed]
- Kariyawasam, K.M.G.M.M.; Lee, N.K.; Paik, H.D. Effect of set-type yoghurt supplemented with the novel probiotic Lantiplantibacillus plantarum 200655 on physicochemical properties and the modulation of oxidative stress-induced damage. Food Sci. Biotechnol. 2023, 32, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Wei, L.; Chiu, Y.S.; Hsu, Y.J.; Tsai, T.Y.; Wang, M.F.; Huang, C.C. Lactobacillus plantarum TWK10 supplementation improves exercise performance and increases muscle mass in mice. Nutrients 2016, 8, 205. [Google Scholar] [CrossRef]
- Lee, K.; Kim, J.; Park, S.D.; Shim, J.J.; Lee, J.L. Lactobacillus plantarum HY7715 ameliorates sarcopenia by improving skeletal muscle mass and function in aged Balb/c mice. Int. J. Mol. Sci. 2021, 22, 10023. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Kang, D.K.; Paik, H.D.; Park, Y.S. Beyond probiotics: A narrative review on an era of revolution. Food Sci. Biotechnol. 2023, 32, 413–421. [Google Scholar] [CrossRef]
- Kuo, T.; Harris, C.A.; Wang, J.C. Metabolic functions of glucocorticoid receptor in skeletal muscle. Mol. Cell. Endocrinol. 2013, 380, 79–88. [Google Scholar] [CrossRef]
- Wang, B.Y.H.; Hsiao, A.W.T.; Wong, N.; Chen, Y.F.; Lee, C.W.; Lee, W.Y.W. Is dexamethasone-induced muscle atrophy an alternative model for naturally aged sarcopenia model? J. Orthop. Transl. 2023, 39, 12–20. [Google Scholar] [CrossRef]
- Stitt, T.N.; Drujan, D.; Clarke, B.A.; Panaro, F.; Timofeyva, Y.; Kline, W.O.; Gonzalez, M.; Yancopoulos, G.D.; Glass, D.J. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 2004, 14, 395–403. [Google Scholar] [CrossRef]
- Clarke, B.A.; Drujan, D.; Willis, M.S.; Murphy, L.O.; Corpina, R.A.; Burova, E.; Rakhilin, S.V.; Stitt, T.N.; Patterson, C.; Latres, E.; et al. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007, 6, 376–385. [Google Scholar] [CrossRef]
- Polge, C.; Heng, A.E.; Jarzaguet, M.; Ventadour, S.; Claustre, A.S.; Combaret, L.; Béchet, D.; Matondo, M.; Uttenweiler-Joseph, S.; Monsarrat, B.; et al. Muscle actin is polyubiquitinylated in vitro and in vivo and targeted for breakdown by the E3 ligase MuRF1. FASEB J. 2011, 25, 3790–3802. [Google Scholar] [CrossRef] [PubMed]
- Lokireddy, S.; Wijesoma, I.W.; Sze, S.K.; McFarlane, C.; Kambadur, R.; Sharma, M. Identification of atrogin-1-targeted proteins during the myostatin-induced skeletal muscle wasting. Am. J. Physiol. Cell. Physiol. 2012, 303, C512–C529. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kang, M.; Yoo, J.; Lee, S.; Kang, M.; Yun, B.; Kim, J.N.; Moon, H.; Chung, Y.; Oh, S. Lactobacillus rhamnosus JY02 Ameliorates sarcopenia by anti-atrophic effects in a dexamethasone-induced cellular and murine model. J. Microbiol. Biotechnol. 2023, 33, 915. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M. Signaling in muscle atrophy and hypertrophy. Physiology 2008, 23, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Brault, J.J.; Schild, A.; Cao, P.; Sandri, M.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007, 6, 472–483. [Google Scholar] [CrossRef]
- Jaiswal, N.; Gavin, M.; Loro, E.; Sostre-Colón, J.; Roberson, P.A.; Uehara, K.; Rivera-Fuentes, N.; Neinast, M.; Arany, Z.; Kimball, S.T.; et al. AKT controls protein synthesis and oxidative metabolism via combined mTORC1 and FOXO1 signalling to govern muscle physiology. J. Cachexia Sarcopenia Muscle 2022, 13, 214–495. [Google Scholar] [CrossRef]
- Egerman, M.A.; Glass, D.J. Signaling pathways controlling skeletal muscle mass. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 59–68. [Google Scholar] [CrossRef]
- Furukawa, K.; Kousaka, M.; Jia, H.; Kato, H. Suppressive effects of turmeric extract on muscle atrophy in dexamethasone-treated mice and myotubes. Nutrients 2022, 14, 3979. [Google Scholar] [CrossRef] [PubMed]
- Macedo, A.G.; Krug, A.L.O.; Souza, L.M.; Martuscelli, A.M.; Constantino, P.B.; Zago, A.S.; Rush, J.W.E.; Santos, C.F.; Amaral, S.L. Time-course changes of catabolic proteins following muscle atrophy induced by dexamethasone. Steriods 2016, 107, 30–36. [Google Scholar] [CrossRef]
- Livingstone, I.; Johnson, M.A.; Mastaglia, F.L. Effects of dexamethasone on fibre subtypes in rat muscle. Neuropathol. Appl. Neurobiol. 1981, 7, 381–398. [Google Scholar] [CrossRef]
- Augusto, V.; Padovani, C.R.; Campos, G.E.R. Skeletal muscle fiber types in C57BL6J mice. Braz. J. Morphol. Sci. 2004, 21, 89–94. [Google Scholar]
- Brancaccio, P.; Lippi, G.; Maffulli, N. Biochemical markers of muscular damage. Clin. Chem. Lab. Med. 2010, 48, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.B.; Ahn, Y.; Suh, H.J.; Jo, K. Yeast hydrolysate ameliorates dexamethasone-induced muscle atrophy by suppressing MuRF-1 expression in C2C12 cells and C57BL/6 mice. J. Func. Foods 2022, 90, 104985. [Google Scholar] [CrossRef]
- Jackson, E.R.; Kilroy, C.; Joslin, D.L.; Schomaker, S.J.; Pruimboom-Brees, I.; Amacher, D.E. The early effects of short-term dexamethasone administration on hepatic and serum alanine aminotransferase in the rat. Drug Chem. Toxicol. 2008, 31, 427–445. [Google Scholar] [CrossRef] [PubMed]
- Short, K.R.; Wair, K.S. The effect of age on protein metabolism. Curr. Opin. Clin. Nutr. Metab. Care 2000, 3, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, Y.I.; Kim, M.J.; Hahm, J.H.; Seo, H.D.; Ha, T.Y.; Jung, C.H.; Ahn, J. Castor oil plant (Ricinus communis L.) leaves improve dexamethasone-induced muscle atrophy via Nrf2 activation. Front. Pharmacol. 2022, 13, 891762. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.M.; Loenneke, J.P.; Jo, E.; Wilson, G.J.; Zourdos, M.C.; Kim, J.S. The effects of endurance, strength, and power training on muscle fiber type shifting. J. Strength Cond. Res. 2012, 26, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Mula, J.; Lee, J.D.; Liu, F.; Yang, L.; Peterson, C.A. Automated image analysis of skeletal fiber cross-sectional area. J. Appl. Physiol. 2013, 114, 148–155. [Google Scholar] [CrossRef]
- McCarthy, J.J.; Esser, K.A. Anabolic and catabolic pathways regulating skeletal muscle mass. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 230. [Google Scholar] [CrossRef]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; Yancopoulos, G.D. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef]
- Ticinesi, A.; Nouvenne, A.; Cerundolo, N.; Catania, P.; Prati, B.; Tana, C.; Meschi, T. Gut microbiota, muscle mass and function in aging: A focus on physical frailty and sarcopenia. Nutrients 2019, 11, 1633. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, J.; Liang, H.; Ye, L.; Lan, L.; Lu, F.; Wang, Q.; Lei, T.; Yang, X.; Cui, P.; et al. Differences in alpha diversity of gut microbiota in neurological diseases. Front. Neurosci. 2022, 16, 879318. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Cheng, Y.; Deng, Y.; Ren, G.; Wang, J. Composition of gut microbiota involved in alleviation of dexamethasone-mediated muscle atrophy by whey protein. NPJ Sci. Food 2023, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Ishizu, Y.; Honda, T.; Ito, T.; Imai, N.; Nakamura, M.; Kawashima, H.; Kitaura, Y.; Ishigami, M.; Fujishiro, M. Patients with low muscle mass have characteristic microbiome with low potential for amino acid synthesis in chronic liver disease. Sci. Rep. 2022, 12, 3674. [Google Scholar] [CrossRef] [PubMed]
- Frampton, J.; Murphy, K.G.; Frost, G.; Chambers, E.S. Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function. Nat. Metab. 2020, 9, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.E.; Bhattacharya, A.; Sataranatarajan, K.; Qaisar, R.; Sloane, L.; Rahman, M.M.; Kinter, M.; Van Remmen, H. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell 2015, 14, 957–970. [Google Scholar] [CrossRef]
- Polansky, O.; Sekelova, Z.; Faldynova, M.; Sebkova, A.; Sisak, F.; Rychlik, I. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl. Environ. Microbiol. 2016, 82, 1569–1576. [Google Scholar] [CrossRef] [PubMed]
- Han, D.S.; Wu, W.K.; Liu, P.Y.; Yang, Y.T.; Hsu, H.C.; Kuo, C.H.; Wu, M.S.; Wang, T.G. Differences in the gut microbiome and reduced fecal butyrate in elders with low skeletal muscle mass. Clin. Nutr. 2022, 41, 1491–1500. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.; Rodriguez-Palacios, A. The genus Alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Ferreira-Halder, C.V.; de Sousa Faria, A.V.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef]
- Byeon, H.R.; Jang, S.Y.; Lee, Y.; Kim, D.; Hong, M.G.; Lee, D.; Shin, J.H.; Seo, J.G. New strains of Akekermansia muciniphila and Faecalibacterium prausnitzii are effective for improving the muscle strength of mice with immobilization-induced muscular atrophy. J. Med. Food 2022, 25, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.Q.; Lin, X.; Shen, H.; Liu, H.M.; Qiu, X.; Li, B.Y.; Shen, W.D.; Ge, C.L.; Lv, F.Y.; Shen, J.; et al. Human gut microbiome impacts skeletal muscle mass via gut microbial synthesis of the short chain fatty acid butyrate among healthy menopausal woman. J. Cachexia Sarcopenia Muscle 2021, 12, 1860–1870. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Yang, L.; Jiang, J.; Ni, Y.; Zhu, J.; Zheng, X.; Wang, Q.; Lu, X.; Fu, Z. Chronic glucocorticoid treatment induced circadian clock disorder leads to lipid metabolism and gut microbiota alterations in rats. Life Sci. 2018, 192, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Veliça, P.; Bunce, C.M. A quick, simple and unbiased method to quantify C2C12 myogenic differentiation. Muscle Nerve 2011, 44, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Im, J.Y. Antiobesity effect of tricin, a methylated cereal flavone, in high-fat-diet-induced obese mice. J. Agric. Food Chem. 2018, 66, 9989–9994. [Google Scholar] [CrossRef]
- Mas-Lloret, J.; Obon-Santacana, M.; Ibanez-Sanz, G.; Guino, E.; Pato, M.L.; Rodriguez-Moranta, F.; Mata, A.; Garcia-Rodriguez, A.; Moreno, V.; Pimenoff, V.N. Gut microbiome diversity detected by high-coverage 16S and shotgun sequencing of paired stool and colon sample. Sci. Data 2020, 7, 92. [Google Scholar] [CrossRef]

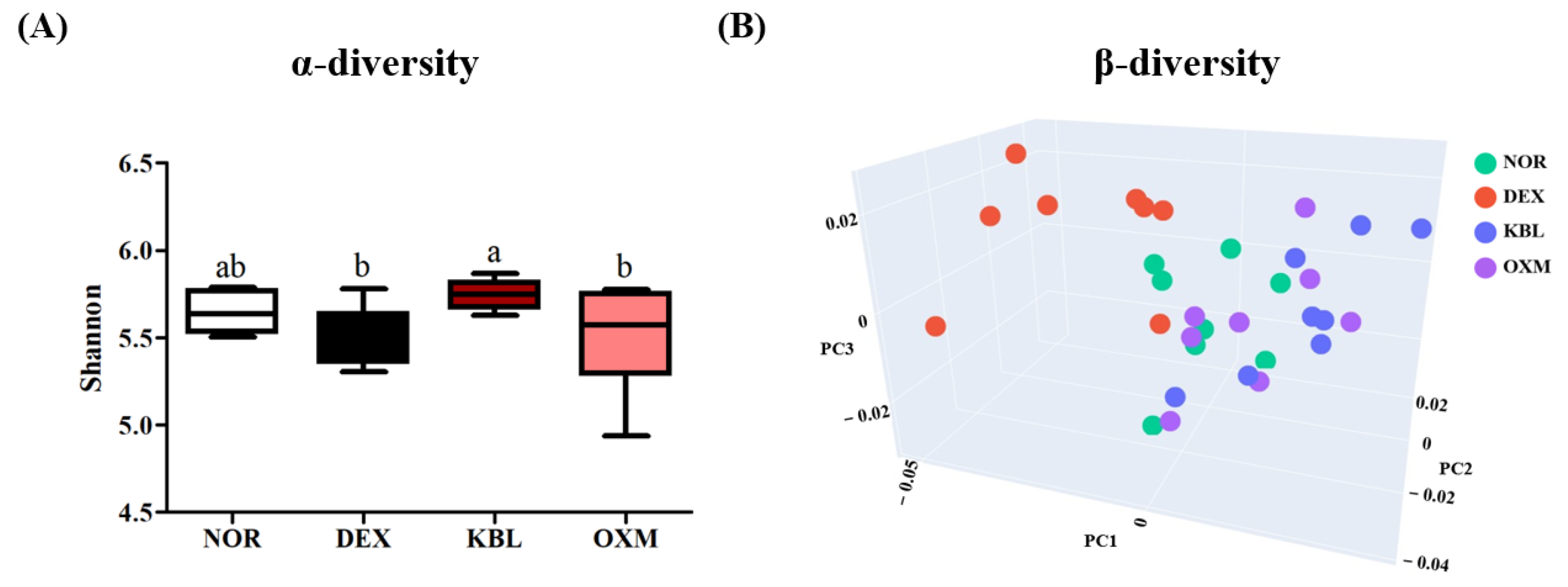
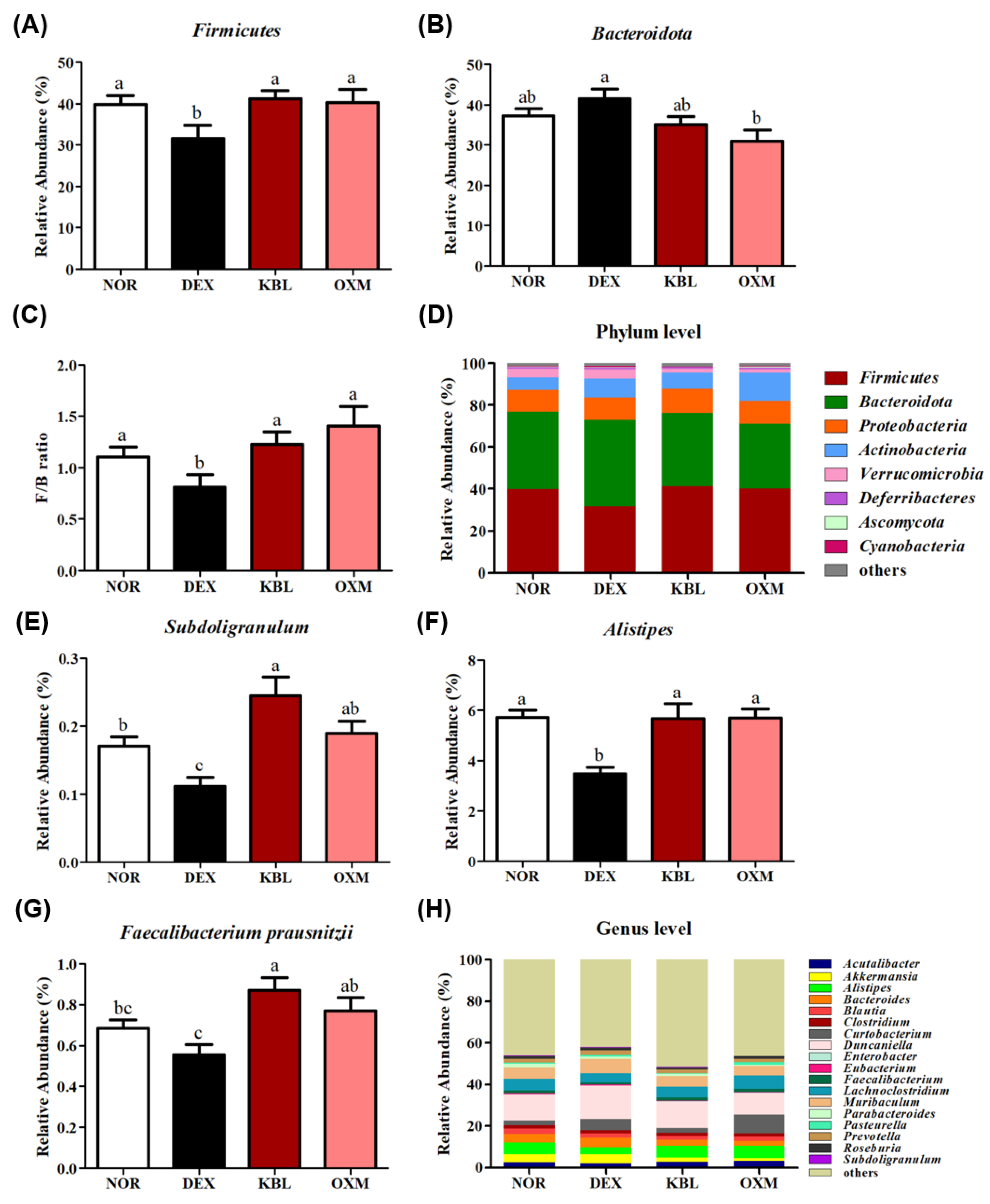
| Group | CPK (U/L) | LDH (U/L) | ALT (U/L) | AST (U/L) | BUN (mg/dL) |
|---|---|---|---|---|---|
| NOR | 187.00 ± 62.34 b | 288.50 ± 73.19 a | 25.25 ± 6.88 c | 38.88 ± 9.31 b | 24.60 ± 2.76 a |
| DEX | 340.88 ± 154.51 a | 295.63 ± 65.91 a | 31.13 ± 2.30 bc | 59.25 ± 10.07 a | 24.15 ± 1.57 a |
| KBL | 147.50 ± 101.33 b | 99.25 ± 15.74 c | 29.00 ± 5.35 bc | 52.88 ± 10.22 a | 21.39 ± 1.13 b |
| KBH | 199.63 ± 56.45 b | 137.00 ± 18.59 c | 32.00 ± 3.42 b | 62.75 ± 12.28 a | 22.90 ± 1.09 ab |
| OXM | 99.13 ± 35.13 b | 211.38 ± 46.99 b | 43.75 ± 7.96 a | 58.88 ± 11.73 a | 24.74 ± 1.47 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, Y.-J.; Kim, J.-H.; Jung, Y.-J.; Kwak, M.-S.; Sung, M.-H.; Imm, J.-Y. KL-Biome (Postbiotic Formulation of Lactiplantibacillus plantarum KM2) Improves Dexamethasone-Induced Muscle Atrophy in Mice. Int. J. Mol. Sci. 2024, 25, 7499. https://doi.org/10.3390/ijms25137499
Jeong Y-J, Kim J-H, Jung Y-J, Kwak M-S, Sung M-H, Imm J-Y. KL-Biome (Postbiotic Formulation of Lactiplantibacillus plantarum KM2) Improves Dexamethasone-Induced Muscle Atrophy in Mice. International Journal of Molecular Sciences. 2024; 25(13):7499. https://doi.org/10.3390/ijms25137499
Chicago/Turabian StyleJeong, Yu-Jin, Jong-Hoon Kim, Ye-Jin Jung, Mi-Sun Kwak, Moon-Hee Sung, and Jee-Young Imm. 2024. "KL-Biome (Postbiotic Formulation of Lactiplantibacillus plantarum KM2) Improves Dexamethasone-Induced Muscle Atrophy in Mice" International Journal of Molecular Sciences 25, no. 13: 7499. https://doi.org/10.3390/ijms25137499
APA StyleJeong, Y.-J., Kim, J.-H., Jung, Y.-J., Kwak, M.-S., Sung, M.-H., & Imm, J.-Y. (2024). KL-Biome (Postbiotic Formulation of Lactiplantibacillus plantarum KM2) Improves Dexamethasone-Induced Muscle Atrophy in Mice. International Journal of Molecular Sciences, 25(13), 7499. https://doi.org/10.3390/ijms25137499






